Abstract
Cytomegalovirus (CMV) may be a relevant cause of morbidity in patients displaying various inflammatory diseases. In this study, it was investigated whether CMV DNA is detected in the lower respiratory tract and the systemic compartment in pediatric patients with chronic or recurrent bronchopulmonary diseases. A total of 42 lower respiratory tract specimens and 11 paired plasma samples from 42 patients were analyzed for the presence of CMV DNA by real‐time PCR. The respiratory specimens were also screened for the presence of respiratory viruses and human herpesvirus 6 (HHV‐6) and 7 (HHV‐7) by PCR methods. Quantitative bacterial and fungal cultures were performed. IL‐6 levels in the respiratory specimens were quantified using ELISA. CMV DNA was detected either in the lower respiratory airways, in plasma, or both in 54.5% of CMV‐seropositive patients. The levels of IL‐6 were significantly higher in these patients than in those with no detectable levels of CMV DNA. HHV‐6 and HHV‐7 DNA were detected in three and one patients, respectively. Respiratory viruses were detected in 13 of the 42 patients. Significant growth of one or more bacterial species was observed in 17 patients. No significant association was found between the presence of CMV DNA and the detection of other microorganisms. The data indicated that the presence of CMV DNA in the lower respiratory tract is a frequent finding in children with chronic or recurrent bronchopulmonary diseases. Further, prospective observational studies are needed to assess the impact of this phenomenon, if any, on the clinical course of these patients. J. Med. Virol. 85:888–892, 2013. © 2013 Wiley Periodicals, Inc.
Keywords: cytomegalovirus, children, bronchopulmonary diseases, respiratory viruses, IL‐6, human herpesvirus‐6
INTRODUCTION
Cytomegalovirus (CMV) infection is a leading cause of morbidity and mortality in severely immunosuppressed patients, such as those undergoing allogeneic hematopoietic stem cell or solid organ transplantation [Solano and Navarro, 2010]. Nevertheless, recent data suggest that CMV may also be a relevant cause of morbidity in patients lacking canonical immunosuppression and displaying various inflammatory processes, including cardiovascular, autoimmune, and chronic bowel diseases [Söderberg‐Nauclér, 2008; Núñez et al., 2010, 2012]. In these patients, active CMV infection is detected frequently in either the inflamed tissues or even the blood compartment. Furthermore, active CMV infection, either restricted to the lower respiratory tract or involving both the lower respiratory airways and the systemic compartment, has been shown to occur frequently during critically illness in adult CMV‐seropositive patients [Chilet et al., 2010; Blanquer et al., 2011], and has been associated with prolonged intensive care unit hospitalization, extended periods of mechanical ventilation, higher rates of nosocomial infection, and overall mortality [for review, see Kalil and Florescu, 2009]. From a pathogenetic perspective, data obtained in the murine CMV model indicate that a pro‐inflammatory state is a critical factor in promoting CMV replication [Cook et al., 2002], and that TNF‐α, a cytokine known to transactivate the expression of CMV immediate‐early genes, plays a critical role in triggering CMV reactivation [Prösch et al., 1995]. In this context, active CMV infection may occur in pediatric patients with chronic or recurrent bronchopulmonary diseases in which inflammation plays potentially a major pathogenic role. The current study was undertaken to investigate this hypothesis.
PATIENTS AND METHODS
Patients and Specimens
A total of 42 lower respiratory tract specimens (bronchoaspirate, n = 22; bronchoalveolar lavage, n = 19; tracheal aspirate, n = 1) and 11 paired plasma samples from 42 children (22 females and 20 males; median age, 4 years old; range, 6 months to 18 years old) attended at the Pneumology, Unit of the University Clinic Hospital of Valencia were analyzed. Patients had a previous history of chronic/recurrent lower tract respiratory symptoms and/or persistent/recurrent radiographic abnormalities (infiltrates, a telectasis, or air trapping images). Fibrobronchoscopies were performed between May 2009 and July 2011, either for primary diagnosis, for diagnosis re‐evaluation in patients responding unsatisfactorily to conventional therapies or as a complementary diagnostic procedure or for assessing the eradication of Pseudomonas aeruginosa in cystic fibrosis patients with negative sputum cultures after a 1‐year‐long course of antibiotic therapy, according to local protocols. No patient had acute upper or lower respiratory tract signs or symptoms at the time of the fibrobronchoscopy. The patients had the following underlying bronchopulmonary conditions: atelectasis rounded or middle lobe syndrome (n = 14), cystic fibrosis (n = 11), bronchiectasis (n = 5), chronic asthma not responding to conventional treatments (n = 3), tracheomalacia (n = 3), bronchiolitis obliterans (n = 2), chronic interstitial pneumopathy (n = 2), bronchopulmonary dysplasia (n = 1), or bronchial hypoplasia (n = 1). All patients lacked a known cause of immunosuppression.
