Abstract
The discovery of human Metapneumovirus (hMPV) and human Bocavirus (hBoV) identified the etiological causes of several cases of acute respiratory tract infections in children. This report describes the molecular epidemiology of hMPV and hBoV infections observed following viral surveillance of children hospitalized for acute respiratory tract infections in Milan, Italy. Pharyngeal swabs were collected from 240 children ≤3 years of age (130 males, 110 females; median age, 5.0 months; IQR, 2.0–12.5 months) and tested for respiratory viruses, including hMPV and hBoV, by molecular methods. hMPV‐RNA and hBoV‐DNA positive samples were characterized molecularly and a phylogenetical analysis was performed. PCR analysis identified 131/240 (54.6%) samples positive for at least one virus. The frequency of hMPV and hBoV infections was similar (8.3% and 12.1%, respectively). Both infections were associated with lower respiratory tract infections: hMPV was present as a single infectious agent in 7.2% of children with bronchiolitis, hBoV was associated with 18.5% of pediatric pneumonias and identified frequently as a single etiological agent. Genetically distinct hMPV and hBoV strains were identified in children examined with respiratory tract infections. Phylogenetic analysis showed an increased prevalence of hMPV genotype A (A2b sublineage) compared to genotype B (80% vs. 20%, respectively) and of the hBoV genotype St2 compared to genotype St1 (71.4% vs. 28.6%, respectively). Interestingly, a shift in hMPV infections resulting from A2 strains has been observed in recent years. In addition, the occurrence of recombination events between two hBoV strains with a breakpoint located in the VP1/VP2 region was identified. J. Med. Virol. 83:156–164, 2011. © 2010 Wiley‐Liss, Inc.
Keywords: pediatric respiratory infections, phylogenetic analysis, human Metapneumovirus, human Bocavirus, pediatric hospitalization, viral recombination
INTRODUCTION
Acute respiratory tract infections are an important cause of morbidity and mortality worldwide. These infections are ubiquitous and contagious, affecting repeatedly individuals of all ages, particularly children younger than 5 years of age [World Health Organization, 2009] and many infections have unknown etiologies. The recent discovery of the human Metapneumovirus (hMPV) in 2001 [van den Hoogen et al., 2001] and of the human Bocavirus (hBoV) in 2005 [Allander et al., 2005] identified the etiological causes of several respiratory infections. hMPV is a common cause of respiratory disease in children under 5 years old and often accounts for severe clinical manifestations, including bronchiolitis or pneumonia [Foulongne et al., 2006]. hMPV is a single‐stranded negative RNA virus of the Paramyxoviridae family, Pneumovirinae subfamily [van den Hoogen et al., 2001; Broor et al., 2008]. Molecular analysis identified two major genotypes (A and B) with genetic lineages A1, A2 and B1, B2, respectively [van den Hoogen et al., 2004]. Lineage A2 has been recently divided into A2a and A2b sublineages [Huck et al., 2006].
hBoV is detected frequently in the respiratory and gastrointestinal tracts, suggesting its pathologic involvement at both sites [Lindner et al., 2008; Schildgen et al., 2008]. hBoV belongs to the Bocavirus genus of the Parvoviridae family. It is a non‐enveloped virus and contains a linear single‐stranded DNA molecule. The two major hBoV genotypes correspond to the original St1 (Stockholm 1) and St2 (Stockholm 2) isolates [Allander et al., 2005].
This report describes the molecular epidemiology of hMPV and hBoV identified following surveillance for respiratory viruses in children hospitalized for acute respiratory tract infections in Milan, Italy. A detailed genetic analysis was carried out using the hMPV and hBoV sequences identified.
PATIENTS AND METHODS
Study Population
Pediatric patients (N = 240; 130 males and 110 females; median age, 5.0 months; interquartile range, 2.0–12.5 months) admitted to the San Carlo Borromeo Hospital in Milan between 2004 and 2008 with a diagnosis of acute respiratory infection were enrolled in the study. Lower respiratory tract infections (i.e., bronchitis, bronchiolitis, pneumonia, or asthma) were observed in 66.7% (95% CI: 60.5–72.4) of children. Bronchiolitis and bronchitis were the most frequent outcomes (60.6%, 95% CI: 52.9–68.0 and 37.5%, 95% CI: 30.3–45.2, respectively), pneumonia and asthma being less frequent (16.9%, 95% CI: 11.7–23.3 and 9.4%, 95% CI: 5.5–14.7).
At the time of hospital admission pharyngeal swabs were collected from each patient for the identification and characterization of respiratory viruses using Plain Swabs and ITM‐RT (Copan Diagnostics, Murrieta, CA). Informed consent was obtained from parents of eligible children and children were divided into four age groups: ≤6 months (60%, 95% CI: 53.7–66.1), 7–12 months (15%, 95% CI: 10.9–19.9), 13–24 months (18.8%, 95% CI: 14.2–24.1), and 25–36 months (6.2%, 95% CI: 3.7–9.9).
The seasonal distribution of the samples collected is as follows: 35.4% (95% CI: 29.6–41.6) of samples were collected during autumn (October–December), 52.5% (95% CI: 46.2–55.8) during winter (January–March), and 12.1% (95% CI: 8.4–16.7) during spring (April–June).
