Abstract
In 2005, human coronavirus HKU1 (HCoV‐HKU1) was isolated and identified from a 71‐year‐old man with pneumonia in Hong Kong. To identify and classify genotypes of HCoV‐HKU1 in Korea, a sensitive, specific, and quantitative real‐time polymerase chain reaction (PCR) assay was developed and analyzed the sequences of HCoV‐HKU1 isolated in Korea. A total of 1,985 respiratory specimens taken from patients with acute respiratory illness were tested for HCoV‐HKU1 from January 2007 to May 2008. The major clinical symptoms associated with HCoV‐HKU1 infection were examined statistically and sequence variations of the RNA‐dependent RNA polymerase (RdRp), spike, and nucleocapsid genes were also analyzed. Fifty cases (2.5%) HCoV‐HKU1 were identified by real‐time PCR and viral loads ranged from 6.7 × 104 to 1.6 × 109 copies/ml. The clinical symptoms of HCoV‐HKU1 infection included rhinorrhea (72%), cough (64%), nasal congestion (56%), fever (32%), sputum (30%), sore throat (18%), chills (16%), postnasal discharge (14%), and tonsillar hypertrophy (10%). There was a seasonal distribution of HCoV‐HKU1 infection, peaking in winter and spring. Both genotypes A and B were detected but no recombination between them was found. This is the first report on the identification and genotyping of HCoV‐HKU1 as a causative agent of acute respiratory illness in Korea. The data suggest that at least two genotypes, A and B, of HCoV‐HKU1 with scattered silent mutations were circulating in Korea from 2007 to 2008. J. Med. Virol. 85:309–314, 2013. © 2012 Wiley Periodicals, Inc.
Keywords: human coronavirus HKU1, real‐time PCR, acute respiratory illness, genotype
INTRODUCTION
Human coronavirus (HCoV) 229E and HCoV‐OC43 account for 5–30% of cases of acute respiratory tract infection [McIntosh et al., 1969]. HCoV‐NL63 was discovered in 2004 as a minor pathogen of HCoV infections in humans [van der Hoek et al., 2004]. A new human coronavirus‐HKU1 (HCoV‐HKU1) has been reported from a patient with pneumonia in Hong Kong. This species was classified into family Coronaviridae, genus Betacoronavirus, and further as a lineage A [Woo et al., 2005c].
The genome contains the genes for replicase polyproteins 1a/1b (pp1a/1b), hemagglutinin esterase (HE), spike (S), envelope (E), membrane (M), and nucleocapsid (N) in the same order as in other members of the genus Betacoronavirus [Pyrc et al., 2007; Woo et al., 2009]. HCoV is associated frequently with community acquired upper respiratory tract infections showing typical clinical symptoms including a runny nose, fever, coughing, and wheezing [Labret et al., 2005; Esper et al., 2006; Gose et al., 2011]. However, other disease manifestations including bronchiolitis and pneumonia can occur [Bosis et al., 2007; Gerna et al., 2007; Vabret et al., 2007]. The respiratory symptoms of HCoV‐HKU1 infection are usually similar to infections with other HCoVs, but HCoV‐HKU1 can cause death in elderly people [Ren et al., 2011] and upper and lower respiratory tract infections in children [Woo et al., 2005b; Jin et al., 2012].
In this study, a Taq‐Man®‐based real‐time polymerase chain reaction (PCR) method that targets the HCoV‐HKU1 open reading frame (ORF) 1a and ORF 1b genes with high sensitivity and specificity was developed and evaluated. Sequence information for the RdRp, S, and N genes of HCoV‐HKU1 was also analyzed to classify the genotype prevalence of the virus in Korea [Woo et al., 2005a; Woo et al., 2006]. This study is the first to identify and classify the genotypic predisposition of HCoV‐HKU1 as a causative agent of acute respiratory illness in Korea.
