Abstract
Background and Aim: Response to interferon therapy and disease progression in hepatitis C virus (HCV) infected patients differs among individuals, suggesting a possibility of a contribution of host genetic factors. 2′‐5′‐oligoadenylate synthetase 1 (OAS1), an important component of the innate immune system with a proven antiviral function, may therefore have a relationship with the response to interferon therapy and clinical course of HCV disease. Our aim was to determine the frequency of single nucleotide polymorphism (SNP) at exon 7 splice acceptor site (SAS) of the OAS1 gene in relation to the interferon response and status of HCV infection.
Methods: A 203 bp fragment containing exon 7 SAS was amplified in 70 HCV chronic patients and 50 healthy controls. SNP was examined using restriction fragment length polymorphism (RFLP) genotyping method. Correlations of SNP genotypes with response to interferon and clinical status of patients were statistically analyzed.
Results: There was an increasing trend of response from AA to AG to GG genotypes (P = 0.007). Genotype AA was associated with non‐response to interferon and higher degree of liver fibrosis (P = 0.05). Multivariate analysis showed this SNP as independent and a significant determinant of the outcome of interferon therapy (odds ratio 4.913 [95% confidence interval 1.365–8.2], P = 0.006).
Conclusions: This is the first study to show a significant association between the functional SNP at exon 7 SAS of OAS1 gene and the viral response to interferon in chronic HCV patients. Patients with AA genotype were associated with progressive HCV disease and viral resistance to interferon therapy. This OAS SNP is a potential bio‐marker to predict IFN response in chronic hepatitis C patients.
Keywords: Hepatitis C virus, liver fibrosis, OAS1, response to interferon therapy, single nucleotide polymorphism
Introduction
The current recommended treatment for chronic hepatitis C virus (HCV) infection is the combination of pegylated alpha interferon (IFN) and ribavirin given for 24 or 48 weeks. 1 , 2 , 3 Both viral and host factors may contribute to the phenomenon that some patients with chronic hepatitis C respond well to IFN therapy but others do not. 4 , 5 Specifically, viral factors, such as genotype 1 and high HCV RNA levels are associated with a higher incidence of resistance to IFN therapy. Also, host factors, such as old age, advanced fibrosis, long duration of disease and host genetic factors can adversely influence the response to IFN treatment. 6 , 7 , 8 The host genetic factors mediate the vigor of the immune response of the host through the control of inter‐individual differences in the production of intracellular antiviral proteins or some cytokines. 9 , 10 These inter‐individual differences may be attributable to single nucleotide polymorphisms (SNPs) in the human genome, which significantly influence the progression of chronic HCV infection and response to therapy. 11 , 12 Biological activity of IFN therapy is mediated by the induction of intracellular antiviral proteins, such as 2′–5′ oligoadenylate synthetase (OAS), dsRNA activated protein kinase (PKR) and MxA protein. 6 , 13 , 14 The 2′–5′ OAS protein has selective activity against several viruses. It requires double stranded (ds) RNA structures, such as viral genomes, to become activated. The activated enzyme then catalyzes the polymerization of adenosine triphosphate (ATP) into 2′,5′ linked oligoadenylates, and these, in turn, bind to and activate latent ribonuclease L (RNaseL), which degrades viral RNA and inhibits protein synthesis. The 2′‐5′ OAS‐RNaseL system may also be involved in growth and apoptosis. 15 , 16 The 2′‐5′‐oligoadenylate synthetase genes comprise a gene family (OAS1, OAS2, and OAS3). The OAS1 gene, located on human chromosome 12q24.2, encodes the p42, p44, p46, and p48 isoforms. 17 The importance of OAS1 for clearing viral infections has been demonstrated in different studies. 18 , 19 , 20 In humans, it was reported that basal activity of OAS1 varies among individuals and is associated with susceptibility to diabetes mellitus type 1, whose pathogenesis is thought to involve enterovirus infections. 21 Furthermore, previous analysis of human OAS1 response to yellow fever vaccine revealed a highly significant correlation between basal enzyme activity and virus‐stimulated activity measured 7 days after vaccination. 22 Actually, in HCV patients, several findings were reported regarding OAS1 (i) HCV core protein specifically activated the 40/46‐kDa OAS1 gene promoter in a dose‐dependent manner in different human hepatocyte derived cell lines. (ii) The activation by core protein was further enhanced in the cells treated with alpha interferon. 23 (iii) The induced levels of OAS1 were lower in poor‐responder patients than in marked‐ or intermediate‐responder patients. 24 Thus, any genetic variants that affect OAS1 activity could be important determinants of susceptibility/resistance to antiviral treatment and to virus‐related disease progression. Although OAS1 is critical in antiviral interaction, its sequence is not strictly conserved. There are eight SNPs in exons, 24 SNPs in introns, three in promoters and one in the 5′‐untranslated region (UTR) of the OAS1 gene. It was reported that more than 12 genetic markers in OAS1 were strongly associated with enzyme activity. 21 , 25 The strongest genetic association with OAS1 activity occurred at exon 7 SAS of the OAS1 gene, in which the sequence AG is required for normal splicing at this site. The G allele at this acceptor site retains the splice site and is associated with high enzyme activity p46, whereas the A allele (AA at acceptor site) ablates the splice site and is associated with low enzyme activity p48 and p52. 25 Ghosh et al. 26 reported that the basal OAS1 activity is higher in persons carrying the G (p46) allele than in those having only the A (p48/p52) allele. These findings suggest strong genetic control of OAS1 basal activity and the genetic polymorphism at exon 7 SAS makes OAS1 an excellent candidate for a human gene that can significantly influence the host susceptibility to viral infection, affect the disease progression and the response to IFN therapy in patients with hepatitis C or hepatitis B viral infection. 10 , 27 , 28 In the present study, we investigated 70 Egyptian HCV genotype 4a infected patients and 50 healthy individuals as controls to determine if the genetic SNP in the exon 7 SAS of the human OAS1 gene is associated with reduced response to IFN based therapy.