Nucleic Acid Extraction and Amplification
Nucleic acids were extracted using the RNeasy Mini Kit and QIAmp DNA Mini Kit (Qiagen GmbH, Hilden, Germany). For RNA virus detection cDNA was synthesized with pd(N)6 random hexamer primers (Amersham Biosciences, Little Chalfont, Buckinghamshire, UK) using a M‐MLV reverse transcriptase (Invitrogen Tech‐Line, Paisley, UK). Viral detection was performed by PCR for the identification and typing of both classic viral respiratory pathogens i.e., influenza virus A and B (FluA/B), parainfluenza viruses 1–4 (hPIV1‐4), respiratory syncytial virus A and B (RSVA/B), coronavirus 229E and OC43 (hCoV229E/OC43), rhinovirus (hRV), adenovirus (hAdV), and hMPV and hBoV (Table IA).
Table I.
Primers Used in This Study
| Primer | Sequence 5′ → 3′ | Gene | Amplicon size (bp) | Reference | |
|---|---|---|---|---|---|
| Primers used for respiratory infection screeninga | |||||
| [0,1‐6]hAdV nested‐PCR | |||||
| I step | ADHEX1f | AACACCTAYGASTACATGAAC | Hexon | 473 |
Modified from [Avellón et al., 2001 ] |
| ADHEX1r | ATGGGGTARAGCATGTTRGC | ||||
| II step | ADHEX2f | AAYCCMTTYAACCACCACC | 170 | ||
| ADHEX2r | ACATCCTTYCKGAAGTTCCA | ||||
| [0,1‐6]hBoV nested‐PCR | |||||
| I step | 162f | CCAGCAAGTCCTCCAAACTCACCTGC | NP‐1 | 399 |
[Manning et al., 2006 ] |
| 561r | GGAGCTTCAGGATTGGAAGCTCTGTG | ||||
| II step | 188f | GAGCTCTGTAAGTACTATTAC | 354 |
Modified from [Allander et al., 2005 ] |
|
| 542r | CTCTGTGTTGACTGAATACAG | ||||
| [0,1‐6]hCoV multiplex nested‐PCR | |||||
| [0,1‐6]I step | |||||
| hCoV‐229E | CORO9 | GCACAGGACCCCATAAAGATGC | N | 444 |
Modified from [Dessau et al., 2001 ] |
| CORO10 | GAGAACGAGCAAGACTCTTGGCA | ||||
| hCoV‐OC43 | CORO11 | GCAATCCAGTAGTAGAGCGTCC | 445 | ||
| CORO12 | TTGACATCAGCCTGGTTRCTAGCG | ||||
| [0,1‐6]II step | |||||
| hCoV‐229E | CORO1 | AGGCGCAAGAATTCAGAACCAGAG | 308 | ||
| CORO2 | AGCAGGACTCTGATTACGAGAAGG | ||||
| hCoV‐OC43 | CORO5 | CCCAAGCAAACTGCTACYTCTCAG | 228 | ||
| CORO7 | GCAGCARTTGACGCTGGTTG | ||||
| [0,1‐6]Flu A/B multiplex real time PCR | |||||
| [0,1‐6]FluA | |||||
| FLUAV_FOR | ACAAGACCAATCCTGTCACCTCT | M | 108 |
[Valle et al., 2006 ] |
|
| FLUAV_REV | GGCATTTTGGACAAAGCGTCTAC | ||||
| FLUAV_TM | FAM‐CAGTCCTCGCTCACTGGGCACGGT(p)‐BHQ1 | ||||
| [0,1‐6]FluB | |||||
| FLUBV_FOR | CCAGTGGGACAACCAGA | NP | 89 | ||
| FLUBV_REV | TGCTCTTTCCGGGGATG | ||||
| FLUBV_TM | JOE‐ATCATCAGACCAGCAACCCTTGCC(p)‐BHQ1 | ||||
| [0,1‐6]hMPV nested‐PCR | |||||
| I step | HMPV‐M1f | GAGTCCTAYCTRGTAGACACC | M | 247 | This study |
| HMPV‐M1r | AGTACAGACATDGCWGCACC | ||||
| II step | HMPV‐M2f | GACCWGCTGTTCAAGTTG | 151 | ||
| HMPV‐M2r | YTGTGATGYAGCATACAGAG | ||||
| [0,1‐6]hPIV1‐4 multiplex nested‐PCR | |||||
| [0,1‐6]I step | |||||
| hPIV‐1 | PIP1+ | CYTTAAATTCAGATATGTAT | HN | 480 |
Modified from [Echevarría et al., 1998 ] |
| PIP1− | GATAAATARTWATTGATACG | ||||
| hPIV‐2 | PIP2+ | AACAATCTGCTGCAGSRTTT | HN | 507 | |
| PIP2− | ATGTCAGAYAATGGRCAAAT | ||||
| hPIV‐3 | PIP3+ | CTGTAAACTCAGACTTGGTA | HN | 477 | |
| PIP3− | TTTARGCCYTTGTCAACAAC | ||||
| hPIV‐4 | PI4P+ | CTGAACGGTTGCATTCAGGT | P | 441 |
[Aguilar et al., 2000 ] |
| PI4P− | TTGCATCAAGAATGAGTCCT | ||||
| [0,1‐6]II step | |||||
| hPIV‐1 | PIS1+ | CCGGHAATYTCTCATACCTATG | HN | 316 |
Modified from [Echevarría et al., 1998 ] |
| PIS1− | CYTTGGAGCGGAGTTGTTAWG | ||||
| hPIV‐2 | PIS2+ | CCATTTACCTAAGTGATGGAAT | HN | 203 | |
| PIS2− | GCCCTGTTGTATTTGGAAGAGA | ||||
| hPIV‐3 | PIS3+ | ACTCCCAARGTTGATGAAAGAT | HN | 102 | |
| PIS3− | TAAATCTTGTTGTTGAGATTG | ||||
| hPIV‐4 | PI4S+ | AAAGAATTAGGTGCAACCAGTC | P | 244 |
[Aguilar et al., 2000 ] |