MATERIALS AND METHODS
Clinical Specimens
Throat swab specimens were collected from patients with an acute respiratory illness. Enrollment of 1,985 patients was performed under the acute respiratory tract infection surveillance (ARI‐Net) program in the Korea Centers for Disease Control and Prevention (KCDC) from January 2007 to May 2008. All clinical specimens were collected under regulation of Korea National Institute of Health institutional review boards (IRBs) and written informed consents were obtained from patients or their parent/guardian.
Real‐Time PCR Assay for HCoV‐HKU1
To design primer and probes for real‐time PCR, full genome sequence of HCoV‐HKU1 was retrieved from GenBank accession no. NC006577. HCoV‐HKU1 ORF 1a and ORF 1b gene were analyzed using Primer Express software version 2.0 (Applied Biosystems, Foster City, CA) to allocate target sites which could be used as primers and probes. Total RNAs from 140 µl of clinical specimens were extracted using QIAamp viral RNA mini kit (Qiagen, Hilden, Germany) and directly used as templates. cDNA synthesis was performed using random primers and murine mammal leukemia virus (M‐MLV) reverse transcriptase (Promega, Madison, WI) in accordance with manufacturer's instruction. Real‐time PCR assays targeting the ORF 1a and ORF 1b sequences were performed using 10 µl aliquots of mixtures containing 1 µl cDNA or a standard plasmid, 5 µl Taq‐Man® universal PCR Master Mix (Applied Biosystems) containing ROX as a passive reference dye, 900 nM of forward and reverse primers and 250 nM of probe (Table I). Amplification and detection were performed using an ABI PRISM 7900HT sequence detection system (Applied Biosystems) under the following conditions: uracil‐N‐glycosylase was activated at 50°C for 2 min, Taq polymerase was activated at 95°C for 10 min and 40 cycles of amplification (95°C for 15 sec, 60°C for 1 min) were run [Choi et al., 2008; Lu et al., 2008]. Positive values were decided when the copy number exceeded 10 copies/µl and the two target genes (ORF 1a and ORF 1b) were detected simultaneously. The viral load was calculated and determined from mean threshold cycle (Ct) values of specimens and a standard curve prepared from a known concentration of the DNA encoding the RdRp gene. All quantification experiments were performed using triple repetitions.
Table I.
Primer and Probe Sequences of HCoV‐HKU1 Used in Real‐Time PCR Assay
| Gene | Primer/Probe | Sequence (5′–3′) | Position |
|---|---|---|---|
| ORF 1a | Forward | GTTGGTTGTATGATGCGTTTGTTCT | 7,922–7,946 |
| Reverse | TCTACAAATAAACTAGCATCAACATCATCGT | 7,971–8,001 | |
| Probe | FAM‐CACGCTGTCCATCTCT‐NFQ | 7,952–7,967 | |
| ORF 1b | Forward | CCTAACTGTGATGTGAGTGATGTCA | 16,516–16,540 |
| Reverse | ACAAACCAAAGACCATACCATTCATAACT | 16,608–16,636 | |
| Probe | FAM‐CATACCGCCCAAATAT‐NFQ | 16,548–16,563 |
FAM, 6‐carboxylfluorescein; NFQ, non‐fluorescent quenchers.
Statistical Analysis of Clinical Symptoms
The characteristics of clinical symptoms of HCoV‐HKU1 infection were investigated retrospectively from list of clinical records which were compiled up through ARI‐Net. Regression analysis was performed with 95% confidential level to identify relationship between HCoV‐HKU1 infection and clinical symptoms using the SAS program (version 9.2).