Methods
Subjects
Seventy patients with proven chronic hepatitis C who received pegylated IFN + ribavirin treatment and 50 healthy controls (negative for HCV Ab and RNA as well as hepatitis B surface antigen [HBsAg]) were enrolled in this study. Subjects were all Egyptians and unrelated to each other. Informed consent was obtained from each subject before collecting blood samples. All HCV patients were positive for both anti‐HCV antibody and serum HCV RNA by reverse transcription‐polymerase chain reaction (RT‐PCR). All patients were infected with HCV genotype 4a as detected by (INNO‐LiPA HCV; Innogenetics, Gent, Belgium). Viral RNA quantifications were performed using (Copus Amplicore HCV monitor test, Roche, Pleasanton, CA, USA). IFN treatment and clinical follow‐ups were performed at the Department of Tropical Medicine, Kasr El Ainy Hospital, Cairo University. Prior to the start of the IFN therapy, patients had persistently elevated serum alanine aminotransferase (ALT) levels for at least 6 months and none of them had evidence of metabolic liver diseases, alcohol‐induced, drug‐induced or autoimmune hepatitis, thyroid hypo‐ or hyper‐function, or active schistosomiasis infection. A pretreatment liver biopsy was carried out for histopathological examination and assessment of the stages of fibrosis according to the Metavir Score. 29 Patients were treated with interferon and ribavirin combination therapy for 48 weeks. They received 20‐kDa linear pegylated (PEG) IFNα‐2a derived from Hansenula Polymorpha expression system (MinaPharm‐Rhein Company, 10th of Ramadan City, Egypt), taken once a week at 160 µg s.c., in addition to oral ribavirin taken daily in a dose of 1000–1200 mg (according to body weight). Patients were followed for at least 6 months after completion of therapy. Sustained virological responders (SVR; n = 31) were defined as patients who tested negative for HCV RNA (using both nested RT‐PCR and real‐time PCR) for at least 6 months after end of treatment (ETR) to combined therapy. All other patients, who did not show SVR, were considered non‐responders (NR; n = 39).
Nested and real‐time PCR of HCV
Disappearance of HCV viremia was confirmed by a variety of tests including RT‐PCR using nested primers derived from the highly conserved HCV 5′UTR 30 , 31 and real‐time PCR. HCV RNA was extracted using BIOZOL‐total RNA extraction reagent kit (Hangzhou Bioer Technology, Hangzhou, China).
Hepatitis C virus quantification was performed using Fluorescence Quantitative Detection kit (Hangzhou Bioer Technology, Hangzhou, China). HCV RNA was reverse transcribed and a specific fragment was amplified with specific primers in a one‐step RT‐PCR reaction. The products were detected by using a specific probe from Taqman‐MGB Probes (Hangzhou Bioer Technology, Hangzhou, China). The real‐time PCR reaction was done according to the manufacturer's recommendations. All PCR tubes were placed in the Real Time PCR Detection Instrument (Stratagene, La Jolla, CA, USA).
Amplification of exon 7 SAS of human OAS1gene
Extraction of DNA
Blood samples from all healthy controls and HCV‐infected patients undergoing IFN based treatment were collected. Genomic DNA was extracted from the peripheral blood mononuclear cells (PBMC) after digestion with proteinase K and subsequent salting out of cellular proteins using sodium chloride followed by ethanol precipitation and final storage at −20°C until required. 32
Primer design and PCR conditions
A pair of primers that amplified the SNP region and provided 203 bp PCR fragments that cover exon 7 AG splice‐acceptor sites was designed using Primer3 Input (http://www.embnet.sk/cgi-bin/primer3_www.cgi). We optimized the PCR protocol and thermal cycling program as follows: PCR amplification was carried out in 25 µL, containing 100–500 ng DNA, 2.5 mM MgCl2 (Promega, Madison, WI, USA), 500 nM of each primer, 200 mM dNTP's (Promega), 1× PCR buffer (Promega), and 1U Go Taq DNA polymerase (Promega). Thermal cycling in an MJ Research cycler included denaturation at 94°C for 5 min followed by 35 cycles each of denaturation at 94°C for 30 s; annealing at 58°C for 30 s; and extension at 72°C for 1 min. Cycling was followed by a final extension step at 72°C for 10 min. Primer sequences were as follows OAS1 5′‐tgcaatgcaggaagactcc‐3′ as a forward primer and OAS2 5′‐tgcaggtccagtcctcttct‐3′ as a reverse primer.
Determination of exon7 SAS‐SNP in human OAS1 gene
A restriction fragment length polymorphism (RFLP) analysis was constructed for detection of AG allelic nucleotides at exon 7 SAS of the OAS1 gene. Using a theoretical restriction map program at http://www.NEBcutter.com, we identified a restriction endonuclease, AluI, which recognizes the sequence of the SNP within the exon 7 SAS. The 203‐bp long PCR fragments from all subjects were digested with Alu I (Promega). The AluI restriction digestion was performed on 8 µL of the PCR product for at least 4 h in a total volume of 20 µL with 5U of AluI according to the manufacturer's recommendations. Then, 10 µL of the digested products were run on a 2% of agarose gel stained with ethidium bromide and observed under UV transillumination.