| PI4S− | GTGTCTGATCCCATAAGCAGC | ||||
| [0,1‐6]hRV eminested‐PCR | |||||
| I step | PR1f | CGGACACCCAAAGTAG | 5'UTR | 380 |
[Ireland et al., 1993 ] |
| PR2r | GCACTTCTGTTTCCCC | ||||
| II step | PR1f | CGGACACCCAAAGTAG | 202 | ||
| PR3r | GGCAGCCACGCAGGCT | ||||
| [0,1‐6]RSV multiplex nested‐PCR | |||||
| [0,1‐6]I step | |||||
| RSV‐A/B | RSVAB1 | ATGGAGYTGCYRATCCWCARRRCAARTGCAAT | F | 737 |
[Coiras et al., 2003 ] |
| RSVAB2 | AGGTGTWGTTACACCTGCATTRACACTRAATTC | ||||
| [0,1‐6]II step | |||||
| RSV‐A | RSVA3 | TTATACACTCAACAATRCCAAAAAWACC | 363 | ||
| RSVA4 | AAATTCCCTGGTAATCTCTAGTAGTCTGT | ||||
| RSV‐B | RSVB3 | ATCTTCCTAACTCTTGCTRTTAATGCATTG | 611 | ||
| RSVB4 | GATGCGACAGCTCTGTTGATTTACTATG | ||||
| [0,1‐6]B. Primers used for the molecular characterization of hbov and hmpv strainsa | |||||
| [0,1‐6]hBoV sequencing nested‐PCR | |||||
| I step | VPs1 | GCACTTCTGTATCAGATGCCTT | VP1/2 | 903 |
[Smuts and Hardie, 2006 ] |
| VPas1 | CGTGGTATGTAGGCGTGTAG | ||||
| II step | VPs2 | CTTAGAACTGGTGAGAGCACTG CGTGGTATGTAGGCGTGTAG | 850 | ||
| VPas1 | |||||
| [0,1‐6]hMPV sequencing nested‐PCR | |||||
| I step | F‐f | GTYAGCTTCAGTCAATTCAAC | F | 532 |
Modified from [Peret et al., 2002 ] |
| MPVF1r | GTCTTCCTGTGCTAACTTTG | ||||
| II step | MPVF1f | CTTTGGACTTAATGACAGATG | 445 |
[Peret et al., 2002 ] |
|
| F‐r | CCTGTGCTGACTTTGCATG |
Modified from [Huck et al., 2006 ] |
|||
Primers used for the molecular surveillance for acute respiratory tract infections (A) and for the molecular characterization of hBoV and hMPV strains (B).
Sequencing of Identified hMPV and hBoV Strain Sequences
Molecular characterization was performed by sequence analysis of a 450 bp amplicons of the fusion (F) gene (spanning nucleotides 653–1,102) for hMPV and an 850 bp amplicon of the VP1/VP2 region (spanning nucleotides 1,084–1,933) for hBoV (Table IB).
Following PCR amplification, amplicons were purified with NucleoSpin® Extract II (Macherey‐Nagel GmbH & Co. KG, Duren, Germany). Nucleotide sequences were obtained by automated DNA sequencing on the ABI PRISM 3100 genetic analyzer (Applied Biosystems, Foster City, CA). The hMPV‐F gene and hBoV‐VP1/VP2 region nucleotide sequences obtained were deposited into GenBank.
Molecular Characterization and Phylogenetic Analysis
Multiple nucleotide sequences were aligned using ClustalX version 2.0 [Thompson et al., 1997]. Phylogenetic trees were constructed by means of the Neighbor‐Joining method and Kimura 2‐Parameter model using the MEGA package, version 4.0 [Kimura, 1980; Saitou and Nei, 1987; Tamura et al., 2007]. A bootstrap resampling analysis was performed (1,000 replicates) to test tree robustness [Felsenstein, 1985].
Mean evolutionary distances over all sequence pairs between and within groups were calculated using the Kimura 2‐Parameter model in MEGA software [Kimura, 1980; Tamura et al., 2007]. To detect recombination events, a bootscanning analysis was performed using the Simplot software version 3.5.1 (Kimura 2‐Parameter model; window size: 200 bp; step size: 20 bp; 1,000 bootstrap replicates, Neighbor‐Joining tree analysis), [Lole et al., 1999]. Predicted protein sequences were obtained by means of BioEdit software [Hall, 1999].
Statistical Analysis
Data were expressed as median (interquartile range, IQR) and percentages (95% confidence intervals, 95% CI) as appropriate. Comparisons between groups were performed using the Chi‐squared test or Fisher's exact test. A P‐value <0.05 was considered statistically significant (two‐tailed test). All statistical analyses were performed using the OpenEPI software, version 2.2.1 [Dean et al., 2009].