Reverse Transcription (RT)‐PCR and Sequencing of RdRp, S, and N Genes of HCoV‐HKU1 and Phylogenetic Analysis
The RdRp, S, and N genes of HCoV‐HKU1 were amplified by RT‐PCR with specific primers (Table II). cDNA synthesis was performed as described above using M‐MLV reverse transcriptase (Promega). The PCR assays were performed in a 50 µl reaction volumes containing 5 µl cDNA, 5 µl 10 × PCRbuffer, 2 µl dNTPmix (final concentration of 400 mM of each dNTP), 1 µl SP Taq (Cosmo Gentech, Seoul, Korea), 1 µl of each primer (10 pM), and nuclease‐free water to 50 µl. The PCR reaction was carried out at 94°C for 3 min, 35 cycles of amplification (30 sec at 94°C; 30 sec at 48°C for RdRp, 58°C for S, 56°C for N; 30 sec at 72°C), and a final extension step at 72°C for 10 min. The PCR products were run on a 2% agarose gel, stained with SYBR safe DNA gel stain dye (Invitrogen, Carlsbad, CA) and visualized under UV light. Sequencing reactions were performed using BigDye® Terminators version 3.1 Cycle Sequencing Kits (Applied Biosystems) using the manufacturer's instructions on a DNA analyzer (model 3730, Applied Biosystems). Phylogenetic tree construction was performed using the MEGA4 program (http://www.megasoftware.net/) using the neighbor‐joining method with Clustal W, and bootstrap values calculated from 500 trees.
Table II.
Primer Sequences of HCoV‐HKU1 for Amplification of RdRp, S, and N Gene
| Primer | Sequence (5′–3′) | Position |
|---|---|---|
| RdRp‐F | GGG TAT GAA GTA TCA TCC TA | 14,433–14,452 |
| RdRp‐R | GAT AAT CCC AAC CCA TAA GAA C | 15,400–15,421 |
| S–F | AAC RYG GTG TTA TTA CTA | 23,756–23,773 |
| S–R | AGA WGA TTG CAR AAA RCC AGA ACT | 24,154–24,177 |
| N–F | ACT TGA RCG AAA YYA YCA AAC | 28,409–28,429 |
| N–R | CGY AAA CCT AGT AGG GAT AGC TT | 28,825–28,847 |
Genbank Nucleotide Sequence Accession Numbers
The following nucleotide accession numbers were used (http://www.ncbi.nlm.nih.gov/genbank/): reference genes; AU884001, AY597011, DQ415899, DQ415911, DQ415912, DQ415914, DQ437616, EF507775, RdRp genes; JN234449, JN234450, JN234453, JN234454, JN234455, JN234456, JN234458, JN234459, JN234460, JN234461, JN234462, S genes; JN234465, JN234466, JN234468, JN234469, JN234470, JN234471, JN234473, JN234474, JN234475, JN234476, JN234477, N genes; JN234478, JN234479, JN234482, JN234483, JN234484, JN234487, JN234489, JN234490, JN234492, JN234493, and JN234494.
RESULTS
Development of Real‐Time PCR for Detecting HCoV‐HKU1
The sensitivity and specificity of this real‐time PCR were evaluated. Tenfold serial dilutions of the standard HCoV‐HKU1 plasmid DNA ranging from 1 to 1 × 1010 copies/µl were used as templates for comparing the sensitivity between real‐time PCR and conventional PCR. The detection limit of real‐time PCR assays was as low as 10 copies/µl. The sensitivity of the real‐time PCR assays used in this study was 1 × 105 times higher than conventional PCR (Fig. 1). Cross reactivity was also evaluated with cDNA of influenza viruses A/B, parainfluenza viruses 1/2/3, respiratory syncytial virus, human rhinovirus and enterovirus, DNA of adenovirus, human bocavirus, and respiratory pathogenic bacteria (Haemophilus influenzae, Legionella spp., Streptococcus pneumoniae, Mycoplasma pneumoniae, and Chlamydophila pneumoniae). No non‐specific amplification was detected in either assay system (data not shown).
Figure 1.
