Abstract
Background Diarrheal disease is a major cause of morbidity and mortality in humans and animals, including non human primates. While the diagnostics for gastrointestinal bacterial and parasitic pathogens and their etiological role in disease are well established, little is known about the epidemiology, prevalence and role of viral agents in diarrheal illness among monkeys.
Methods We collected fecal specimens from monkeys with diarrhea that were housed in two primate colonies, the Institute of Laboratory Animal Sciences, Beijing, China and the Yerkes National Primate Research Center, Georgia, USA. We screened these fecal specimens for rotaviruses and enteric adenoviruses 40/41 by using commercial EIA kits (Rotaclone and Adenoclone), enteroviruses by RT‐PCR and Southern blot hybridization, and picobirnaviruses by polyacrylamide gel electrophoresis and silver staining. Some of the specimens were examined by EM for coronaviruses and noroviruses.
Results Of the 92 specimens from China, we found 63 (68%) positive for viruses, including enteroviruses (52%), enteric adenoviruses (21%), rotaviruses (20%), and picobirnaviruses (2%). Coronaviruses were detected in some specimens. Mixed infection of two or more viral agents was seen in 23 (25%) specimens. In the US collection, we detected enteroviruses and enteric adenoviruses in 76% (45/59) and 14% (7/50) of the specimens, respectively. Electron microscopy showed norovirus‐like particles in some specimens from both colonies.
Conclusions Our findings indicate endemic infections with enteric viruses in monkeys of both colonies. The availability of new simian rotaviruses, enteric adenoviruses, enteroviruses, and coronaviruses and the discovery of noroviruses and picobirnaviruses may allow us to develop better diagnostics for these agents and determine which of these agents are clearly associated with gastroenteritis in monkeys.
Keywords: adenovirus, coronavirus, enterovirus, norovirus, picobirnavirus, rotavirus
Introduction
Diarrheal disease is a major cause of morbidity in monkeys and is associated with high mortality in some non‐human primate colonies [19]. Monkeys housed in colonies experience multiple episodes of diarrhea that can affect the maintenance and use of these animals in biomedical research. Bacteria, such as Campylobacter, Shigella and Yersinia enterocolitica species, and parasites, including Cryptosporidium, are common and are recognized causes of enteric infection in monkeys [19, 22], but the role of viral agents has been less clearly defined. No pathogens are identified in up to 69% of fecal specimens from episodes of diarrhea, and the rate of detection is especially low in neonates [19].
While the diagnostics for gastrointestinal bacterial and parasitic pathogens and their etiological role in disease are well established, there are few sensitive, simple, and routine diagnostic assays for most simian enteric viruses. Consequently, little is known about the epidemiology, prevalence and causal role of viral agents in diarrheal disease among monkeys. A surveillance study conducted 28 years ago found that 12% of the samples from captive Kenyan baboons in a primate colony contained enteroviruses and adenoviruses [17]. Despite their widespread prevalence, enteroviruses that grow well in the intestine are rarely associated with disease in monkeys [10], although adenoviruses are recognized as a cause of diarrhea in monkeys [22, 25]. Coronaviruses were common in normal and diarrhea stools of monkeys in two US colonies but no causal relationship with disease has been established [18, 23]. Monkeys have high prevalences of antibodies to rotaviruses, a major cause of infantile dehydrating diarrhea and to noroviruses, which cause major outbreaks of acute gastroenteritis in humans [1, 9, 12, 15]. However, rotaviruses are rarely detected and are not considered a major cause of diarrhea in monkeys although two simian strains, SA11 and RRV, are the most widely used reference strains in laboratories worldwide [11, 13, 19, 22, 25]. Noroviruses have not yet been detected in monkeys.
In this study, we sought to detect viral agents in fecal specimens collected from monkeys with diarrhea in two major primate colonies, one in China and one in the United States. Our goals were to investigate the prevalence and epidemiology of each viral agent, develop better diagnostic tools and examine the genetic or evolutionary relationship between simian and human viruses. We demonstrated a high prevalence of viruses, predominantly enteroviruses, among monkeys in the US colony and found a more diverse group of viral agents, including enteroviruses, enteric adenoviruses, rotaviruses, coronaviruses and picobirnaviruses, among monkeys in China.