RESULTS
Viral Surveillance in Children With Acute Respiratory Tract Infections
One hundred and thirty‐one out of 240 (54.6%, 95% CI: 48.3–60.8) samples collected from hospitalized children were PCR‐positive for at least one respiratory virus. Forty‐six samples (19.2%) were positive for RSVA/B, 11.7% for hAdV, 10% for hRV, 4.6% for Flu A/B, 2.9% for hCoV229E/OC43, and 0.4% for hPIV3. hMPV was detected in 8.3% and hBoV in 12.1% of samples examined (Table IIA). Mixed infections were detected in 22.1% of acute viral respiratory tract infections (Table IIB). Respiratory viruses were detected more frequently in children younger than 2 years of age (52.8%, 61.1%, and 66.7% for children ≤6, 7–12, and 13–24 months of age, respectively) in comparison to children 25–36 months of age (20%, P < 0.05, Table IIB).
Table II.
Molecular Detection of Respiratory Viruses in Pharyngeal Swabs Collected From Hospitalized Children With Acute Respiratory Tract Infections and Children With Viral Respiratory Infections
| Total 240 | 0–6 months 144 (60; 53.7–66.1) | 7–12 months 36 (15; 10.9–19.9) | 13–24 months 45 (18.8; 14.2–24.1) | 25–36 months 15 (6.3; 3.7–9.9) | |
|---|---|---|---|---|---|
| A. Number (95% CI) of infections and age distribution children (N = 240) hospitalized with acute respiratory tract infections | |||||
| Viral agents | |||||
| RSV | 46 (19.2; 14.6–24.5) | 35 (24.3; 17.8–31.8) | 4 (11.1; 3.6–24.7) | 7 (15.5; 7.1–28.4) | 0 |
| hAdV | 28 (11.7; 8.1–16.2) | 13 (9; 5.1–14.6) | 7 (19.4; 8.9–34.7) | 7 (15.5; 7.1–28.4)) | 1 (6.7; 0.3–28.7) |
| hRV | 24 (10; 6.7–14.3) | 14 (9.7; 5.6–15.4) | 5 (13.9; 5.3–28.1) | 4 (8.9; 2.9–20.1) | 1 (6.7; 0.3–28.7) |
| FLU | 11 (4.6; 2.4–7.8) | 5 (3.5; 1.3–7.5) | 2 (5.5; 0.9–17.2) | 4 (8.9; 2.9–20.1) | 0 |
| CoV | 7 (2.9; 1.3–5.7) | 3 (2.1; 0.5–5.6) | 0 | 3 (6.7; 1.7–17.1) | 1 (6.7; 0.3–28.7) |
| hPIV | 1 (0.4; 0.02–2.0) | 0 | 0 | 1 (2.2; 0.1–10.5) | 0 |
| hMPV | 20 (8.3; 5.3–12.4) | 13 (9; 5.1–14.6) | 3 (8.3; 2.2–21) | 4 (8.9; 2.9–20.1) | 0 |
| hBoV | 29 (12; 18.4–16.7) | 14 (9.7; 5.6–15.4) | 4 (11.1; 3.6–24.7) | 11 (24.4;13.6–38.5)a | 0 |
| B. Number (95% CI) of single and co‐infections in patients (N = 131) in the context of age. | |||||
| Total | 131 (54.6; 48.3–60.8) | 76 (52.8; 44.6–60.8)b | 22 (61.1; 44.6–75.9)b | 30 (66.7; 52–79.2)b | 3 (20; 5.4–45.4) |
| Single infections | 102 (77.9; 70.2–84.4) | 58 (76.3; 65.8–84.9) | 19 (86.4; 67.2–96.4) | 22 (73.3; 55.6–86.8) | 3 |
| Co‐infections | 29 (22.1; 15.6–29.9) | 18 (23.7; 15.2–34.2) | 3 (13.6; 3.6–32.8) | 8 (26.7; 13.2–44.4) | 0 |
P < 0.05, prevalence of hBoV infections versus that observed in children 0–6 months and 7–12 months of age.
P < 0.05, N (%) viral infections versus N (%) of children 25–36 months of age.
Viral pathogens were associated with 60.6% (95% CI: 52.9–68.0) of lower respiratory‐tract infections, particularly in bronchiolitis cases (60.8%, 95% CI: 50.9–70.2). The most frequently detected viruses were RSV (50.8%, 95% CI: 38.2–63.4), hBoV (20.3%, 95% CI: 11.5–32.0), hRV (16.9%, 95% CI: 8.9–28.1), hAdV (16.9%, 95% CI: 8.9–28.1), and hMPV (11.9%, 95% CI: 5.3–22.1).