Statistical analysis
Frequency of exon 7 SAS‐ SNP was compared in the control group versus the chronic HCV patient group. Statistical differences between groups were determined by χ2 test. Independent predictive factors associated with SVR to IFN treatment were studied using stepwise multinomial logistic regression analysis. All statistical analyses were performed using the SPSS 9.0 statistical software program. The odds ratio (OR) and 95% confidence interval (CI) were calculated to assess the relative risk confidence. A two tailed P‐value ≤ 0.05 was considered statistically significant. Statistical analysis allowed us to determine the degree at which this (SNP) can influence the outcome of HCV infection and response to therapy.
Results
Cohort description
All subjects (patients and controls) were descendents of Egyptian origin representing most of the governorates of the country. To explore whether any of the clinical, pathological, biochemical or virological parameters is associated with specific response pattern, a comparison between SVR and NR was outlined in Table 1. Among a total of 70 chronic HCV patients, the univariate analyses of all factors showed that there were no significant differences between both patient populations in age, sex, body mass index (BMI) and viral loads. On the other hand alpha fetoprotein (AFP) levels were significantly higher in NR than SVR patients (P = 0.014). Also, lower stages of liver fibrosis (F0–F1) were associated with a higher probability of achieving SVR (P = 0.0001), whereas higher stages of hepatitis activity (A2–A3) and fibrosis (F2–F4) were higher among the NR patient population (Table 1). The hepatitis activity, which is the amount of inflammation (specifically, the intensity of necro‐inflammatory lesions), is graded on a 4‐point scale from A0 to A3. A1 indicates mild activity whereas A2–A3 indicate moderate to severe activity.
Table 1.
Laboratory findings in 70 chronic hepatitis C virus (HCV) 4a patients treated with peg interferon plus Ribavirin therapy
| Sustained virological responders (SVR) (n = 31) | Non‐responders (NR) (n = 39) | |
|---|---|---|
| Age (years) | 37.7 ± 9.9 | 41.51 ± 18.87 |
| Sex (female/male) | 5/26 | 11/28 |
| BMI (kg/m2) | 34.17 ± 4.45 | 31.1 ± 6.73 |
| Baseline alanine aminotransferase (IU/L) | 34 ± 23.8 | 45.9 ± 32.5 |
| Alpha fetoprotein level (ng/mL) | 3.5 ± 1.9 | 9.35 ± 13.49 |
| HCV RNA | 3.97 × 105 ± 6.47 × 105 | 4.77 × 105 ± 9.1 × 105 |
| level (copies/mL) | ||
| Histological stage | ||
| F(0–1) | 26/31 | 5/39 |
| F(2–4) | 5/31 | 34/39 |
| A(0–1) | 25/31 | 21/39 |
| A(2–4) | 6/31 | 18/39 |
The univariate analysis of clinical and virological factors was compared in NR versus SVR patients. Data of this table showed that AFP levels were significantly higher in NR patients than SVR patients (P = 0.014). Also, lower stages of liver fibrosis (F0–F1) were associated with a higher probability of achieving sustained response (P = 0.0001).
P‐value < 0.05 considered statistically significant.
RFLP analysis of OAS1 at exon 7 SAS
The presence of AA genotype at the exon 7 SAS was indicated by complete digestion of the 203 bp PCR product using AluI (AGCT) into two fragments of 150 bp and 53 bp. While GG genotype was indicated by absence of AluI site where A to G polymorphism generated an indigestible sequence (GGCT) with a retained intact 203 bp PCR fragment. In cases of heterozygosity (AG), three fragments of 203 bp, 150 bp and 53 bp appeared on agarose gel electrophoresis as shown in Figure 1.
Figure 1.
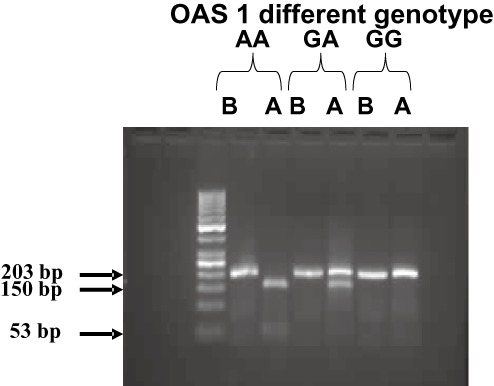
Exon 7 splice acceptor site (SAS) single nucleotide polymorphism (SNP) analysis. Total genomic DNA was extracted, polymerase chain reaction (PCR) amplified, digested with AluI enzyme and run on 2% agarose gel. At the exon 7 splice acceptor site, the presence of homozygote (AA) was indicated by AluI digestion of the 203 bp PCR product into two fragments of 150 bp and 53 bp. While homozygote (GG) was indicated by absence of AluI site and retaining of the 203 bp PCR fragment intact. If both alleles exist as in heterozygotes (GA), three fragments of 203 bp, 150 bp and 53 bp appear. Lane B represents the PCR product before digestion with Alu1, whereas lane A represents PCR products after digestion with Alu1.
