Abstract
There have been recent, significant advances about the role of mRNA turnover in controlling gene expression in immune cells. Post‐transcriptional regulation of gene expression contributes to the characteristics of many of the processes underlying the immune response by ensuring early, rapid, and transient action. The emphasis of this review is on current work that deals with the regulation of mRNA decay during innate immunity against microbes and T cell activation as a model of the adaptive response.
Keywords: mRNA stability, post‐trancriptional regulation, cytokines
INTRODUCTION
In large cities, “rapid transit” is a term that refers to the efficient movement of people allowing rapid entry and exit from a transportation system. A rapid transit scheme also occurs in the immune system to allow immediate activation of the immune cells toward microbes, transient mode of action, and swift recovery. Post‐transcriptional regulation contributes significantly to this rapid transit by several mechanisms, including mRNA stability modulation and translational control; collectively, they aim to control the expression of key gene products involved in the immune response. Sequence elements, mainly in the 3′ untranslated region (3′UTR), and modulation by RNA‐binding proteins can affect mRNA stability and protein translation. Many of the gene products involved in the immune system, such as cytokines, harbor destabilization sequence elements in the mRNA, mostly adenylate‐uridylate (AU)‐rich elements (AREs), which comprise a heterogeneous group of sequence classes that can affect protein interactions with the mRNAs and therefore influence the mRNA decay characteristics [ 1 , 2 ].
Large‐scale studies involving profiling of mRNAs that are bound to RNA‐binding proteins indicate that many of these genes are involved in the early response to stimuli, host defense, and regulation of RNA metabolism and translation [ 3 , 4 , 5 ]. The stabilization of cytokine mRNA and other immune response gene products can occur by the activity of mRNA stabilization‐promoting proteins such as human antigen (HuR) protein or by inactivation of RNA decay‐promoting proteins such as the zinc finger protein, tristetraprolin (TTP). HuR is one of the most characterized RNA‐binding proteins; it is a mammalian homologue of embryonic, lethal, abnormal vision proteins, which was first described in Drosophila. It is implicated in the stabilization of many AU‐rich mRNAs including those that participate in immunity and inflammation such as TNF‐α, IL‐3, IL‐8, and the urokinase activator, urokinase‐type plasminogen activator [ 6 , 7 , 8 ]. Among RNA‐binding proteins that promote mRNA decay is TTP. It is an endotoxin‐inducible, early response gene product, which destabilizes several proinflammatory cytokine mRNAs including TNF‐α, GM‐CSF, and IL‐2 by binding to a specific class of AREs in the 3′UTR. In addition to TTP, K‐homology splicing‐regulatory protein (KSRP [ 9 ]), butyrate response factor (BRF1 [ 10 ]), and certain gene products of the ARE RNA‐binding protein 1 [ARE/poly(U)‐binding/degradation factor 1 (AUF1)] family [ 2 ] are considered negative‐feedback mediators. TTP and several other RNA‐binding proteins appear to function by targeting AU‐rich mRNA to the exosome, a multiprotein complex with 3′–5′ exoRNase activity representing a major degradation machine in eukaryotes [ 11 ]. ARE‐binding proteins can also function by recruitment of degradation factors other than the exosome [ 12 , 13 ]. An alternative pathway of 5′–3′ decay can also occur during degradation of AU‐rich mRNAs in cytoplasmic RNA foci—processing bodies [ 14 ]. After a brief introduction about the major signaling pathway regulating mRNA turnover, the review is structured in such a way that reflects key functions of the immune cells, including recognition by innate immunity and T cell activation.
THE p38 MAPK PATHWAY AS A UNIVERSAL REGULATOR OF CYTOKINE mRNA TURNOVER
Signaling pathways that tend to stabilize ARE‐mRNAs are multiple and include p38 MAPK, ERK, c‐Jun N‐terminal kinase, PI‐3K‐AKT, and Wnt/β‐catenin pathways [ 10 , 15 , 16 , 17 ]. The p38 MAPK pathway is one of the most common pathways that regulates mRNA stability in multiple cell types. Recent work extends the role of the p38 MAPK pathway and its downstream target kinase, MAPK‐activated protein kinase 2 (MK2) [ 18 , 19 , 20 , 21 , 22 ]. Activation of the p38 MAPK pathway drives the accumulation of TTP via the stabilization of TTP mRNA and protein [ 18 , 19 , 20 ]. Although this remains controversial [ 22 ], MK2‐mediated phosphorylation of TTP has been reported to impair its function, possibly as a result of reduced binding affinity with the TNF‐α ARE region [ 18 , 23 ]. Another p38 MAPK downstream target with structural and functional similarity to MK2 is MK3; it is thought to cooperate with MK2 in TNF‐α mRNA stabilization [ 24 ]. TTP and other RNA decay‐promoting proteins, namely BRF1, KSRP, and AUF1, particularly when phosphorylated, are complexed with 14‐3‐3 proteins that influence their subcellular localization and mRNA decay activity [ 10 , 16 , 25 ]. Another player in the post‐transcriptional regulation of the immune system by the p38 MAPK pathway is the downstream MAPK signal‐integrating kinase, mnk1; this kinase is thought to phosphorylate the eukaryotic initiation factor 4E (eIF4E) and heterogeneous nuclear ribonucleoprotein A1 [ 26 , 27 ].