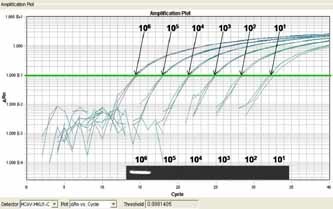
Comparison of the sensitivity of the real‐time polymerase chain reaction (PCR) assay for the human coronavirus HKU1 (HCoV‐HKU1) ORF 1b gene with conventional PCR. The RT‐PCR assay detected up to 10 copies/µl of standard DNA, whereas the detection limit of conventional PCR was 1 × 106 copies/µl.
Detection and Clinical Characteristics of HCoV‐HKU1 Infection
Fifty HCoV‐HKU1‐positive cases were detected (2.5%) out of 1,985 clinical specimens using real‐time PCR assays targeting ORF 1a and ORF 1b. The amount of RNA detected in positive specimens ranged from 6.7 × 104 to 1.6 × 109 copies/ml. Based on analysis of multiple alternative clinical records, rhinorrhea (72% of patients), cough (64%), and nasal congestion (56%) were the major symptoms of patients infected with HCoV‐HKU1. The other basic respiratory symptoms were fever (32%), sputum discharge (30%), sore throat (18%), chills (16%), postnasal discharge (14%), and tonsillar hypertrophy (10%; Table III). There were no positive cases associated with severe respiratory symptoms such as pneumonia and bronchiolitis. Seasonal distribution of HCoV‐HKU1 infection was found, with the main prevalence from early winter to late spring with an 8% mean positive rate (Fig. 2).
Table III.
Proportion of Clinical Symptoms of HCoV‐HKU1 Infected or Non‐Infected Patients
| No. (%) of HCoV‐HKU1a | No. (%) of non‐HCoV‐HKU1a | |
|---|---|---|
| Rhinorrhea | 36 (72.0) | 1,547 (79.9) |
| Cough | 32 (64.0) | 1,318 (68.1) |
| Stuffy nose | 28 (56.0) | 1,215 (62.8) |
| Fever | 16 (32.0) | 622 (32.1) |
| Sputum | 15 (30.0) | 493 (25.5) |
| Sore throat | 9 (18.0) | 267 (13.8) |
| Chill | 8 (16.0) | 230 (11.9) |
| Postnasal discharge | 7 (14.0) | 332 (17.2) |
| Tonsillar hypertrophy | 5 (10.0) | 114 (5.9) |
| Throat flare | 4 (8.0) | 235 (12.1) |
| Otalgia | 3 (6.0) | 19 (1.0) |
| Pain | 3 (6.0) | 52 (2.7) |
| Hoarseness | 2 (4.0) | 78 (4.0) |
| Eye discharge | 1 (2.0) | 37 (1.9) |
| Otarrhea | 1 (2.0) | 10 (0.5) |
| Wheeze | 0 (0.0) | 37 (1.9) |
| Dyspnea | 0 (0.0) | 38 (2.0) |
Clinical symptoms were scored as multiple‐alternative manner.
Figure 2.
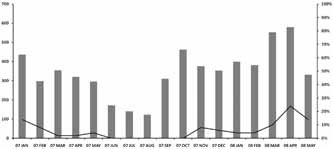
Seasonal distribution of HCoV‐HKU1 infection from January 2007 to May 2008. The HCoV‐HKU1 prevailed from early winter to late spring in Korea. Bars indicate the monthly number of clinical specimens. The line represents the monthly proportion of positive cases as a whole.
Sequencing and Phylogenetic Analysis of HCoV‐HKU1
RdRp, S, and N genes and the putative amino acid sequences of 11 cases, of which the target genes were amplified by conventional PCR, were analyzed. Eight cases were HCoV‐HKU1 genotype B (73%) and three were HCoV‐HKU1 genotype A (27%) when queried through a public database [Benson et al., 2005]. Phylogenetic analysis also indicated that HCoV‐HKU1 isolates in this study consisted of genotypes A and B, whereas genotype C was not identified (Fig. 3).
Figure 3.