Frequency of exon 7 SAS‐ SNP in human OAS1 gene in a pilot sample of Egyptian population
To explore whether the tested SNP represents a significant genetic components of the Egyptian population, 50 non‐infected subjects (healthy controls) from both sexes were exposed to RFLP analysis at the exon 7 SAS. To examine whether there is an allele association with chronic HCV infection, we compared the frequency of the tested SNP in both groups. The human OAS1 genotypes in the control group were 52% GG, 38% GA and 10% AA. The results indicated that G allele tends to be more frequent than A allele in both control and infected populations. However, in controls, genotype GG is slightly higher than in chronic HCV patients (52% vs 45%) as shown in Figure 2.
Figure 2.
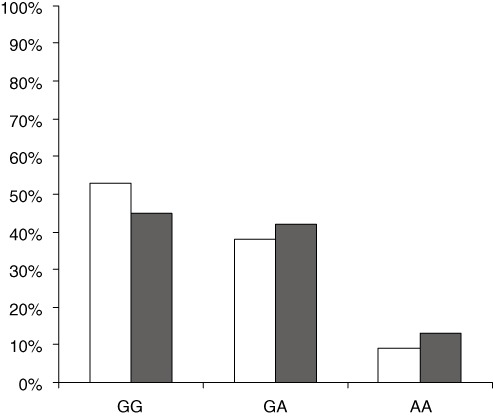
Distribution of oligoadenylate synthetase 1 (OAS1) exon 7 splice‐acceptor site (SAS) genotypes in Control and hepatitis C virus (HCV) infected patients. Seventy chronic HCV patients and 50 healthy controls were subjected to polymerase chain reaction‐restriction fragment length polymorphism (PCR‐RFLP) analysis at the exon7 SAS of OAS1 gene. OAS1 genotypes were compared in both HCV patient (black) and control (white) groups. □, Control;  , HCV patients.
, HCV patients.
Influence of different OAS1 genotypes on response rates and liver fibrosis stages
The frequency of different OAS1 genotypes in SVR patients showed a trend of having more G than A allele as 11% were AA, 37.9% were AG and 59.4% were GG (P = 0.007). The AA genotype was detected only in 11% of SVR versus 89% of NR patients as shown in Figure 3. On the other hand, patients with AA and GA genotypes had significantly higher fibrosis scores (88.9% and 62%) than those with GG (43.7%) genotype (P = 0.05) as shown in Table 2.
Figure 3.
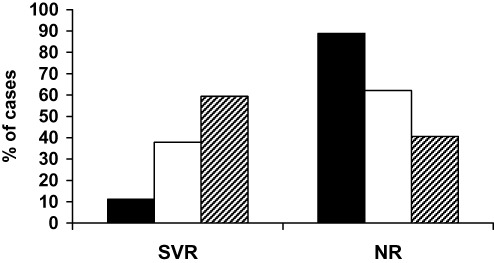
Effect of different oligoadenylate synthetase (OAS) genotypes on the hepatitis C virus (HCV) response rate to IFN based therapy. Chronic HCV patients received interferon (IFN) + ribavirin for 48 weeks and followed for 24 weeks after end of treatment. Total genomic DNA was extracted and analyzed for genotyping at exon7 SAS of OAS1 gene. Frequency of AA (black), AG (white) and GG (hatched) were represented in sustained virological responder (SVR) and non‐responder (NR) groups.  , AA; □, AG;
, AA; □, AG;  , GG.
, GG.
Table 2.
Effect of different oligoadenylate synthetase (OAS) genotypes on response rate, liver activity and liver fibrosis
| Factor | OAS | P‐value* | ||
|---|---|---|---|---|
| AA | AG | GG | ||
| n = 9 | n = 29 | n = 32 | ||
| n (%) | n (%) | n (%) | ||
| Fibrosis | ||||
| F1 | 1 (11.1) | 11 (37.9) | 18 (56.3) | |
| F2–F4 | 8 (88.9) | 18 (62.1) | 14 (43.7) | 0.05 |
| Activity | ||||
| A1 | 4 (44.4) | 19 (65.5) | 24 (75.0) | |
| A2–A3 | 5 (55.6) | 10 (34.5) | 8 (25.0) | 0.220 |
| HCV | ||||
| Responders | 1 (11.1) | 11 (37.9) | 19 (59.4) | |
| Non responders | 9 (88.9) | 18 (62.1) | 13 (40.6) | 0.007 |
P‐values ≤ 0.05 are considered significant.
The effect of OAS single nucleotide polymorphism (SNP) on liver fibrosis, liver activity and response rate to therapy was statistically analyzed. HCV, hepatitis C virus.
Comparison of genotypes between SVR and NR patients
To investigate whether the tested SNP does exist more frequently in either the SVR or NR groups of patients, GG genotype was detected in 32 patients, GA in 29 patients and AA in nine patients as shown in Table 3. The GG genotype was detected more in the SVR group than in the NR group (61.29% vs 33.3%). Frequencies of genotypes and alleles at exon 7 SAS were compared between SVR and NR patients. Analyses of the allele pool in the studied sample revealed that the number of A allele was 47 (nine patients were AA and 29 patients were AG) whereas the number of G allele was 93 (32 patients were GG and 29 patients were GA). The NR patients had higher frequency of A allele (72.3%) as much as the SVR (27.6%) patients (P = 0.003) as shown in Table 3.
Table 3.