Materials and Methods
Primate colony and specimen collection
The Yerkes National Primate Research Center consists of a main laboratory on the campus of Emory University in Atlanta, Georgia, and a field station in Lawrenceville, Georgia. The Center houses over 3000 animals in seven species. From February 1999 to April 1999, fecal specimens were collected from 41 rhesus and 12 pigtailed macaques housed in the field station. All had diarrhea at the time of specimen collection. The rhesus and pigtailed macaques ranged in age from 10 months to 25 years and from 6 months to 21 years, respectively.
Between February 2002 and January 2003, fecal or rectal swab specimens were collected from rhesus, cynomolgus and pigtailed monkeys with diarrhea housed in the monkey farm of the Institute of Laboratory Animal Sciences, Chinese Academy of Medical Sciences, near Beijing, China. These monkeys ranged in age from 18 months to 14 years. All specimens were stored at −70°C before testing.
Enzyme Immunoassay (EIA) and electron microscopy (EM)
After each fecal specimen was diluted 10‐fold in PBS, the suspension was clarified by centrifugation at 2000 g for 10 minute in an AllegraTM 6R centrifuge (Beckman Coulter, Fullerton, CA, USA). The supernatant was then transferred to a fresh tube and centrifuged at 5220 g for 10 minute in an Eppendorf microfuge (model 5415; Westbury, NY, USA). Fecal extracts were then tested for rotaviruses and enteric adenoviruses using commercial EIA kits (Rotaclone and Adenoclone 40/41; Meridian Diagnostics, Cincinnati, OH, USA) in accordance with the manufacturer's instructions. Rotaclone and Adenoclone 40/41 kits are validated and highly specific diagnostic assays that have a sensitivity of 5 × 105 and 8 × 103 particles/ml for rotaviruses and enteric adenovirus serotypes 40 and 41 in stools, respectively. Some specimens were examined by EM for the presence of enteric viruses. Briefly, glow‐discharge pre‐treated formvar‐carbon coated grids were placed on drops (3–5 μl) of clarified stool suspension and incubated at room temperature for 10 minute. The grids were blotted on edge with filter paper and placed onto a 10‐μl drop of 2% phosphotungstic acid in water (pH 6.5) for 30 s. The grids were then blotted again with filter paper and viewed in a Philips 201 electron microscope (Philips Electron Optics Inc., Einhoven, The Netherlands).
Polymerase chain reaction (PCR) and polyacrylamide gel electrophoresis (PAGE)
Viral RNA or DNA was extracted from clarified stool suspension using guanidine isothiocyanate and silica as previously described [6]. RNA was examined for viral agents using a one‐step RT‐PCR kit (QIAGEN Inc., Valencia, CA, USA). For enteroviruses, primer pairs 299 (5′‐ACCGGATGGCCAATCCAA‐3′) at positions 642–625 and 300 (5′‐CCTCCGGCCCCTGAATG‐3′) at positions 448–464 were used [21]. RT was conducted at 42°C for 40 minute and PCR at 95°C for 15 minute, followed by 35 cycles at 95°C for 30 s, 42°C for 30 s and 72°C for 1.5 minute, and finally at 72°C for 7 minute. PCR products were electrophoresed in a 1.5% agarose gel and stained with ethidium bromide. Southern hybridization and chemiluminescent detection were performed as previously described using a digoxigenin‐labeled oligonucleotide probe BMJ192 (5′‐TCGTAATGGGTAACTCTG ‐ 3′) at positions 515–532 [20]. Viral RNA or DNA samples were also analyzed by 10% polyacrylamide gels, followed by silver staining according to the manufacturer's protocol (Bio‐Rad, Hercules, CA, USA).
Results
The EIA screening of fecal specimens for rotaviruses and enteric adenoviruses showed that the prevalence of these viruses varied in the two colonies (Table 1). Of the 92 fecal specimens from China, 18 (20%) tested positive for rotaviruses and 19 (21%, the same 18 plus another specimen) tested positive for enteric adenoviruses 40 and 41. The prevalence of rotaviruses and enteric adenoviruses was similar in rhesus and cynomolgus macaques. Of the specimens collected from monkeys at the Yerkes colony, seven (19%) of 36 from rhesus monkeys tested positive for enteric adenoviruses 40 and 41, whereas none of the nine specimens from pigtailed macaques were positive for enteric adenoviruses. No rotaviruses were detected in rhesus and pigtailed macaques in the Georgia facility.