Epidemiologic and Molecular Features of hMPV and hBoV Infections
hMPV or hBoV were detected in 37.4% (95% CI: 29.4–45.9) of respiratory virus‐positive samples but only in children ≤24 months old. hMPV prevalence was similar in the first three age groups (range: 8.3–9.0%) while hBoV was identified more frequently in the 13–24 months old group compared to the other groups (24.4% vs. 9.7% and 11.1%, P < 0.05; Table IIA).
hMPV was identified more often as a single pathogen and only 30% of cases were associated with mixed infections (P < 0.05). hBoV was detected as a mixed infection in 58.6% (17/29) of cases, mainly in association with RSV (N = 10), hRV (N = 5) and hAdV (N = 4). Among these, two triple‐infection (hBoV‐RSV‐hRV) cases were detected. The frequency of co‐infections was similar in all age groups but different viruses were involved. The median age of hBoV co‐infected children was 2.0 months (IQR: 1.5–50.0 months), for hBoV‐RSV 8.5 months (IQR: 3.5–13.0 months), and 14 months for both hBoV‐hRV and hBoV‐hAdV (IQR: 11.5–17.0 months). No hMPV‐hBoV co‐infections were identified.
hMPV was found as the single source of infection in 7.2% (95% CI: 3.2–13.8) of bronchiolitis cases and part of co‐infections in 11.1% of pneumonia (95% CI: 2.9–27.3) and 13.3% of asthma cases (95% CI: 2.3–37.5). hBoV was identified in 12.4% of bronchiolitis cases (95% CI: 6.9–20.1), mainly as a hBoV‐RSV co‐infection. In addition, a high prevalence of hBoV infections (18.5%, 95% CI: 7.1–36.4) were seen in association with pneumonia, most often as a single viral agent.
Phylogenetic Analysis of hMPV and hBoV Sequences
Ten out of 20 hMPV‐positive samples were characterized at the molecular level and a phylogenetic tree of the hMPV F gene was generated (Fig. 1A). The identity range between tree sequences was 81.1–100%. Mean evolutionary distances for all sequence pairs are reported in Figure 1B. Phylogenetic analysis showed a prevalence of genotype A strains (8/10) compared to genotype B strains (2/10). In the context of genotype A, six hMPV‐F sequences shared similarity with the A2b sublineage, one with A2a and one with the A1 group that presented with a single amino acid change (N358T). Genotype B sequences belonged exclusively to lineage B1.
Figure 1.
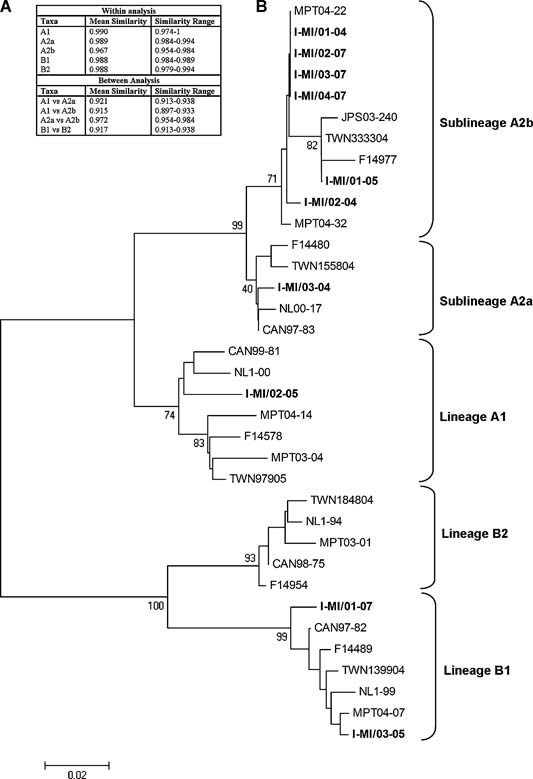
Genetic analysis of hMPV. A: Phylogenetic tree of the Italian I‐MI hMPV sequences (boldface) demonstrating the phylogenetic relationship of the I‐MI sequences based on partial F gene nucleotide sequences. Bootstrap values (in addition to A1, A2a, A2b, B1, and B2 group/sublineages) are indicated. Reference sequences correspond to the following GenBank accession numbers: A1) AF371337, AY145294, DQ453017, AJ867506, AJ867520, EF612444; A2a) AY304360, AY145296, DQ453014, EF612451; A2b) AY530095, DQ453021, AJ867528, AJ867538, EF612457; B1) AY304361, AY145294, DQ453015, AJ867513, EF612440; B2) AY304362, AY145289, DQ452995, AJ867503, EF612441. B: Evolutionary mean distances over all sequence pairs within and between groups.
Sixteen out of 29 hBoV‐positive samples were analyzed and the hBoV VP1/VP2 region described (Fig. 2). All sequences shared a high degree of similarity with reference strains and other defined sequences (identity range: 97.3–100%). Most sequences (14/16) were genetically similar to genotypes St1 (4/14) and St2 (10/14). A third cluster (bootstrap value: 92), including two Italian sequences (I‐MI/04‐08 and I‐MI/02‐07, identity 100%) and the Asian HK7 isolate, were identified between the two main branches. The boot scanning analysis revealed a recombination event with a breakpoint located at nucleotide position 1,302 of the VP1/VP2 region (Fig. 3). The 5′ region of these two Italian sequences was more similar to the MPT‐8 strain (identity 99% vs. 97.6%) and the 3′ portion more similar to the HK7 strain (identity 97.8% vs. 99%; Fig. 3 inset).
Figure 2.