The p38 MAPK pathway is central to the stabilization of many immune system mRNAs such as those of cytokines during innate and adaptive immune responses. Cytokines can regulate immune response via positive‐ and negative‐feedback circuits. In general, when cytokines amplify a specific immune response, they tend to stabilize mRNAs that participate in such a process. Examples of those cytokines are the proinflammatory cytokines, IL‐1α and TNF‐α, which have been found to stabilize a significant number of cytokines and chemokines, including IFN‐γ‐inducible protein 10 (IP‐10), growth‐related oncogene proteins, and fractalkine (SCYD1), and several genes involved in regulation of inflammatory responses such as TNF‐α‐induced protein 3, NF‐κBαI, and mannose‐binding lectin 2 (MBL2) [ 28 , 29 , 30 ]. Cytokines use p38 MAPK signaling and notably, its target MK2 to trigger mRNA stabilization. For example, using this pathway, IFN‐γ amplifies immune responses such as increasing NK activity and up‐regulating MHC expression [ 31 ]. In several inflammatory conditions, other cytokines can amplify inflammatory responses via post‐transcriptional mechanisms. IL‐17A, in a p38 MAPK‐dependent manner, can increase the stability of TNF‐α‐induced IL‐8 mRNA in human airway smooth muscle, a process that is important in development of asthma [ 32 , 33 ]. IL‐1, in addition to activated platelets, triggers stabilization of cyclooxygenase 2 (COX‐2) mRNA with concomitant cytoplasmic accumulation and increased binding of HuR to the AREs of COX‐2 mRNA in monocytes [ 33 ]. In general, stabilization of the mRNA targets seems to occur after translocation of HuR from the nucleus to the cytoplasm upon cellular activation [ 34 ].
RECOGNITION AND RESPONSE IN INNATE IMMUNITY
Innate immunity is the first line of defense against microbes and requires early and rapid gene expression of effector mediators to control infection effectively. Recognition and effector mechanisms of innate immunity encompass post‐transcriptional gene regulation, which amplifies the much‐needed, early response and enforces the transient response. Efficient recognition of the foreign antigens in microbes, such as bacterial surface molecules and viral dsRNA, is accomplished by cells of the innate immunity, notably macrophages and dendritic cells (DC). Members of the TLR family [ 35 , 36 ], which have gained considerable attention in recent years, are responsible for triggering the efficient response of the host cell to microbes.
Traditionally, post‐transcriptional regulation in innate immunity has been studied in response to the bacterial endotoxin, LPS, which binds CD14 in a complex with TLR4 on the surface of neutrophils and macrophages and initiates a cascade of signals that causes cell activation, the inflammatory response, and phagocytosis [ 35 ]. Recent work gives further insights into LPS‐induced pathways. Upon activation of TLR4 by LPS, the adaptor protein MyD88 is recruited to the receptor and couples with IL‐1 receptor–associated kinase (IRAK)1 and ‐4, causing IRAK activation and recruitment into a complex with TNF receptor‐associated factor 6 (TRAF6). This pathway ultimately culminates in a cascade of signaling events including NF‐κB activation, MAPK activation, and transcriptional activation of proinflammatory cytokines [ 37 ]. Engagement of LPS with TLR4 can itself lead to stabilization of its own mRNA, resulting in a LPS amplification response. Although this has been shown in the case of vascular smooth muscle cells when exposed to LPS [ 38 ], it is likely to occur with cells of the innate immune system. TLR4 activation causes HuR translocation to the cytoplasm, enhanced binding of HuR to the 3′UTR, and increased stability of TLR4 mRNA [ 38 ]. The adaptor molecule MyD88, which transduces a signal from TLR, causes stabilization of cytokine mRNAs, TNF‐α, IL‐8, and IP‐10 in macrophages when stimulated by IFN‐γ [ 29 ]. MyD88 bridges IFN‐γ receptor 1 and mixed linkage kinase 3 domains, leading to p38 MAPK activation [ 31 ]. LPS‐induced TLR4 activation can increase formyl peptide receptor 1 (FPR1) mRNA stability via a PI‐3K pathway in phagocytic leukocytes such as neutrophils and monocytes, leading to infiltration to sites where the invading bacteria possess the formyl peptide ligands [ 39 , 40 ]. Thus, TLR4 activation encompasses several post‐transcriptional, positive regulatory events, which lead to amplification of LPS‐induced signaling and ultimately, production of proinflammatory cytokines ( Fig. 1 ). These cytokines and other mediators, including vascular endothelial growth factor, the solute carrier family 11 member 1, and NO, can be up‐regulated at the level of mRNA stability in LPS‐stimulated, monocytic cells [ 30 , 41 , 42 , 43 , 44 ]. Cytokines such as TNF‐α, IL‐1, IL‐6, and IL‐8 not only contribute to effector mechanisms of innate immunity but also help the activation of adaptive immunity.