Table 1.
Detection of viral agents in fecal specimens of monkeys with diarrhea by location and species
| Location, species | No. of specimens | % Prevalence of viruses (no. positive/no. tested) | ||||
|---|---|---|---|---|---|---|
| RV | EAV | EV | PV | CV | ||
| Monkey Farm, Beijing, China | ||||||
| Rhesus | 79 | 20 (16/79) | 22 (17/79) | 44 (35/79) | 3 (2/79) | 25 (3/12) |
| Cynomolgus | 10 | 20 (2/10) | 20 (2/10) | 100 (10/10) | 0 | ND |
| Pigtail | 3 | 0 | 0 | 100 (3/3) | 0 | 100 (1/1) |
| Total | 92 | 20 (18/92) | 21 (19/92) | 52 (48/92) | 2 (2/92) | 31 (4/13) |
| Yerkes, Georgia, USA | ||||||
| Rhesus | 50 | 0 (0/48) | 19 (7/36) | 81 (35/43) | ND | ND |
| Pigtail | 12 | 0 (0/12) | 0 (0/9) | 67 (8/12) | ND | ND |
| Total | 62 | 0 (0/60) | 16 (7/45) | 78 (43/55) | ||
RV, rotavirus; EAV, enteric adenovirus; EV, enterovirus; PV, picobirnavirus; CV, coronavirus; ND, not done.
We tested RNA samples by RT‐PCR and Southern hybridization for enteroviruses (Fig. 1) and found they were common in monkeys with diarrhea in both colonies (Table 1). Of 92 fecal specimens from China, 48 (52%) tested positive (prevalence of 100% in cynomolgus and pigtailed monkeys and 44% in rhesus macaques). Forty‐three (78%) of 55 fecal specimens from the Yerkes colony tested positive, with a similar high prevalence in rhesus and pigtailed macaques.
Figure 1.

Detection of enteroviruses by RT‐PCR and Southern hybridization. PCR products were analyzed on 1.5% agarose gels, stained with ethidium bromide, transferred to nylon membranes and hybridized with a digoxigenin‐labeled oligonucleotide probe as described in the text. Lanes are as follows: 1, molecular size markers; 2, H2O control; and 3–13, fecal specimens from 11 monkeys with diarrhea.
To determine whether virus infections are age dependent, we analyzed the prevalence of rotaviruses, enteric adenoviruses, and enteroviruses in monkeys from China by age group (Table 2). Rotaviruses and enteric adenoviruses were slightly more common in monkeys aged 1–5 years than in those ≥5 years of age. The prevalence of enteroviruses was high (47–67%) in all monkeys regardless of age.
Table 2.
Detection of viral agents in fecal specimens of monkeys with diarrhea in a Beijing colony by age
| Age (year) | No. of specimens | % Prevalence (no. positive/no. tested) | ||
|---|---|---|---|---|
| Enterovirus | Enteric adenovirus | Rotavirus | ||
| 1–2 | 9 | 67 (6/9) | 22 (2/9) | 22 (2/9) |
| 2–3 | 28 | 54 (15/28) | 25 (7/28) | 25 (7/28) |
| 3–4 | 24 | 50 (12/24) | 25 (6/24) | 25 (6/24) |
| 4–5 | 12 | 50 (6/12) | 17 (2/12) | 17 (2/12) |
| ≥5 | 19 | 47 (9/19) | 11 (2/19) | 5 (1/19) |
| Total | 92 | 52 (48/92) | 21 (19/92) | 20 (18/92) |
Selected fecal specimens from China were also examined by EM for the presence of enteric viruses (Fig. 2). Rotavirus‐like particles, adenoviruses, coronaviruses, norovirus‐like particles, enteroviruses and picobirnaviruses were observed in fecal specimens of some monkeys with diarrhea. Of note, rotavirus‐like particles appeared empty and atypical. Enteric adenoviruses, enteroviruses and norovirus‐like particles were also observed in fecal specimens of some monkeys with diarrhea in the Yerkes colony but no rotaviruses and coronaviruses were seen.