The lower respiratory tract specimens were processed using conventional quantitative bacterial and fungal cultures. Growths of ≥104 or ≥105 CFU/ml of bacterial or fungal species in bronchoaspitates or bronchoalveolar lavages respectively were considered significant. Microbial identification was accomplished by conventional procedures. For some patients, molecular detection of respiratory viruses was also performed shortly after fibrobronchoscopy. Bronchoalveolar lavages and bronchial or tracheal aspirates pre‐treated with 0.1% dithiothreitol (in the ratio of 2 ml to 1 g bronchoaspirate) were vortexed and centrifuged at 462g for 5 min at 4°C to pellet the cells. Cell supernatants were stored at −20°C and were retrieved for further molecular analyses and cytokine measurements. Serum specimens for the assessment of CMV‐serostatus were available from 26 patients. These specimens were obtained either prior to, or at the time of, fibrobronchoscopy. Sera were frozen at −20°C and retrieved for serological testing.
This study conformed to the ethical guidelines of the Declaration of Helsinki and was approved by the local ethics committee. Written informed consent was obtained from parents.
Molecular Analyses
CMV DNA load was quantified by real‐time PCR with an Abbott CMV PCR kit (produced by Qiagen GmbH, Hilden, Germany, for Abbott Diagnostics, Des Plaines, IL), as previously described [Gimeno et al., 2008; Chilet et al., 2010; Bravo et al., 2011]. This assay is able to reliably quantify CMV DNA loads of >10 copies/ml [Chilet et al., 2010]. DNA extractions were performed from a volume of 500 µl (plasma, or lower respiratory tract specimens) using an Abbott mSample preparation system DNA kit on an m24 SP instrument (Abbott Molecular, IL). Detection and quantification of human herpesvirus‐6 (HHV‐6) and human herpesvirus‐7 (HHV‐7) was performed in respiratory specimens by real‐time PCR (Realquality RS‐HHV6 and RS‐HHV7; AB Analitica, Padova, Italy). The detection limit of these assays is approximately 25 copies/ml. DNA was extracted from respiratory specimens (400 µl) using an EZ1 Virus 2.0 kit (Qiagen, Valencia, CA) on the BioRobot EZ1 extraction platform (Qiagen), following the manufacturers' instructions.
Detection of respiratory viruses was performed by multiplex PCR using an RVP Fast assay (Luminex Molecular Diagnostics, Toronto, Canada), following the manufacturer's instructions. The analysis was performed using an xMAP 100 IS instrument (Luminex Molecular Diagnostics). The RVP Fast assay simultaneously detects: influenza A virus, H1, H3, or H5 subtypes; influenza B virus; RSV‐A and ‐B; PIV‐1, ‐2, ‐3, and ‐4; adenovirus; human metapneumovirus; coronaviruses 229E, NL63, OC43, and HKU1; enterovirus/rhinovirus; and human bocavirus. Nucleic acid extractions were performed on 400 µl respiratory samples with an EZ‐1 virus 2.0 Kit on a BioRobot EZ‐1.
Cytokine Measurements
A commercial capture immunoassay was used to quantify IL‐6 in respiratory specimens (Human IL‐6 ELISA Ready‐set Go; eBioscience, San Diego, CA), following the manufacturer's instructions. For bronchoalveolar lavage specimens, epithelial lining fluid volume was calculated using urea as a dilution marker [Zedtwitz‐Liebenstein et al., 2005]. The detection limit of this assay is 2 pg/ml.