Figure 1.
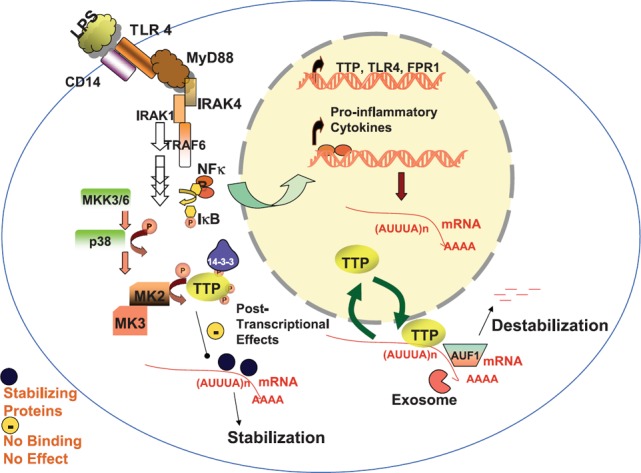
mRNA decay model in innate immune response to endotoxin. LPS initiates a signaling cascade in macrophages, which engages transcriptional induction such as NF‐κB‐mediated gene expression and post‐transcriptional mechanisms, involving activation of the p38 MAPK, central in mRNA stabilization of many of the proinflammatory cytokines. This pathway affects the activity and subcellular localization of several RNA‐binding proteins such as TTP, which promote mRNA decay. Details are given in the text. MKK3/6, MAPK kinases 3/6.
Post‐transcriptional regulation of the inflammatory response as a result of endotoxin also offers a negative‐feedback control, as excessive proinflammatory cytokine production, notably, TNF‐α, can lead to deleterious, local and systemic effects including septic shock. TTP knockout mice having macrophages and neutrophils, which release higher levels of TNF‐α and GM‐CSF, display diseases associated with TNF‐α over secretion such as inflammatory arthritis [ 45 ]. Some of these diseases are similar to mice with genetic knockout of the TNF‐α ARE region [ 46 , 47 ].
Like TTP, AUF1 acts as another negative‐feedback mediator of the innate response to endotoxin; it has been demonstrated recently that LPS challenge in AUF1‐knockout mice displays symptoms of severe endotoxic shock as a result of overproduction of TNF‐α and IL‐1β [ 48 ]. This is attributed to the inability to degrade the mRNAs efficiently as a result of absence of AUF1. Although HuR is an RNA stabilization protein, it can have a negative modulatory function in specific situations. This effect has been shown to suppress specific, proinflammatory mRNA targets such as TNF‐α and COX‐2, using in vivo models with conditional overexpression in macrophage subpopulations [ 49 ]. These three animal models of TTP, AUF1, and HuR offer direct in vivo evidence for the role of post‐transcriptional control in innate immunity.
In addition to LPS, another bacterial component that is an important stimulus of innate immunity is the unmethylated CpG DNA. It binds TLR9 on DC, neutrophils, monocytes, and also, in airway epithelia cells triggering a signal transduction cascade similar to those in response to LPS [ 50 , 51 , 52 ]. Cytokines such as IL‐1α and TNF‐α synergize with CpG DNA to induce IL‐8 production in airway epithelia, mainly by p38 MAPK activation and stability of IL‐8 mRNA [ 51 ]. CpG DNA can augment DC function and maturation using post‐transcriptional mechanisms. For example, CpG DNA increases MHC I mRNA stability in DC by an IFN‐α/β‐dependent process [ 52 ]. HuR binds to a defined, secondary RNA element in the DC mRNA of CD83 and increases its protein expression [ 53 ]. However, this effect does not involve mRNA stabilization, probably as a result of lack of AREs in CD83 3′UTR [ 1 ] but most likely because of enhanced, chromosome region maintenance 1‐mediated translocation of CD83 mRNA to the cytoplasm by HuR [ 53 ]. Mature DC, which express CD83, are efficient in the stimulation of naive CD4+ and CD8+ T cells. So, these post‐transcriptional, regulation events contribute to the efficient maturation of DC—cells, which are important APCs—and further, aid bridging innate immunity to adaptive immunity.