Figure 2.

Electron micrographs of enteric viruses in fecal specimens of monkeys with diarrhea in China. Specimens were examined by EM for the presence of viruses as described in the text. Shown are a rotavirus‐like particle (A), adenovirus (B), coronavirus (C), norovirus‐like particle (D), enterovirus (E), and picobirnavirus (F). Bar = 100 nm.
Nucleic acids extracted from fecal specimens of monkeys in the Chinese colony were further analyzed by PAGE to detect enteric viruses (Fig. 3). Two (2%) of 92 samples had a genome of two segments, with size similar to those of human and rabbit picobirnaviruses [4, 7]. No RNA profiles typical of rotavirus electropherotype were seen in any of the specimens tested (data not shown).
Figure 3.

Genomic profiles of picobirnaviruses from monkeys with diarrhea in China. Viral RNA was analyzed by PAGE and silver staining. Lanes are as follows: 1, rotavirus RRV control and 2 and 3, picobirnavirus in fecal specimens of two monkeys with diarrhea.
Mixed infections with different enteric viruses were relatively common in monkeys with diarrhea (Fig. 4). Of the 92 specimens from China that were examined for rotaviruses, enteric adenoviruses, enteroviruses, picobirnaviruses or coronaviruses, 41 samples (45%) had one virus, 15 (16%) had two different viruses and six (7%) had three different viruses. One monkey had a mixed infection with four different viruses. No virus was detected in 29 specimens (31%).
Figure 4.
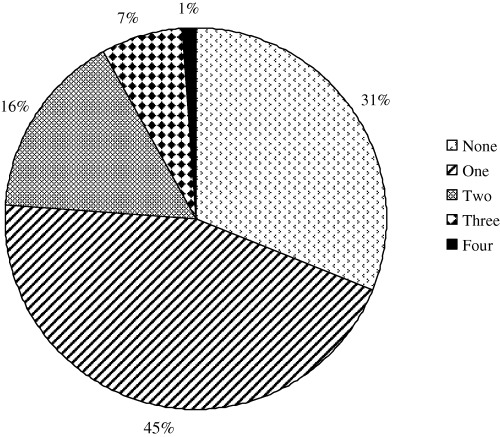
Profiles of virus infections in monkeys with diarrhea. Fecal specimens from monkeys with diarrhea in China were examined for enteric viruses as described in the text. Shown are the percentages of specimens that tested positive for single or multiple enteric viruses. No viruses were detected in 31% of the specimens.
Discussion
The continuing and expanded use of monkey models in biomedical research requires working with animals whose exposures to infectious agents are limited and well defined [14, 16, 26, 29, 30]. Because acute and chronic diarrhea is common in monkeys and poses a major problem for the management of primate facilities, we surveyed viral agents in fecal specimens of monkeys with diarrhea in the two major breeding and research colonies in China and the United States. We found a relatively high prevalence of enteroviruses and enteric adenoviruses in both colonies. These results are similar to those of an earlier study in which infections with enteroviruses and enteric adenoviruses were found to be common in captive normal and sick baboons in a Texas primate facility [17]. We did not detect rotaviruses in fecal specimens of monkeys with diarrhea in Georgia (Yerkes colony). This finding, together with results of our recent 2‐year survey [9], indicates that rotavirus is not a major cause of diarrhea among older moneys in the colony. A previous study also failed to identify rotavirus as a major cause of gastroenteritis in infant, juvenile and adult monkeys in a primate colony in California [25]. In contrast, rotaviruses were relatively common among monkeys with diarrhea in the Chinese colony and were exclusively associated with the presence of enteric adenoviruses in fecal specimens. Furthermore, by EM, rotaviruses appeared as empty and severely damaged particles, an observation that is supported by our failure to detect viral RNA by PAGE. It is an interesting and novel observation that group A rotaviruses appeared unstable and atypical in stools of monkeys with diarrhea. Whether this lack of particle stability explains the discrepancy in the detection of rotavirus and antibody in monkeys needs further investigation.