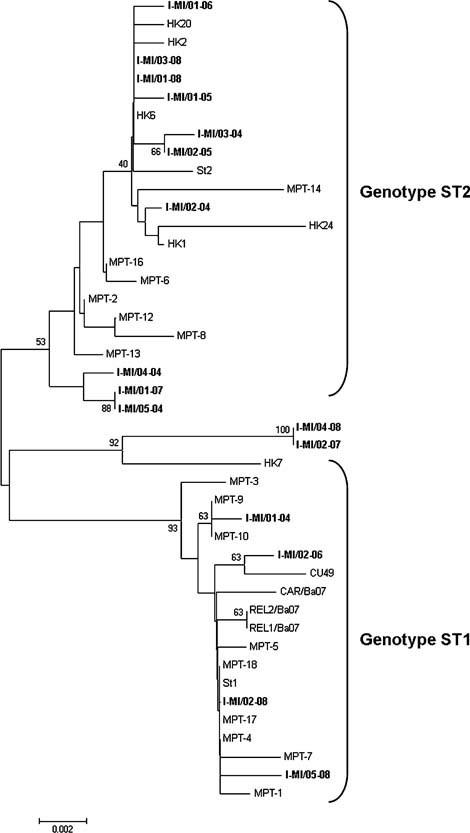
Genetic analysis of hBoV sequences. Phylogenetic tree of the Italian I‐MI human Bocavirus sequences (boldface) based on partial VP1/VP2 region nucleotide sequences. Bootstrap values (in addition to St1 and St2 genotype clusters) are indicated. Reference sequences correspond to the following GenBank accession numbers: St‐1) DQ000497, AM160609, AM160611–AM160613, AM160615, AM689298, AM689299, AM689306, AM689307, EU069434, EU069436, EU069437, EF203921; St‐2) DQ000495, AM160610, AM160614, AM689297, AM689301‐AM689303, AM689305, EF450717, EF450718, EF450722, EF450723, EF450736, EF450740.
Figure 3.
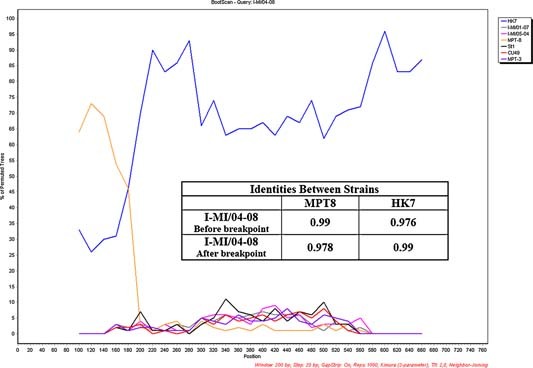
Recombination recognition of the Italian I‐MI/04‐08 sequence: bootscan analysis and identities values between the recombinant sequence and the two reference sequence (MPT8 and HK7) before and after the breakpoint (inset).
Six amino acid substitutions were found: G415S, N474S, F540Y, N546H, A555T, and G566R. Mutations G415S, F540Y, and N546H were identified in the I‐MI/04‐08 and I‐MI/02‐07 sequences.
DISCUSSION
A molecular‐epidemiologic survey of acute respiratory infections was conducted in children younger than 3 years old hospitalized between 2004 and 2008 in Milan, Italy. Both hospitalization and positive viral detection were more frequent in children <2 years of age compared to older patients.
The prevalence of both hMPV and hBoV infections (8.3% and 12.1%, respectively) were similar to rates reported previously [Maggi et al., 2003; Bastien et al., 2006; Foulongne et al., 2006; Allander et al., 2007]. Both hMPV and hBoV infections were associated with lower respiratory tract infections. hMPV was present as a single infectious agent in 7.2% of children aged ≤12 months with bronchiolitis. The involvement of hBoV at the onset of bronchiolitis was difficult to asses due to the frequent association with other viruses, particularly RSV. In this study, hBoV was associated with 18.5% of pediatric pneumonias and frequently identified as a single agent.
Twenty percent of hospitalized children had dual or triple co‐infections and hBoV was involved in approximately 60% of mixed infections. The extended persistence of hBoV in airways (up to 7 months) increased patient's chances of acquiring a co‐infection, explaining the high frequency of co‐infections with this virus [Brieu et al., 2008; Lindner et al., 2008; von Linstow et al., 2008]. The seasonal distribution of hBoV, with a peak incidence between December and January, resembled that observed for RSV, hRV, and hAdV. This suggested that the overlapping distribution of these viruses during the seasons could have facilitated hBoV co‐infections. The epidemiologic model of hMPV infections was distinct from many other respiratory viruses, with peak incidence rates in late spring, mostly between March and April. This observation may account for the absence of hMPV‐hBoV co‐infections.
Co‐circulation of both hMPV genotypes was observed, with predominance of genotype A (particularly the A2b sublineage), supporting previous reports demonstrating an increase in A2 strain infections in recent years [Boivin et al., 2004; Wang et al., 2008; Herbert et al., 2005]. Guant et al. [Gaunt et al., 2006] suggested a further potential back‐shift to genotype B in Scotland during the winter season of 2007/2008. Unfortunately, no samples from this study were available for sequencing to confirm this hypothesis. As suggested by Herbert et al. [2005], the complex circulation pattern of hMPV may be the reason this strain can escape pre‐existing host immune responses, suggesting that additional molecular surveillance studies need to be carried out to confirm genotype B back shift circulation and to clarify the existence of a seasonal distribution of hMPV strains. Phylogenetic analyses demonstrated that both hBoV genotypes co‐circulated during the study period and that genotype St2 predominated. Interestingly, evidence of genetic recombination between the two hBoV strains (with a breakpoint site in the VP1/VP2 region) was found. The two mosaic sequences (identity: 100%) were identified in two different years (2007 and 2008), indicating that this variant had been circulating in the population since at least 2007. Genetic recombination has been reported to occur in Parvoviruses [Shackelton et al., 2007; Ohshima and Mochizuki, 2009] and in different hBoV species [Arthur et al., 2009; Kapoor et al., 2009, 2010]. To date, only Lin et al. [2009] have reported an intra‐species recombination event involving hBoV1, with a breakpoint located in the NS1 gene. Taken together, these data suggested that intra‐species recombination could occur in hBoV1 at different positions of its genome, playing a major role in the evolutionary history of this virus. Further studies will be necessary to identify breakpoint patterns in these viruses and to identify their potential association with viral spread among populations.