CMV Serological Testing
CMV IgGs were detected using a DiaSorin LIAISON® CMV IgG assay (DiaSorin, Saluggia, Italy).
Statistical Analysis
The data were analyzed with the aid of the SPSS statistical package, version 17.0 (SPSS, North Chicago, IL). Comparisons were undertaken using the χ2‐test for categorical variables and the non‐parametric Mann–Whitney U‐test for unpaired continuous data. Two‐sided exact P values are presented. A P value <0.05 was considered statistically significant.
RESULTS
Detection of CMV DNA in the Lower Respiratory Tract and in Plasma
Eleven patients were CMV‐seropositive at the time of fibrobronchoscopy, 15 were CMV‐seronegative, and the CMV‐serostatus of 15 was unknown. The presence of CMVDNA in the lower respiratory tract specimens was assessed for all patients. It was detected in six of the 11 (54.5%) CMV‐seropositive patients (median, 258 copies/ml; range, 27–815 copies/ml), and in none of the remaining patients. The presence of CMV DNA in the blood compartment (plasma) was investigated exclusively in CMV‐seropositive patients. Plasma CMV DNAemia was detected in two patients (18.8%). In both these patients, CMV DNA was also detected in the lower respiratory tract. Clinical and microbiological features of these patients are shown in Table I. It is noteworthy that two of the three patients with cystic fibrosis who were CMV‐seropositive at the time of fibrobronchoscopy had CMV DNA detected in their lower respiratory tract. Despite the fact that five of the six patients with CMV DNA detected in the lower respiratory tract were under corticosteroid treatment at the time of fibrobronchoscopy (Fluticasone), no significant association was found between the use of corticosteroids and the detection of CMV DNA (P = 0.435).
Table I.
Clinical and Microbiological Data of Six Cytomegalovirus (CMV)‐Seropositive Patients With CMV DNA Detected in the Lower Respiratory Tract
| Underlying disease | CMV/HHV‐6 DNA in LRT copies/mla | CMV DNA in plasma copies/ml | Respiratory virusesb | Bacterial and fungal culturesc |
|---|---|---|---|---|
| Cystic fibrosis | 334/2,350 | NDd | ND | S. aureus |
| Atelectasis | 182/ND | 123 | ND | Negative |
| Cystic fibrosis | 27/126 | ND | Rhinovirus | S. aureus |
| Atelectasis | 425/ND | 18 | ND | Negative |
| Bronchiolitis obliterans | 69/ND | ND | ND | Negative |
| Atelectasis | 1,285/ND | ND | Bocavirus | Negative |
CMV DNA load was quantitated by real‐time PCR with the Abbott CMV PCR kit. Detection and quantitation of human herpesvirus‐6 was performed by real‐time PCR (Realquality RS‐HHV6; AB Analitica, Padova, Italy).
Detection of respiratory viruses was performed by multiplex PCR using the RVP Fast assay (Luminex Molecular Diagnostics Inc., Toronto, Canada).
Growth of ≥104 or ≥105 CFU/ml of bacterial or fungal species in bronchoaspitates or bronchoalveolar lavages respectively was considered significant.
Not detected.
Microbiological Features of Patients With or Without Detectable CMV DNA
A comprehensive microbiological analysis of the lower respiratory tract specimens from the patients was performed. The presence of respiratory viruses was investigated by multiplex PCR, while the occurrence of common bacterial and fungal species was assessed by conventional culture methods. Respiratory viruses were detected in 13 of the 42 patients (rhinovirus, n = 6; respiratory syncytial virus, n = 2; bocavirus, n = 2; metapneumovirus, n = 1; coronaviruses HKN1, n = 1; and adenovirus, n = 1). Two of these 13 patients tested positive for CMV DNA in the lower respiratory tract. Significant growth of one or more bacterial species was observed in 17 of the 42 patients (Haemophilus influenzae biotypes VIII or III, n = 7; Staphylococcus aureus, n = 5; Streptococcus pneumoniae, n = 3; Moraxella catharralis, n = 2; Pseudomonas aeruginosa, n = 1; and Achromobacter xyloxosidans, n = 1). Candida parapsilosis was isolated from three patients. Two out of the six patients with detectable CMV DNA had a positive bacterial culture. No significant correlation (P = > 0.5) was found between the presence of CMV DNA infection and that of respiratory viruses, or bacterial or fungal species, either when all patients were considered collectively, irrespective of their CMV serostatus, or when the subgroup of CMV‐seropositive patients was analyzed separately.