We recently documented the high prevalence of norovirus antibody in serum of monkeys in the Yerkes colony [9]. The present study identified the presence of norovirus‐like particles in some fecal specimens of monkeys with diarrhea in both the Chinese and American colonies. We tried to confirm noroviruses by RT‐PCR using primers specific for human noroviruses [3, 8] but were not successful. Because of the lack of sensitive and specific diagnostic assays (e.g. EIA and PCR) for simian noroviruses, the prevalence of noroviruses in monkeys remains unknown. Given the sensitivity of 105 to 106 particles/ml by EM [5], the concentration of this simian virus in original stools should be at least 106 viral particles. The detection of noroviruses is highly significant because we now can perform biochemical and molecular characterization of the virus and attempt to isolate it in cell culture. Furthermore, the availability of this simian virus may allow us to develop a challenge model to study the pathogenesis and immunity of norovirus diarrhea and to test potential human norovirus vaccines, similar to our recent rotavirus studies [28, 29].
To our knowledge, this study is the first to detect picobirnaviruses in fecal specimens of monkeys with diarrhea. This finding further expands the host range of this virus from rats, rabbits, chickens, pigs, calves and humans. Future studies will examine the genetic and antigenic relatedness of this simian virus with other picobirnaviruses and to see if this virus might be pathogenic in monkeys. We also observed by EM coronaviruses in fecal specimens of monkeys with diarrhea from China. However, because we did not test all of the specimens by EM we were not able to reliably assess the prevalence of this virus in the primate colony. Of note, people with severe acute respiratory syndrome (SARS) that were first reported in China had diarrhea as a common clinical symptom [2, 24]. SARS coronavirus has been detected in the sewage from hospitals where SARS patients were treated [27]. Whether these simian coronaviruses are genetically related to human SARS coronaviruses needs to be investigated. Availability of a simian coronavirus will greatly facilitate the study of infection and disease in the gastrointestinal or respiratory tract and the development of vaccines against this virus in a monkey challenge model.
Our study is subject to several limitations. We determined the prevalence of rotaviruses, enteric adenoviruses and enteroviruses utilizing the two relatively sensitive and specific commercial EIA kits and molecular diagnostic tools adapted from a previous study (21). While EIA Rotaclone detects both human and animal rotaviruses, Adenoclone‐Type 40/41 is intended for the detection of enteric adenovirus serotypes 40 and 41 in human fecal specimens. This means we might have missed other or previously unidentified simian adenoviruses. Because of the relatively high rate of mixed virus infection and the lack of fecal specimens from clinically healthy monkeys, we were not able to establish a causal relationship between a specific viral agent and diarrhea in these juvenile and adult monkeys. Additional studies should address the prevalence of these enteric viruses and their role in diarrheal disease among infant and young monkeys. We could not examine the seasonality of virus infection because fecal specimens were not collected every month. Whether our observation of a less diverse group of viral agents in the Yerkes colony reflects the shorter period of sample collection remains to be determined. Despite these limitations, this pilot study indicates that multiple enteric viruses circulate in monkeys with diarrhea in two major research primate colonies. The high prevalence of these viruses poses a serious problem for the management of breeding colonies and may have a negative and unforeseen influence on the nature and quality of data resulting from the use of these monkeys in biomedical research. Studies are under way to further characterize these diverse enteric viruses, including determination of species, serotypes, or genotypes for enteroviruses, enteric adenoviruses and noroviruses. There is also a need for sensitive and specific diagnostics that will further increase our understanding of the biology and evolution of simian enteric viruses and may lead to better management and prevention of acute and chronic diarrhea among monkeys in primate facilities.
Acknowledgments
We dedicate this paper to Dr Harold M. McClure who was informed of the preparation of this manuscript before he passed away in October 2004. We thank W. Zhong for technical assistance, staff at the Institute of Laboratory Animal Sciences, Beijing and the Yerkes National Primate Research Center for help in specimen collection, and C. Chesley for editorial assistance.
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the funding agency. This work was supported in part by a Cooperative Research and Development Agreement with Aventis Pasteur and by Yerkes Base Grant RR00165.