In conclusion, this study showed that both hMPV and hBoV circulated in Italy and were involved in the pathogenesis of lower respiratory tract diseases in children. Data presented in this report provided evidence for both a shift in the incidence of hMPV A2 strain infections in recent years and the occurrence of recombination events in hBoV strains. Therefore, continuous molecular surveillance is important for the detection of new viral strains as a means of detecting their spread and assessing their epidemiological impact.
Acknowledgements
We thank Dr. D. Colzani for skilful technical assistance.
Conflict of interest: None of the authors have financial or other conflicting interests with regard to the information described in this manuscript.
REFERENCES
- Aguilar JC, Pérez‐Breña MP, García ML, Cruz N, Erdman DD, Echevarría JE. 2000. Detection and identification of human parainfluenza viruses 1, 2, 3, and 4 in clinical samples of pediatric patients by multiplex reverse transcription‐PCR. J Clin Microbiol 38: 1191–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allander T, Tammi MT, Eriksson M, Bjerkner A, Tiveljung‐Lindell A, Andersson B. 2005. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc Natl Acad Sci USA 102: 12891–12896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allander T, Jartti T, Gupta S, Niesters HGM, Lehtinen P, Österback R, Vuorinen T, Waris M, Bjerkner A, Tiveljung‐Lindell A, van den Hoogen BG, Hyypiä T, Ruuskanen O. 2007. Human Bocavirus and acute wheezing in children. Clin Infect Dis 44: 904–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur JL, Higgins GD, Davidson GP, Givney RC, Ratcliff RM. 2009. A novel bocavirus associated with acute gastroenteritis in Australian children. PLoS Pathog 5: e1000391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avellón A, Pérez P, Aguilar JC, Lejarazu R, Echevarría JE. 2001. Rapid and sensitive diagnosis of human adenovirus infections by a generic polymerase chain reaction. J Virol Methods 92: 113–120. [DOI] [PubMed] [Google Scholar]
- Bastien N, Brandt K, Dust K, Ward D, Li Y. 2006. Human Bocavirus infection, Canada. Emerg Infect Dis 12: 848–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boivin G, Mackay I, Sloots TP, Madhi S, Freymuth F, Wolf D, Shemer‐Avni Y, Ludewick H, Gray GC, LeBlanc E. 2004. Global genetic diversity of human metapneumovirus fusion gene. Emerg Infect Dis 10: 1154–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brieu N, Guyon G, Rodière M, Segondy M, Foulongne V. 2008. Human bocavirus infection in children with respiratory tract disease. Pediatr Infect Dis J 27: 969–973. [DOI] [PubMed] [Google Scholar]
- Broor S, Bharaj P, Chahar HS. 2008. Human metapneumovirus: A new respiratory pathogen. J Biosci 33: 483–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coiras MT, Pérez‐Breña P, García ML, Casas I. 2003. Simultaneous detection of influenza A, B, and C viruses, respiratory syncytial virus, and adenoviruses in clinical samples by multiplex reverse transcription nested‐PCR assay. J Med Virol 69: 132–144. [DOI] [PubMed] [Google Scholar]
- Dean AG, Sullivan KM, Soe MM. 2009. OpenEpi: Open Source Epidemiologic Statistics for Public Health, Version 2.3. Update 2009/20/05. Available at: http://www.openepi.com.