Detection of Other Beta‐Herpesviruses in the Lower Respiratory Tract
The presence of HHV‐6 and HHV‐7 DNA in the lower respiratory tract was assessed in the 11 CMV‐seropositive patients. HHV‐6 DNA was detected in three patients (35, 52, and 2,350 copies/ml), two of which had CMV DNA present in the lower respiratory tract. HHV‐7 DNA was detected in one patient (408 copies/ml) who had no CMV DNA detected in the lower respiratory tract.
IL‐6 Levels in the Lower Respiratory Tract in Patients With or Without Detectable CMV DNA
IL‐6 has been shown to be a reliable marker of inflammation of the lower respiratory airways. The level of IL‐6 in lower respiratory tract specimens was quantified for all 42 patients. As shown in Figure 1, significantly higher levels of IL‐6 (P = 0.001) were found in patients with detectable CMV DNA (median, 99.8 pg/ml; range, 9.6–590 pg/ml) compared to patients without it (median, <2 pg/ml; range, <2–1,200 pg/ml). No differences in IL‐6 levels were found between CMV‐seropositive patients with no detectable CMV DNA and patients known to be CMV‐seronegative (P = 0.34).
Figure 1.
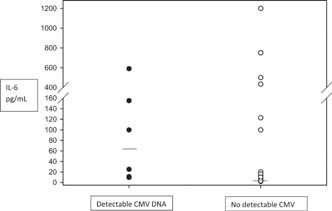
IL‐6 levels (pg/ml) in the lower respiratory tract of patients with or without detectable CMV DNA, as determined by a commercial capture immunoassay. Bars represent median values.
DISCUSSION
Active CMV infection is known to occur frequently in adult patients lacking canonical immunosuppression with inflammatory diseases, and may lead to increased morbidity [Söderberg‐Nauclér, 2008]. The current study investigated whether CMV DNA was detectable in the lower respiratory tract or plasma in children with various underlying bronchopulmonary diseases characterized by the presence of a chronic or recurrent inflammation of the lower respiratory airways.
Overall, CMV DNA was detected in the lower respiratory tract of 14.2% patients. However, this percentage increased to 54.6% when only patients known to be CMV‐seropositive at the time of fibrobronchoscopy were considered. As the lungs are a major site of CMV latency [Cook et al., 2002], it would be interesting to know the rate of CMV DNA detection in the lower respiratory tract in healthy CMV‐seropositive children; It is highly unlikely though this being as high as that found in this study. Involvement of the systemic compartment was demonstrated in two of the six patients with detectable CMV DNA in the lower respiratory tract. Low CMV DNA loads were usually observed in both compartments. The above scenario resembles that seen in critically ill patients subjected to mechanical ventilation. In these patients, it was demonstrated that a rapid expansion of CMV‐specific functional CD8+ T cells occurs upon CMV reactivation, keeping viral loads at low levels in both compartments and eventually controlling the episode [Chilet et al., 2010; Blanquer et al., 2011].
Particularly noteworthy, was the discovery that CMV DNA was detectable in the lower respiratory tract in two of three CMV‐seropositive cystic fibrosis patients. The levels of IL‐6 in the lower respiratory tract were significantly higher in patients with detectable CMV DNA than in those without it, further supporting the possibility of a bidirectional pathogenic relationship between CMV and inflammation [Söderberg‐Nauclér, 2008].
HHV‐6 replication was also detected in the lower respiratory tract in 50% of patients with active CMV infection. In turn, active HHV‐7 infection was detected in 1 out of the 11 CMV‐seropositive patients. This is consistent with the assumption that pro‐inflammatory states also may trigger reactivation of other beta‐herpes viruses which may in turn result in an increase in the net inflammatory state in the lower respiratory tract [Flamand et al., 2010].