Statistical analyses of the oligoadenylate synthetase (OAS) gene exon 7 splice acceptor site (SAS) genotypes & alleles in non‐responder (NR) and sustained virological responder (SVR) patients
| Non responders (NR = 39) | Responders (SVR = 31) | Odds ratio | 95% confidence intervals | P‐value | |
|---|---|---|---|---|---|
| AA (n = 9) | 8 (20.51%) | 1 (3.23%) | 8.000 | 1.001–63.963 | 0.05 |
| AG (n = 29) | 18 (46.15%) | 11 (35.48%) | 1.636 | 0.773–3.465 | 0.198 |
| GG (n = 32) | 13 (33.33%) | 19 (61.29%) | 0.684 | 0.338–1.385 | 0.292 |
| A (n = 47) | 34 (72.34%) | 13 (27.66%) | 2.615 | 1.380–4.956 | 0.003 |
| G (n = 93) | 44 (47.31%) | 49 (52.69%) | 0.898 | 0.598–1.349 | 0.604 |
P‐values > 0.05 = Not significant; P‐values ≤ 0.05 = Significant.
Frequencies of genotypes and alleles at exon 7 SAS were compared between SVR and NR patients (columns 1 and 2). Odds ratio, confidence intervals and P‐value were represented in columns 3, 4 and 5. Genotype AA represents the lowest frequency among the studied sample of patients where it is mostly associated with NR (odds ratio = 8.00).
Data are expressed as number (%).
Multivariate analysis of predictive factors for interferon treatment among all HCV patients
The significance of several host factors in determining the response to IFN therapy was investigated by stepwise logistic regression analysis. Histological pattern (liver fibrosis and activity), different OAS genotypes at exon 7 SAS and AFP levels (less than 5 ng/mL) independently and significantly influenced the outcome of IFN therapy (Table 4). In order to study the effect of a single factor after controlling for the effect of all other factors, a stepwise logistic regression analysis was performed. HCV patients with higher fibrosis stage (F2–F4) had a significantly higher risk of no response than those with low fibrosis stage (F1), they had 23‐fold more chance of no response (95% confidence 6.607 to 82.275, P = 0.0001). On the other hand, A allele at OAS 1 exon 7 SAS increased the risk of no response by 4.9‐fold more than those with G allele (95% confidence 1.365 to 8.214, P = 0.006).
Table 4.
Stepwise logistic regression for the effect of prognostic factors on the interferon response in hepatitis C virus (HCV) infected patients
| Regression coefficient | Standard error | Odds ratio | 95% CI for odds ratio | P‐value | ||
|---|---|---|---|---|---|---|
| Lower | Upper | |||||
| Fibrosis (F0–F1 vs F2–F4) | 3.149 | 0.643 | 23.314 | 6.607 | 82.275 | 0.0001 |
| Activity (A0–A1 vs A2–A3 | 1.495 | 0.584 | 4.457 | 1.418 | 14.015 | 0.011 |
| AFP (<5 ng/mL vs >5 ng/mL) | 1.257 | 0.512 | 3.514 | 1.287 | 9.593 | 0.014 |
| OAS Genotype (GG vs GA or AA) | 1.053 | 0.403 | 4.867 | 1.301 | 8.320 | 0.009 |
| OAS Alleles (A vs G) | 1.069 | 0.387 | 4.913 | 1.365 | 8.214 | 0.006 |
P‐values > 0.05 = Not significant; P‐values ≤ 0.05 = Significant.
Multivariate analyses of the different prognostic factors in both sustained virological responder (SVR) and non‐responder (NR) patients including liver fibrosis, liver activity, alpha fetoprotein (AFP), oligoadenylate synthetase (OAS) genotypes and OAS alleles. Data are expressed as number (%).
Discussion
Individuals infected with HCV may exhibit a spectrum of possible outcomes; some (15–25%) are able to clear the virus naturally, whereas others 75–85% develop persistent infections. During the chronic infection, some patients have minimal progression of disease, whereas others develop severe liver disease such as elevated transaminase levels, chronic hepatitis, steatosis, liver fibrosis and cirrhosis. 33 Both host and viral factors are thought to influence HCV disease outcome and response to IFN treatment. Since more than 93% of patients in Egypt are infected with HCV genotype 4 34 it seemed more applicable to carry out the current study on patients infected with genotype 4a who receive the pegylated IFN + ribavirin at doses mentioned in the methods section. Host genetic factors that affect the outcome of IFN treatment in chronic hepatitis C have been investigated. Several SNPs in IFN‐inducible genes leading to altered gene expression or function were found to be associated with divergent responses to IFN treatment. 28 , 35 , 36 , 37 Understanding which patients are likely to develop persistent infection, progressive liver disease or not responding significantly to interferon therapy would be of utmost importance in disease control programs. In this study we showed that a specific human mutation that results in reduced function in a known antiviral gene OAS1 is associated with resistance to interferon therapy in HCV infected patients. OAS1 codes for a component of the type I interferon signaling system; its homolog in mice has been shown previously to be important for flavivirus susceptibility. 38 Our observations suggested that polymorphisms at exon 7 splice‐acceptor site in the OAS1 gene may be involved in determining the outcome of HCV infection. Patients harboring GG genotype at this site tend to achieve sustained response to interferon therapy (59.4%) suggesting that the G allele conferred a better signaling towards viral clearance. On the other hand, a clear association exists between both allele A containing genotypes, that is GA and AA and resistance to interferon therapy (62% and 89%, respectively). These findings confirmed earlier reports that the G allele at this acceptor site retained the splice site and was associated with a higher OAS enzyme activity (P46), whereas the A allele (AA at acceptor site) ablated the splice site and resulted in a larger molecular weight protein with a lower OAS enzyme activity (P48). 25 , 26 The role of OAS enzyme activity in combating other viral infections has been previously documented, a SNP in exon 3 of OAS1 was related to the development of severe acute respiratory syndrome (SARS, a coronavirus infection). 10 Recently, Lim et al. 39 reported that individuals carrying two copies of the hypofunctional variant of OAS1 were found more frequently than expected among both asymptomatic and symptomatic West Nile Virus (WNV) positive individuals and further proved that OAS1 activity influences the probability of WNV initial infection upon exposure in man.