- Dessau RB, Lisby G, Frederiksen JL. 2001. Coronaviruses in brain tissue from patients with multiple sclerosis. Acta Neuropathol 101: 601–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echevarría JE, Erdman DD, Swierkosz EM, Holloway BP, Anderson LJ. 1998. Simultaneous detection and identification of human parainfluenza viruses 1, 2, and 3 from clinical samples by multiplex PCR. J Clin Microbiol 36: 1388–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. 1985. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39: 783–791. [DOI] [PubMed] [Google Scholar]
- Foulongne V, Guyon G, Rodière M, Segondy M. 2006. Human metapneumovirus infection in young children hospitalized with respiratory tract disease. Pediatr Infect Dis J 25: 354–359. [DOI] [PubMed] [Google Scholar]
- Gaunt E, McWilliam‐Leitch EC, Templeton J, Simmonds P. 2006. Incidence, molecular epidemiology and clinical presentations of human metapneumovirus; assessment of its importance as a diagnostic screening target. J Clin Virol 46: 318–324. [DOI] [PubMed] [Google Scholar]
- Hall TA. 1999. BioEdit: A user‐friendly biological sequence alignment editor and analysis program for windows95/98/nt. Nucl Acids Symp 41: 95–98. [Google Scholar]
- Herbert PL, Abed Y, van Niekerk N, Boivin G, Klugman KP, Madhi SA. 2005. Human metapneumovirus genetic variability, South Africa. Emerg Infect Dis 11: 1074–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huck B, Scharf G, Neumann‐Haefelin D, Puppe W, Weigl J, Falcone V. 2006. Novel human metapneumovirus sublineage. Emerg Infect Dis 12: 147–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ireland DC, Kent J, Nicholson KG. 1993. Improved detection of rhinoviruses in nasal and throat swabs by seminested RT‐PCR. J Med Virol 40: 96–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor A, Slikas E, Simmonds P, Chieochansin T, Naeem A, Shaukat S, Alam MM, Sharif S, Angez M, Zaidi S, Delwart E. 2009. A newly identified bocavirus species in human stool. J Infect Dis 199: 196–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor A, Simmonds P, Slikas E, Li L, Bodhidatta L, Sethabutr O, Triki H, Bahri O, Oderinde BS, Baba MM, Bukbuk DN, Besser J, Bartkus J, Delwart E. 2010. Human Bocaviruses are highly diverse, dispersed, recombination prone, and prevalent in enteric infections. J Infect Dis 201: 1633–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. 1980. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16: 111–120. [DOI] [PubMed] [Google Scholar]
- Lin JH, Chiu SC, Lin YC, Chen HL, Lin KH, Shan KH, Wu HS, Liu HF. 2009. Clinical and genetic analysis of human Bocavirus in children with lower respiratory tract infection in Taiwan. J Clin Virol 44: 219–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner J, Karalar L, Schimanski S, Pfister H, Struff W, Modrow S. 2008. Clinical and epidemiological aspects of human bocavirus infection. J Clin Virol 43: 391–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lole KS, Bollinger RC, Paranjape RS, Gadkari D, Kulkarni SS, Novak NG, Ingersoll R, Sheppard HW, Ray SC. 1999. Full‐length human immunodeficiency virus type 1 genome from subtype C‐infected seroconverters in India, with evidence of intersubtype recombination. J Virol 73: 152–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggi F, Pifferi M, Vatteroni M, Fornai C, Tempestini E, Anzilotti S, Lanini L, Andreoli E, Ragazzo V, Pistello M, Specter S, Bendinelli M. 2003. Human Metapneumovirus associated with respiratory tract infections in a 3‐year study of nasal swabs from infants in Italy. J Clin Microbiol 4: 2987–2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning A, Russell V, Eastick K, Leadbetter GH, Hallam N, Templeton K, Simmonds P. 2006. Epidemiological profile and clinical associations of human Bocavirus and other human Parvoviruses. J Infect Dis 194: 1283–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohshima T, Mochizuki M. 2009. Evidence for recombination between feline Panleukopenia virus and canine Parvovirus type 2. J Vet Med Sci 71: 403–408. [DOI] [PubMed] [Google Scholar]
- Peret TC, Boivin G, Li Y, Couillard M, Humphrey C, Osterhaus AD, Erdman DD, Anderson LJ. 2002. Characterization of human metapneumoviruses isolated from patients in North America. J Infect Dis 185: 1660–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M. 1987. The neighbor‐joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425. [DOI] [PubMed] [Google Scholar]
- Schildgen O, Müller A, Allander T, Mackay IM, Völz S, Kupfer B, Simon A. 2008. Human Bocavirus: Passenger or pathogen in acute respiratory tract infections? Clin Microbiol Rev 21: 291–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackelton LA, Hoelzer K, Parrish CR, Holmes EC. 2007. Comparative analysis reveals frequent recombination in the parvoviruses. J Gen Virol 88: 3294–3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smuts H, Hardie D. 2006. Human Bocavirus in hospitalized children, South Africa. Emerg Infect Dis 12: 1457–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596–1599. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The CLUSTALX windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle L, Amicizia D, Bacilieri S, Banfi F, Riente R, Durando P, Sticchi L, Gasparini R, Esposito C, Icardi G, Ansaldi F. 2006. Performance testing of two new one‐step real time PCR assays for detection of human influenza and avian influenza viruses isolated in humans and respiratory syncytial virus. J Prev Med Hyg 47: 127–133. [PubMed] [Google Scholar]
- van den Hoogen BG, de Jong JC, Groen J, Kuiken T, de Groot R, Fouchier RAM, Osterhaus ADME. 2001. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat Med 7: 719–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Hoogen BG, Herfst S, Sprong L, Cane PA, Forleo‐Neto E, de Swart RL, Osterhaus ADME, Fouchier RAM. 2004. Antigenic and genetic variability of human Metapneumoviruses. Emerg Infect Dis 10: 658–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Linstow ML, Høgh M, Høgh B. 2008. Clinical and epidemiologic characteristics of human bocavirus in Danish infants: Results from a prospective birth cohort study. Pediatr Infect Dis J 27: 897–902. [DOI] [PubMed] [Google Scholar]
- Wang HC, Huang SW, Wang SW, Tsai HP, Kiang D, Wang SM, Liu CC, Su IJ, Wang JR. 2008. Co‐circulating genetically divergent A2 human metapneumovirus strains among children in southern Taiwan. Arch Virol 153: 2207–2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . 2009. Acute Respiratory Infections (Update February 2009). Available at: http://www.who.int/vaccine_research/diseases/ari/en/index.html.