As a part of the routine systematic analysis of the lower tract specimens from these patients, PCR analyses were performed to detect respiratory viruses, in addition to conventional bacterial and fungal cultures. Despite the stable clinical condition of the patients at the time of fibrobronchoscopy, respiratory viruses were detected in almost one‐third of the patients. Although contamination with viruses present in the upper respiratory tract could not be formally excluded, this feature is consistent with either a delayed clearance of a recent acute viral infection episode or with a persistent viral infection of the lower respiratory airways. The plausibility of the latter explanation is supported by data obtained in experimental animal models, and also in humans for respiratory syncytial virus, rhinovirus, and human metapneumovirus infections [Schwarze et al., 2004; Liu et al., 2009]. Persistent replication of these viruses in the lower respiratory tract induces inflammation [Papadopoulos et al., 2000], and may thereby promote local CMV reactivation. In turn, CMV may also potentiate respiratory virus replication in the lower respiratory tract by virtue of its immunosuppressive capacity [Solano and Navarro, 2010]. Nevertheless, no significant association was found between the presence of CMV DNA in the lower respiratory tract and the occurrence of respiratory viruses. Likewise, the occurrence of active CMV infection in critically ill patients has been related to an increased incidence of bacterial and fungal super infection [Jaber et al., 2005; Chiche et al., 2009]. The current investigation also found no association between the presence of CMV DNA in the lower respiratory tract and the occurrence of bacterial or fungal species. However, this study was not specifically designed to investigate these potential pathogenic interactions, and so no definitive conclusions may be drawn on this issue.
The current study had several limitations. Firstly, this was a cross‐sectional study with a limited sample size. Secondly, it included a highly heterogeneous cohort. Thirdly, given its retrospective nature, the CMV‐serostatus was unknown for a large number of patients. Fourthly, although cell supernatants were used for PCR analyses, the possibility that the detection of CMV DNA reflected a latent rather than an active viral infection in some patients, specially those with low CMV DNA loads, could not be formally ruled out, as neither detection of viral mRNA nor inoculation of conventional cell cultures for recovery of live virus were performed. Nevertheless, the data reported warrant further prospective observational studies to confirm the findings presented, explore the possibility of the existence of pathogenetic interactions between CMV, HHV‐6, and respiratory viruses, or bacterial and fungal species, colonizing the lower respiratory airways of these patients and assess the potential impact of CMV replication in the lower respiratory tract on the clinical course and outcome of the patients. In light of the data presented, children with cystic fibrosis should be the primary focus of these studies.
Acknowledgements
We thank Julia García, Matilde Pastor, and Mónica Reig for their technical assistance.
The authors do not have a commercial or other association that might pose a conflict of interest.
REFERENCES
- Blanquer J, Chilet M, Benet I, Aguilar G, Muñoz‐Cobo B, Tellez A, Costa E, Bravo D, Navarro D. 2011. Immunological insights into the pathogenesis of active CMV infection in non‐immunosuppressed critically ill patients. J Med Virol 83:1966–1971. [DOI] [PubMed] [Google Scholar]
- Bravo D, Clari MA, Costa E, Muñoz‐Cobo B, Solano C, José Remigia M, Navarro D. 2011. Comparative evaluation of three automated systems for DNA extraction in conjunction with three commercially available real‐time PCR assays for quantitation of plasma cytomegalovirus DNAemia in allogeneic stem cell transplant recipients. J Clin Microbiol 49:2899–2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiche L, Forel JM, Roch A, Guervilly C, Pauly V, Allardet‐Servent J, Gainnier M, Zandotti C, Papazian L. 2009. Active cytomegalovirus infection is common in mechanically ventilated medical intensive care unit patients. Crit Care Med 37:1850–1857. [DOI] [PubMed] [Google Scholar]