To our knowledge, there are no previous reports on the effects of different OAS1 genotypes at exon 7 splice acceptor site on the response rate of HCV infected patients to combined interferon + ribavirin therapy. Knapp et al. 37 reported that different genotypes at specific SNPs within the 3′UTR of OAS1 gene were not associated with differential response to IFN therapy but more likely were associated with persistent infection of patients. Moreover, Li et al. 40 showed no relationship of a SNP in exon 3 of OAS1 gene and IFN treatment response. Okuno et al., Solinas et al. and Pawlotsky et al. 41 , 42 , 43 hypothesized that OAS1 plays a greater role in mediating self‐limiting versus persistent HCV infections as a component of the innate immune system and an IFN stimulated gene than its role in exogenously administered IFN treatment. The current data support this hypothesis, since the SNP allele (AA or GA) of the OAS1 gene was slightly overrepresented in HCV patients (13%, 42%) compared with the controls (9%, 38%, respectively). This finding indicates that OAS1 plays a role in persistence of HCV infection.
On the contrary, the splice‐acceptor site specific SNP studied here exhibits a deleterious effect on OAS1 enzyme activity and hence is expected to dramatically influence the IFN mediated signal transduction pathway. It was presumed that GA and AA genotypes could negatively affect the suppressive function of OAS of HCV replication, and may therefore be related to sustained inflammation and progression of liver disease. This hypothesis was supported by results of the present study in which the hepatic fibrosis score was higher in AA and GA genotypes compared with the GG genotype. Chronic HCV patients with positive A allele had significantly higher fibrosis scores (≥ F2 metavir) than those observed in patients without A allele (88.9% in AA and 62% in GA vs 43.7% in GG, P = 0.05). Interestingly, fibrosis scores were higher in OAS variants including allele A at exon 7 SAS in a gene dose‐dependent fashion; that is, the more A allele in OAS exon 7 SAS, the more F score in a patient's liver. On the other hand there was no significant association between different OAS genotypes and liver activity (P = 0.22). This may be explained by the fact that A allele encodes p48 with a weak ability to suppress HCV replication compared with OAS1 having G allele coding for the more active enzyme p46. 25 Thus, the presence here of A allele may lead to a higher degree of necrosis, regeneration and ultimately higher degree of fibrosis. These latter findings support the previous results of Li et al. 40 who found that the SNP of OAS‐1 at exon 3 was associated with disease progression in Japanese patients with HCV infection. In general, the effects of different OAS genotypes on the liver disease progression in HCV patients need to be further studied.
There have been relatively few studies aimed at identifying the human genetic differences that influence progression to chronic viral infection and response to interferon. 44 Recently, a SNP has been identified located upstream of the IL28B gene that encodes IFN‐λ3, which is strongly associated with the sustained virological response to pegIFN and ribavirin in HCV‐infected patients. 45 , 46 Matsushita et al. 35 showed an association between polymorphism of the mannose‐binding lectin gene and IFN responsiveness of Japanese patients with chronic hepatitis C. Moreover, Hijikata et al. 36 showed that a SNP (G/T at nt −88) in the promoter region of the MxA gene was associated with response to IFN in Japanese HCV patients. It is possible that other host proteins with different functions may also be associated, via diverse mechanisms, with the response to IFN in chronic HCV patients. The present multivariate analysis results indicate that the OAS1 gene polymorphism is not the only factor that influences the IFN‐induced antiviral activities of host cells against viral infection. However, multivariate analysis showed that the SNP of OAS‐1 at the exon 7 splice acceptor site as independent and significant determinants of the outcome of IFN therapy (odds ratio 4.913 [95% confidence interval 1.365–8.2], P = 0.006). Taken together the present data and the recent findings from our laboratory on the role of cytomegalovirus (CMV) coinfection as well as the liver fibrosis score on the outcome of viral response to IFN based therapy 47 would be valuable components of a predictive mathematical model for selecting ideal patient cohorts before starting the course of treatment.
In conclusion, the results of the present study may have clinical and pathogenic implications and suggest that the SNP of OAS‐1 at the exon 7 splice acceptor site represents added value to the promising markers for prognosis of disease progression and prediction of drug responses of those patients in clinical practice.
Acknowledgments
The present work was supported by National Research Center, Egypt. Project No. E8041129 to Professor Mostafa El‐Awady and project no. 8041177 to Dr Ashraf.