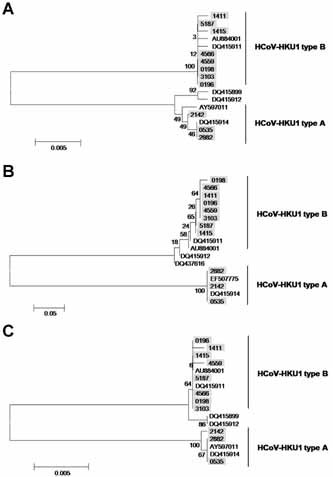
Phylogenetic analysis of the RdRp, S, and N genes of HCoV‐HKU1. Eleven positive cases were classed as genotype A (n = 3) or B (n = 8). There was no HCoV‐HKU1 genotype C detected in Korea during the study period. Each shadowed number indicates the specimen identification number, and reference sequences are marked as GenBank accession numbers. Phylogenetic trees are shown of (A) the RdRp gene (897 neucleotide (nt)), (B) the S gene (309 nt), and (C) the N gene (425 nt). Primer sequences were excluded from phylogenetic analysis.
DISCUSSION
Since HCoV‐HKU1 was first identified in a patient with pneumonia in Hong Kong, clinical manifestations and epidemiological features of infection with this virus have been reported from several countries [Labret et al., 2005; Esper et al., 2006; Gose et al., 2011]. Chung et al. [2007] also tried to identify the virus in patients with acute expiratory wheezing in Korea but evidence of HCoV‐HKU1 infection was not found. These negative results arose from several limitations such as an insensitive diagnostic strategy, selection of samples out of season as well as the relatively small number of patients studied. To circumvent these limitations, a quantitative real‐time PCR was developed and targeted a study group of 1,985 throat swab specimens collected from patients with an acute respiratory illness from January 2007 to May 2008.
The aim of the real‐time PCR assays developed in this study was to detect both of the HCoV‐HKU1 ORF 1a and ORF 1b genes. The assays provided enhanced sensitivity and specificity, which could permit quantitative analysis of HCoV‐HKU1 in clinical specimens. The ORF 1b assay gave more dynamic Ct values than the ORF 1a assay. Therefore, the ORF 1b assay could be more applicable for clinical diagnosis using respiratory specimens from patients with acute respiratory tract infections caused by HCoV‐HKU1.
In this study, HCoV‐HKU1 was found in 2.5% of patients (50 cases) with acute respiratory illness sampled from January 2007 to May 2008 in Korea. A similar incidence was reported previously in Hong Kong [Woo et al., 2009] and China [Jin et al., 2010] but an apparently higher rate was noted in Chinese study because of different clinical practices. Unlikely, apparent lower incidence (0.3%, n = 4,181) of HCoV‐HKU1 in patients with acute respiratory tract in Hong Kong was reported by Lau et al. [2006]. Relatively high incidence of HCoV‐HKU1 in Korea might have resulted from difference of target study group and/or sensitivity of detection strategy. Even though the quantified viral loads varied from 6.7 × 104 to 1.6 × 109 copies/ml in the respiratory disease specimens used, there were no significant correlations between viral load and specific clinical features. Similarly, there were no statistically significant relationships between the HCoV‐HKU1 infection and any particular clinical symptoms. As for seasonal prevalence, the HCoV‐HKU1 infections prevailed in the winter and spring in Korea and this result was consistent with the infection rates of HCoV‐OC43, 229E and with other reports [Lau et al., 2006; Domiquez et al., 2009; Gaunt et al., 2010]. One limitation of this study was that because clinical specimens were chosen from negative cases of acute respiratory illness in the ARI‐Net laboratory surveillance system [Chun et al., 2009], dual or multiple infections of HCoV‐HKU1 with other respiratory viruses could not be detected.