- Chilet M, Aguilar G, Benet I, Belda J, Tormo N, Carbonell JA, Clari MA, Costa E, Navarro D. 2010. Virological and immunological features of active cytomegalovirus infection in nonimmunosuppressed patients in a surgical and trauma intensive care unit. J Med Virol 82:1384–1391. [DOI] [PubMed] [Google Scholar]
- Cook CH, Zhang Y, McGuiness BJ, Lahm MC, Sedmak DD, Ferguson RM. 2002. Intra‐abdominal infection reactivates latent pulmonary cytomegalovirus in immunocompetent mice. J Infect Dis 185:1395–1400. [DOI] [PubMed] [Google Scholar]
- Flamand L, Komaroff AL, Arbuckle JH, Medveczky PG, Ablashi DV. 2010. Human herpesvirus‐6‐basic biology, diagnostic testing, and antiviral efficacy. J Med Virol 82:1560–1568. [DOI] [PubMed] [Google Scholar]
- Gimeno C, Solano C, Latorre JC, Hernández‐Boluda JC, Clari MA, Remigia MJ, Furió S, Calabuig M, Tormo N, Navarro D. 2008. Quantification of DNA in plasma by an automated real‐time PCR assay (CMV PCR Kit, Abbott) for surveillance of active cytomegalovirus infection and guidance of pre‐emptive therapy for allogeneic hematopoietic stem cell transplant recipients. J Clin Microbiol 46:3311–3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaber S, Chanques G, Borry J, Souche B, Verdier R, Perrigault PF, Eledjam JJ. 2005. Cytomegalovirus infection in critically ill patients: Associated factors and consequences. Chest 127:233–241. [DOI] [PubMed] [Google Scholar]
- Kalil AC, Florescu DF. 2009. Prevalence and mortality associated with cytomegalovirus infection in nonimmunosuppressed patients in the intensive care unit. Crit Care Med 37:2350–2358. [DOI] [PubMed] [Google Scholar]
- Liu Y, Haas DL, Poore S, Isakovic S, Gahan M, Mahalingam S, Fu ZF, Tripp RA. 2009. Human metapneumovirus establishes persistent infection in the lungs of mice and is reactivated by glucocorticoid treatment. J Virol 83:6837–6848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Núñez J, Chilet M, Sanchis J, Bodí V, Núñez E, Miñana G, Tormo N, Clari MA, Pellicer M, Chorro FJ, Llàcer A, Navarro D. 2010. Prevalence and prognostic implications of active cytomegalovirus infection in patients with acute heart failure. Clin Sci (Lond) 119:443–452. [DOI] [PubMed] [Google Scholar]
- Núñez J, Chilet M, Blasco ML, Clari MA, Sanjuan R, Muñoz‐Cobo B, Bodí V, Costa E, Bravo D, Sanchis J, Miñana G, Navarro D. 2012. Low rate of detection of active cytomegalovirus (CMV) infection early following acute myocardial infarction. Atherosclerosis 222:295–297. [DOI] [PubMed] [Google Scholar]
- Papadopoulos NG, Bates PJ, Bardin PG, Papi A, Leir SH, Fraenkel DJ, Meyer J, Lackie PM, Sanderson G, Holgate ST, Johnston SL. 2000. Rhinoviruses infect the lower airways. J Infect Dis 181:1875–1884. [DOI] [PubMed] [Google Scholar]
- Prösch S, Wendt CE, Reinke P, Liebenthal C, Stamminger T, Volk HD, Krüger DH. 1995. Stimulation of the human cytomegalovirus IE enhancer/promoter in HL‐60 cells by TNF alpha is mediated via induction of NF‐KappaB. Virology 208:197–206. [DOI] [PubMed] [Google Scholar]
- Schwarze J, O'Donnell DR, Rohwedder A, Openshaw PJ. 2004. Latency and persistence of respiratory syncytial virus despite T cell immunity. Am J Respir Crit Care Med 169:801–805. [DOI] [PubMed] [Google Scholar]
- Söderberg‐Nauclér C. 2008. HCMV microinfections in inflammatory diseases and cancer. J Clin Virol 41:218–223. [DOI] [PubMed] [Google Scholar]
- Solano C, Navarro D. 2010. Clinical virology of cytomegalovirus infection following hematopoietic transplantation. Future Virol 5:111–124. [Google Scholar]
- Zedtwitz‐Liebenstein K, Schenk P, Apfalter P, Fuhrmann V, Stoiser B, Graninger W, Schuster E, Frass M, Burgmann H. 2005. Ventilator‐associated pneumonia: Increased bacterial counts in bronchoalveolar lavage by using urea as an endogenous marker of dilution. Crit Care Med 33:756–769. [DOI] [PubMed] [Google Scholar]