References
- 1. Manns MP, McHutchison JG, Gordon SC et al Peginterferon alfa‐2b plus ribavirin compared with interferon alfa‐2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet 2001; 358: 958–65. [DOI] [PubMed] [Google Scholar]
- 2. Fried MW, Shiffman ML, Reddy KR et al Peginterferon alfa‐2a plus ribavirin for chronic hepatitis C virus infection. N. Engl. J. Med. 2002; 347: 975–82. [DOI] [PubMed] [Google Scholar]
- 3. Strader DB, Wright T, Thomas DL, Seeff LB. Diagnosis, management, and treatment of hepatitis C. Hepatology 2004; 39: 1147–71. [DOI] [PubMed] [Google Scholar]
- 4. Davis GL, Balart LA, Schiff ER. Treatment of chronic hepatitis C with recombinant interferon alfa. A multicenter randomized, controlled trial. Hepatitis Interventional Therapy Group. N. Engl. J. Med. 1989; 321: 1501–6. [DOI] [PubMed] [Google Scholar]
- 5. Hoofnagle JH, DiBisceglie AM. The treatment of chronic viral hepatitis. N. Engl. J. Med. 1997; 336: 347–56. [DOI] [PubMed] [Google Scholar]
- 6. Der SD, Zhou A, Williams BR, Silverman RH. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 1998; 95: 15623–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tsubota A, Chayama K, Ikeda K et al Factors predictive of response to interferon‐alpha therapy in hepatitis C virus infection. Hepatology 1994; 19: 1088–94. [PubMed] [Google Scholar]
- 8. Gale M Jr., Foy EM. Evasion of intracellular host defense by hepatitis C virus. Nature 2005; 436: 939–45. [DOI] [PubMed] [Google Scholar]
- 9. Ben‐Ari Z, Mor E, Papo O et al Cytokine gene polymorphisms in patients infected with hepatitis B virus. Am. J. Gastroenterol. 2003; 98: 144–52. [DOI] [PubMed] [Google Scholar]
- 10. Hamano E, Hijikata M, Itoyama S et al Polymorphisms of interferon‐inducible genes OAS‐1 and MxA associated with SARS in the Vietnamese population. Biochem. Biophys. Res. Commun. 2005; 329: 1234–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Thursz M. Genetic susceptibility in chronic viral hepatitis. Antiviral Res. 2001; 52: 113–20. [DOI] [PubMed] [Google Scholar]
- 12. Suh Y, Vijg J. SNP discovery in associating genetic variation with human disease phenotypes. Mutat. Res. 2005; 573: 41–50. [DOI] [PubMed] [Google Scholar]
- 13. Levy DE, Garcia‐Sastre A. The virus battles: IFN induction of the antiviral state and mechanisms of viral evasion. Cytokine Growth Factor Rev. 2001; 12: 143–56. [DOI] [PubMed] [Google Scholar]
- 14. Sen GC. Viruses and interferons. Annu. Rev. Microbiol. 2001; 55: 255–81. [DOI] [PubMed] [Google Scholar]
- 15. Silverman RH. Implications for RNase L in prostate cancer biology. Biochemistry 2003; 42: 1805–12. [DOI] [PubMed] [Google Scholar]
- 16. Xiang Y, Wang Z, Murakami J et al Effects of RNase L mutations associated with prostate cancer on apoptosis induced by 2′‐5′‐oligoadenylates. Cancer Res. 2003; 63: 6795–801. [PubMed] [Google Scholar]
- 17. Justesen J, Hartmann R, Kjeldgaard NO. Structure and function of the 2′‐5′‐oligoadenylate synthetase family. Cell. Mol. Life Sci. 2000; 57: 1593–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mashimo T, Lucas M, Simon‐Chazottes D et al A nonsense mutation in the gene encoding 2_‐5_‐oligoadenylate synthetase/L1 isoform is associated with West Nile virus susceptibility in laboratory mice. Proc. Natl. Acad. Sci. USA 2002; 99: 11311–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Perelygin AA, Scherbik SV, Zhulin IB, Stockman BM, Li Y, Brinton MA. Positional cloning of the murine flavivirus resistance gene. Proc. Natl. Acad. Sci. USA 2002; 99: 9322–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lucas M, Mashimo T, Frenkiel MP et al Infection of mouse neurones by West Nile virus is modulated by the interferon‐inducible 2′‐5′‐oligoadenylate synthetase 1b protein. Immunol. Cell Biol. 2003; 81: 230–6. [DOI] [PubMed] [Google Scholar]
- 21. Bonnevie‐Nielsen V, Martensen PM, Justesen J et al The antiviral defense system is persistently activated in human type 1 diabetes. Clin. Immunol. 2000; 96: 11–18. [DOI] [PubMed] [Google Scholar]
- 22. Bonnevie‐Nielsen V, Larsen ML, Frifelt JJ, Michelsen B, Lernmark A. Association of IDDM and attenuated response of 2′‐5′‐oligoadenylate synthetase to yellow fever vaccine. Diabetes 1989; 38: 1636–42. [DOI] [PubMed] [Google Scholar]
- 23. Naganuma A, Nozaki A, Tanaka T et al Activation of the interferon‐inducible 2′‐5′‐oligoadenylate synthetase gene by hepatitis C virus core protein. J. Virol. 2000; 74: 8744–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Karino Y, Hige S, Saga A et al Relationship Between Serum 20–50 Oligoadenylate Synthase Activity and the Reduction of HCV‐RNA During Interferon Therapy. Tokyo: Springer, 1994; 631–4. [Google Scholar]