To identify whether there was any recombinant HCoV‐HKU1 isolated from Korean patients, the nucleotide sequence information of the RdRp, S, and N genes was analyzed. Similar to previous study reported by Lau et al. [2006] in Hong Kong, our phylogenetic analysis revealed that 11 cases of HCoV‐HKU1 isolated in Korea comprised two genotypes: A or B. When single reciprocal nucleotide exchanges were analyzed between the RdRp, S, and N genes, no recombination could be identified. Instead, several point mutations were observed at the nucleotide level, resulting in a transition (C to T) or a transversion (C to T or T to C) in the RdRp, S, and N genes. However, these were silent mutations so that no changes occurred at the amino acid level. Interestingly, a silent continuous mutation in the N gene was identified from 6/8 isolates of HCoV‐HKU1 genotype B. Thus, there might have been a unique strain of HCoV‐HKU1 prevailing in Korea at that time.
This is the first report that HCoV‐HKU1 was associated with acute respiratory illness in Korea. Infections with HCoV‐HKU1 peaked from winter through to spring in Korea during 2007–2008. Sequencing and phylogenetic analysis revealed that the 11 HCoV‐HKU1 isolates belonged to two distinct genotypes A and B based on the RdRp, S, and N genes. Unlike a previous study, no gene recombination was identified in HCoV‐HKU1. Instead, a conserved silent mutation was observed in the N gene of the putative Korean strain of HCoV‐HKU1 genotype B from six specimens. Intensive molecular epidemiological investigations would be required to elucidate any possible changes in viral tropism caused by such mutations.
Acknowledgements
Authors are grateful to Mr. H.D. Cheong for excellent statistical analysis of clinical symptoms.
The authors declare no conflict of interest.
REFERENCES
- Benson DA, Karsch‐Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. 2005. GenBank. Nucleic Acids Res 33:34–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosis S, Esposito S, Niesters HG, Tremolati E, Pas S, Principi N, Osterhaus AD. 2007. Coronavirus HKU1 in an Italian pre‐term infant with bronchiolitis. J Clin Virol 38:251–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JH, Chung YS, Kim KS, Lee WJ, Chung IY, Oh HB, Kang C. 2008. Development of real‐time PCR assays for detection and quantification of human bocavirus. J Clin Virol 42:249–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun JK, Lee JH, Kim HS, Cheong HM, Kim KS. 2009. Establishing a surveillance network for severe lower respiratory tract infections in Korean infants and young children. Eur J Clin Microbiol Infect Dis 28:841–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung JY, Han TH, Kim SW, Kim CK, Hwang ES. 2007. Detection of viruses identified recently in children with acute wheezing. J Med Virol 79:1238–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domiquez SR, Robinson CC, Holmes KV. 2009. Detection of four human coronaviruses in respiratory infections in children: A one‐year study in Colorado. J Med Virol 81:1597–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esper F, Weibel C, Ferguson D, Landry ML, Kahn JS. 2006. Coronavirus HKU1 infection in the United States. Emerg Infect Dis 12:775–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaunt ER, Hardie A, Claas EC, Simmonds P, Templeton KE. 2010. Epidemiology and clinical presentations of the four human coronaviruses 229E, HKU1, NL63, and OC43 detected over 3 years using a novel multiplex real‐time PCR method. J Clin Microbiol 48:2940–2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerna G, Percivalle E, Sarasini A, Campanini G, Piralla A, Rovida F, Genini E, Marchi A, Baldanti F. 2007. Human respiratory coronavirus HKU1 versus other coronavirus infections in Italian hospitalized patients. J Clin Virol 38:244–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gose LG, Durigon EL, Campos AA, Hein N, Passos