References
- 1. Awang A, Yap K: Group A rotavirus infection in animals from an animal house and in wild‐caught monkeys. J Diarrhoeal Dis Res 1990; 8:82–86. [PubMed] [Google Scholar]
- 2. Donnelly CA, Ghani AC, Leung GM, Hedley AJ, Fraser C, Riley S, Abu‐Raddad LJ, Ho LM, Thach TQ, Chau P, Chan KP, Lam TH, Tse LY, Tsang T, Liu SH, Kong JH, Lau EM, Ferguson NM, Anderson RM: Epidemiological determinants of spread of causal agent of severe acute respiratory syndrome in Hong Kong. Lancet 2003; 361:1761–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Farkas T, Zhong WM, Jing Y, Huang PW, Espinosa SM, Martinez N, Morrow AL, Ruiz‐Palacios GM, Pickering LK, Jiang X: Genetic diversity among sapoviruses. Arch Virol 2004; 149:1309–1323. [DOI] [PubMed] [Google Scholar]
- 4. Gallimore C, Lewis D, Brown D: Detection and characterization of a novel bisegmented double‐stranded RNA virus (picobirnavirus) from rabbit faeces. Arch Virol 1993; 133:63–73. [DOI] [PubMed] [Google Scholar]
- 5. Gentile M, Gelderblom HR: Rapid viral diagnosis: role of electron microscopy. New Microbiol 2005; 28:1–12. [PubMed] [Google Scholar]
- 6. Gentsch JR, Glass RI, Woods P, Gouvea V, Gorziglia M, Flores J, Das BK, Bhan MK: Identification of group A rotavirus gene 4 types by polymerase chain reaction. J Clin Microbiol 1992; 30:1365–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Grohmann GS, Glass RI, Pereira HG, Monroe SS, Hightower AW, Weber R, Bryan RT: Enteric viruses and diarrhea in HIV‐infected patients. N Eng J Med 1993; 329:14–20. [DOI] [PubMed] [Google Scholar]
- 8. Jiang X, Huang PW, Zhong WM, Farkas T, Cubitt DW, Matson DO: Design and evaluation of a primer pair that detects both Norwalk‐ and Sapporo‐like caliciviruses by RT‐PCR. J Virol Methods 1999; 83:145–154. [DOI] [PubMed] [Google Scholar]
- 9. Jiang B, McClure H, Fankhauser RL, Monroe SS, Glass RI: Prevalence of rotavirus and norovirus antibodies in non‐human primates. J Med Primatol 2004; 33:30–33. [DOI] [PubMed] [Google Scholar]
- 10. Kalter SS: Enteric viruses of nonhuman primates. Vet Pathol 1982; 19 (Suppl. 7): 33–43. [PubMed] [Google Scholar]
- 11. Kalter SS, Smith GC III, Heberling RL: Electron microscopic examination of primate feces for rotaviruses. Lab Anim Sci 1979; 29:516–518. [PubMed] [Google Scholar]
- 12. Kalter SS, Rodriguez AR, Heberling RL: Rotavirus (SA11) antibody in nonhuman primates. Lab Anim Sci 1982; 32:291–293. [PubMed] [Google Scholar]
- 13. Malherbe HH, Strickland‐Cholmley M: Simian virus S.A. 11 and the related O agent. Archiv Gesampte Virusforsch 1967; 22:235–245. [DOI] [PubMed] [Google Scholar]
- 14. McNeal MM, Sestak K, Choi AH, Basu M, Cole MJ, Aye PP, Bohm RP, Ward RL: Development of a rotavirus‐shedding model in rhesus macaques, using a homologous wild‐type rotavirus of a new P genotype. J Virol 2005; 79:944–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Otsyula M, Yee J, Suleman M, Tarara R, Martin J, Woods P, Glass R, Jennings M: Rotavirus infection in African, non‐human primates. Ann Trop Med Parasitol 1996; 90:659–661. [DOI] [PubMed] [Google Scholar]
- 16. Rockx BH, Bogers WM, Heeney JL, Van Amerongen G, Koopmans MP: Experimental norovirus infections in non‐human primates. J Med Virol 2005; 75:313–320. [DOI] [PubMed] [Google Scholar]
- 17. Rodriguez AR, Kalter SS, Heberling RL, Helmke RJ, Guajardo JE: Viral infections of the captive Kenya baboon (Papio cynocephalus): a five‐year epidemiologic study of an outdoor colony. Lab Anim Sci 1977; 27:356–371. [PubMed] [Google Scholar]