- 25. Bonnevie‐Nielsen V, Field L, Lu S et al Variation in antiviral 2′‐5′‐oligoadenylate synthetase (2′‐5′‐AS) enzyme activity is controlled by a single‐nucleotide polymorphism at a splice‐acceptor site in the OAS1 gene. Am. J. Hum. Genet. 2005; 76: 623–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ghosh A, Sarkar SN, Rowe TM, Sen GC. A specific isozyme of 2′‐5′‐oligoadenylate synthetase is a dual function proapoptotic protein of the Bcl‐2 family. J. Biol. Chem. 2001; 276: 25447–55. [DOI] [PubMed] [Google Scholar]
- 27. King JK, Yeh SH, Lin MW et al Genetic polymorphisms in interferon pathway and response to interferon treatment in hepatitis B patients: a pilot study. Hepatology 2002; 36: 1416–30. [DOI] [PubMed] [Google Scholar]
- 28. Suzuki F, Arase Y, Suzuki Y et al Single nucleotide polymorphism of the MxA gene promoter influences the response to interferon monotherapy in patients with hepatitis C viral infection. J. Viral Hepat. 2004; 11: 271–80. [DOI] [PubMed] [Google Scholar]
- 29. Bedossa P, Poynard T. An algorithm for the grading of activity in chronic hepatitis C. The METAVIR Cooperative Study Group. Hepatology 1996; 24: 289–93. [DOI] [PubMed] [Google Scholar]
- 30. El Awady M, Badr El Din N, El Garf W et al Antisense oligonucleotide inhibition of hepatitis C virus genotype 4 replication in HepG2 cells. Cancer Cell Int. 2006; 6: 18–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. El Awady M, Tabll A, El‐Abd Y et al HepG2 cells support viral replication and gene expression of hepatitis C virus genotype 4 in vitro . World J. Gastroenterol. 2006; 12: 4836–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Thakeb F, El‐Serafy M, Zakaria S et al Evaluation of liver tissue by polymerase chain reaction for hepatitis B virus in patients with negative viremia. World J. Gastroenterol. 2005; 11: 6853–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shepard CW, Finelli L, Alter MJ. Global epidemiology of hepatitis C virus infection. Lancet Infect. Dis. 2005; 5: 558–67. [DOI] [PubMed] [Google Scholar]
- 34. Omran MH, Youssef S, El‐Garf W et al Phylogenetic and genotyping of hepatitis C virus in Egypt. Aust. J. Basic Appl. Sci. 2009; 3 (1): 1–8. [Google Scholar]
- 35. Matsushita M, Hijikata M, Matsushita M, Ohta Y, Mishiro S. Association of mannose‐binding lectin gene haplotype LXPA and LYPB with interferon‐resistant hepatitis C virus infection in Japanese patients. J. Hepatol. 1998; 29: 695–700. [DOI] [PubMed] [Google Scholar]
- 36. Hijikata M, Ohta Y, Mishiro S. Identification of a single nucleotide polymorphism in the MxA gene promoter (G/T at nt _88) correlated with the response of hepatitis C patients to interferon1. Intervirology 2000; 43: 124–7. [DOI] [PubMed] [Google Scholar]
- 37. Knapp S, Yee S, Frodsham AJ et al Polymorphisms in interferon‐induced genes and the outcome of hepatitis C virus infection: roles of MxA, OAS‐1 and PKR. Genes Immun. 2003; 4: 411–19. [DOI] [PubMed] [Google Scholar]
- 38. Mashimo T, Simon‐Chazottes D, Guénet JL. Innate resistance to flavivirus infections and the functions of 2′‐5′‐oligoadenylate synthetases. Curr. Top. Microbiol. Immunol. 2008; 321: 85–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lim JK, Lisco A, McDermott DH et al Genetic variation in OAS1 is a risk factor for initial infection with West Nile Virus in man. PLoS Pathog. 2009; 5: e1000321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li C, Kato N, Chang J et al Polymorphism of OAS‐1determines liver fibrosis progression in hepatitis C by reduced ability to inhibit viral replication. Liver Int. 2009; 29: 1413–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Okuno T, Shindo M, Arai K et al Determination of 2′, 5′ Oligoadenylate Synthetase Acitvity in Acute Non‐A, Non‐B Hepatitis Patients, with Special Reference of Histological Outcome. Viral Hepatitis C, D and E. Amsterdam: Elsevier, 1991; 133–5. [Google Scholar]
- 42. Solinas A, Cossu P, Poddighe P et al Changes of serum 2′, 5′‐oligoadenylate synthetase activity during interferon treatment of chronic hepatitis C. Liver 1993; 13: 253–8. [DOI] [PubMed] [Google Scholar]
- 43. Pawlotsky JM, Hovanessian A, Roudot‐Thoraval F et al Activity of the interferon‐induced 20,50‐oligoadenylate synthetase in patients with chronic hepatitis C. J. Interferon Cytokine Res. 1995; 15: 857–62. [DOI] [PubMed] [Google Scholar]
- 44. Peng XM, Lei RX, Gu L, Ma HH, Xie QF, Gao ZL. Influences of MxA gene −88 G/T and IFN‐gamma +874 A/T on the natural history of hepatitis B virus infection in an endemic area. Int. J. Immunogenet. 2007; 34: 341–6. [DOI] [PubMed] [Google Scholar]
- 45. Ge D, Fellay J, Thompson AJ et al Genetic variation in IL28B predicts hepatitis C treatment‐induced viral clearance. Nature 2009; 461: 399–401. [DOI] [PubMed] [Google Scholar]
- 46. Chevaliez S, Hézode C. IL28B polymorphisms and chronic hepatitis C. Gastroenterol. Clin. Biol. 2010; 34: 587–9. [DOI] [PubMed] [Google Scholar]
- 47. Bader El Din NG, Meguid MA, Tabll AA et al Human cytomegalovirus infection inhibits response of chronic hepatitis‐C‐virus‐infected patients to interferon‐based therapy. J. Gastroenterol. Hepatol. 2011; 26: 55–62. (in press). [DOI] [PubMed] [Google Scholar]