SD, Jerez JA. 2011. Coronavirus HKU1 in children, Brazil, 1995. Emerg Infect Dis 17:1147–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Song JR, Xie ZP, GAo HC, Yuan XH, Xu ZQ, Yan KL, Zhao Y, Xiao NG, Hou YD, Duan ZJ. 2010. Prevalence and clinical characteristics of human CoV‐HKU1 in children with acute respiratory tract infections in China. J Clin Virol 49:126–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Zhang RF, Xie ZP, Yan KL, Gao HC, Song JR, Yuan XH, Cheng WX, Hou YD, Duan ZJ. 2012. Newly identified respiratory viruses associated with acute lower respiratory tract infections in Lanzou, China, from 2006 to 2009. Clin Microbiol Infect 18:74–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labret A, Dina J, Guerin S, Petit jean J, Courbet S, Freymuth F. 2005. Detection of the new human coronavirus HKU1: A report of 6 cases. Clin Infect Dis 42:634–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau SK, Woo PC, Yip CC, Tse H, Tsoi HW, Cheng VC, Lee P, Tang BS, Cheung CH, Lee RA, So LY, Lau YL, Chan KH, Yuen KY. 2006. Coronavirus HKU1 and other coronavirus infections in Hong Kong. J Clin Microbiol 44:2063–2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu BJ, Zhang LL, Tan WJ, Zhou WM, Wang Z, Peng K, Ruan L. 2008. Development and comparison of real‐time and conventional RT‐PCR assay for detection of human coronavirus NL63 and HKU1. Bing Du Zue Bao 24:305–311. [PubMed] [Google Scholar]
- McIntosh K, Kapikian AZ, Turner HC, Hartley JW, Parrott RH, Chanock RM. 1969. Seroepidemiologic studies of coronavirus infection in adults and children. Am J Epidemol 91:585–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyrc K, Berkhout B, van der Hoek L. 2007. The novel human coronaviruses NL63 and HKU1. J Virol 81:3051–3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren L, Gonzalez R, Xu J, Xiao Y, Li Y, Zhou H, Li J, Yang Q, Zhang J, Chen L, Wang W, Vernet G, Paranhos‐Baccalà G, Wang Z, Wang J. 2011. Prevalence of human coronaviruses in adults with acute respiratory tract infections in Beijing, China. J Med Virol 83:291–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vabret A, Dina J, Gouarin S, Petitjean J, Tripey V, Brouard J, Freymuth F. 2007. Human (non‐severe acute respiratory syndrome) coronavirus infections in hospitalized children in France. J Paediatr Child Health 44:176–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoek L, Pyrc K, Jebbink MF, Vermeulen‐Oost W, Berkhout RJ, Wolthers KC, Wertheim‐van Dillen P, Kaandorp J, Spaargaren J, Berkhout B. 2004. Identification of a new human coronavirus. Nat Med 10:368–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo PC, Lau SK, Chu CM, Chan KM, Tsoi HW, Huang Y, Wong BH, Poon RW, Cai JJ, Luk WK, Poon LL, Wong SS, Guan Y, Peiris JS, Yuen KY. 2005a. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J Virol 79:884–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo PC, Lau SK, Tsoi HW, Huang Y, Poon RW, Chu CM, Kee RA, Luk WK, Wong GK, Wong BH, Cheng VC, Tang BS, Wu AK, Yung RW, Chen H, Guan Y, Chan KH, Yuen KY. 2005b. Clinical and molecular epidemiological features of coronavirus HKU1‐associated community‐acquired pneumonia. J Infect Dis 192:1898–1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo PC, Lau SK, Huang Y, Tsoi HW, Chan KH, Yuen KY. 2005c. Phylogenetic and recombination analysis of coronavirus HKU1, a novel coronavirus from patients with pneumonia. Arch Virol 150:2299–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo PC, Lau SK, Yip CC, Huang Y, Tsoi HW, Chan KH, Yuen KY. 2006. Comparative analysis of 22 coronavirus HKU1 genomes reveals a novel genotype and evidence of natural recombination in coronavirus HKU1. J Virol 80:7136–7145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo PC, Lau SK, Yip CC, Huang Y, Yuen KY. 2009. More and more a coronaviruses: Human coronavirus HKU1. Viruses 1:57–71. [DOI] [PMC free article] [PubMed] [Google Scholar]