- 18. Russell RG, Brian DA, Lenhard A, Potgieter LN, Gillespie D, Clapp NK: Coronavirus‐like particles and Campylobacter in marmosets with diarrhea and colitis. Dig Dis Sci 1985; 30:72S–77S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Russell RG, Rosenkranz SL, Lee LA, Howard H, DiGiacomo RF, Bronsdon MA, Blakley GA, Tsai CC, Morton WR: Epidemiology and etiology of diarrhea in colony‐born Macaca nemestrina . Lab Anim Sci 1987; 37:309–316. [PubMed] [Google Scholar]
- 20. Sanchez‐Fauquier A, Roman E, Colomina J, Wilhelmi I, Glass RI, Jiang B: First detection of group C rotavirus in children with acute diarrhea in Spain. Arch Virol 2003; 148:399–404. [DOI] [PubMed] [Google Scholar]
- 21. Schwab KJ, De Leon R, Sobsey MD: Concentration and purification of beef extract mock eluates from water samples for the detection of enteroviruses, hepatitis A virus, and Norwalk virus by reverse transcription‐PCR. Appl Environ Microbiol 1995; 61:531–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sestak K, Merritt CK, Borda J, Saylor E, Schwamberger SR, Cogswell F, Didier ES, Didier PJ, Plauche G, Bohm RP, Aye PP, Alexa P, Ward RL, Lackner AA: Infectious agent and immune response characteristics of chronic enterocolitis in captive rhesus macaques. Infect Immun 2003; 71:4079–4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Smith GC, Lester TL, Heberling RL, Kalter SS: Coronavirus‐like particles in nonhuman primate feces. Arch Virol 1982; 72:105–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Srikantiah P, Charles MD, Reagan S, Clark TA, Pletz MW, Patel PR, Hoekstra RM, Lingappa J, Jernigan JA, Fischer M: SARS clinical features, United States, 2003. Emerg Infect Dis 2005; 11:135–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Stuker G, Oshiro LS, Schmidt NJ, Holmberg CA, Anderson JH, Glaser CA, Henrickson RV: Virus detection in monkeys with diarrhea: the association of adenoviruses with diarrhea and the possible role of rotaviruses. Lab Anim Sci 1979; 29:610–616. [PubMed] [Google Scholar]
- 26. Subekti DS, Tjaniadi P, Lesmana M, McArdle J, Iskandriati D, Budiarsa IN, Walujo P, Suparto IH, Winoto I, Campbell JR, Porter KR, Sajuthi D, Ansari AA, Oyofo BA: Experimental infection of Macaca nemestrina with a Toronto Norwalk‐like virus of epidemic viral gastroenteritis. J Med Virol 2002; 66:400–406. [DOI] [PubMed] [Google Scholar]
- 27. Wang XW, Li JS, Guo TK, Zhen B, Kong QX, Yi B, Li Z, Song N, Jin M, Xiao WJ, Zhu XM, Gu CQ, Yin J, Wei W, Yao W, Liu C, Li JF, Ou GR, Wang MN, Fang TY, Wang GJ, Qiu YH, Wu HH, Chao FH, Li JW: Concentration and detection of SARS coronavirus in sewage from Xiao Tang Shan Hospital and the 309th Hospital. J Virol Methods 2005; 128:156–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Westerman LE, Xu J, Jiang B, McClure HM, Glass RI: Experimental infection of pigtailed macaques with a simian rotavirus, YK‐1. J Med Virol 2005; 75:616–625. [DOI] [PubMed] [Google Scholar]
- 29. Westerman LE, McClure HM, Jiang B, Almond JW, Glass RI: Serum IgG mediates mucosal immunity against rotavirus infection. Proc Natl Acad Sci U S A 2005; 102:7268–7273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wu XX, Tu XM, He FQ, Shi HJ, Chen ZW, Wei Q, Jiang H, Shi JD: Study of virological monitoring in rhesus monkeys. Chin J Lab Sci 1991; 1:179–183. [Google Scholar]