INNATE IMMUNITY TO VIRUS AND dsRNA
Different viruses code for different viral proteins, which can control the cell’s machinery for their growth or to evade immune cells [ 54 ]. Viruses can stabilize mRNAs of gene products that are essential to the virus life cycle. HSV infection can lead to stabilization of the ARE‐mRNA, termed immediate early response 3 (IEX‐1), probably by means of its transregulatory protein ICP27, which is essential in HSV early gene expression [ 55 , 56 ]. The latent membrane protein 1 (LMP1) of EBV is a major player in the induction of cytokines, which in turn, support the growth and survival of the EBV‐infected B cells. LMP1 activates the p38 MAPK pathway and stabilizes IP‐10 [ 57 ]. The Kaposi sarcoma (KS) herpes virus (KSHV or HSV‐8), which causes angioproliferative regions in immunocompromised patients such as AIDS and transplant patients, promotes the production of proinflammatory cytokines. Using a two‐hybrid system, the KSHV latent protein kaposin B has been shown to interact with the c‐lobe of the activation domain of MK2 and requires both of the two proline‐rich, 23‐amino acid direct repeats in kaposin B [ 58 , 59 ]. As a result, MK2 becomes phosphorylated, and loss of destabilization of AU‐rich mRNAs such as GM‐CSF and IL‐6 occurs [ 58 ]. IL‐6 is overexpressed in KS tumors and contributes to its pathogenesis [ 60 ].
Replication of hepatitis C virus (HCV) in cultured liver cell lines leads to prolongation of the IL‐8 mRNA half‐life, probably by its nonstructural protein NS5A, and particularly, in the presence of TNF‐α, a potent inducer of IL‐8 [ 28 ]. A possible mechanism is that HCV, in a similar manner to dsRNA, which is an intermediate viral product, may activate the retinoic acid‐inducible gene I (RIG‐I) pathway, leading to IL‐8 mRNA stabilization. RIG‐I along with melanoma differentiation‐associated gene 5, which has been a focus of research in recent years, are RNA helicases, recognizing viral dsRNA and triggering IFN response [ 61 ]. Two recent studies have shown that RIG‐I can distinguish viral, uncapped ssRNA from cellular mRNAs [ 62 , 63 ]. These proteins and their adaptors have been explored largely for their transcriptional induction of IFN regulatory factor 3‐dependent IFN and NF‐κB‐mediated proinflammatory cytokines when compared with post‐transcriptional effects. Recently, it has been shown that a constitutively active RIG‐I protein, RIG‐N, stabilizes TNF‐α‐induced IL‐8 mRNA and increases reporter activity from a construct that is fused with ARE‐bearing, IL‐8 3′UTR [ 64 ]. IL‐8 is a proinflammatory cytokine, which appears to exaggerate HCV‐induced hepatitis. Thus, HCV‐driven stabilization of IL‐8 mRNA may explain, at least in part, the increased IL‐8 levels seen in the sera of HCV patients and those that are nonresponsive to IFN therapy [ 65 ].
For successful takeover of cell machinery for their own growth, viruses have the ability to shut‐off cellular mRNA biogenesis and induce rapid degradation of selected cellular mRNAs, particularly those required for host defense. For example, the nonstructural protein, nsp1, from the severe acute respiratory syndrome coronavirus, suppresses IFN‐β production at a post‐transcriptional level by increasing its mRNA decay [ 66 ]. Cytomegalovirus is able to destabilize IL‐6 mRNA with concomitant translocation of HuR to the cytoplasmic compartment [ 67 ], although this process is usually known to cause up‐regulation of ARE and ARE‐like‐containing mRNA [ 68 ]. The HSV‐1 open‐reading frame UL41, which encodes virion host shut‐off or vhs, can act as an endoRNase or a component of the RNase complex, degrading cellular mRNAs such IEX‐1 mRNA [ 69 ], although this study is in conflict with other reports that HSV‐1 infection leads to IEX‐1 mRNA stabilization [ 55 , 56 ]. This discrepancy may be a result of the presence of alternative forms of transcripts with different regulatory responses owing to inclusion or exclusion of AREs through alternative splicing and polyadenylation [ 70 ].
Can the RNA decay‐promoting activity of the RNA‐binding proteins be harnessed as an antiviral therapeutic strategy? Two recent reports show that this is a possibility [ 9 , 71 ]. KSRP has been tethered to the HIV Type 1 Rev protein, essential in the HIV life cycle, and binds a specific response element in HIV RNA. The KSRP‐Rev is able to bind to HIV RNA and triggers its degradation, leading to a dramatic reduction in HIV replication [ 9 ]. Likewise, TTP can be used in the same manner, probably with an additional advantage: TTP binds directly to the AU‐rich HIV‐1 RNA and enhances multiple splicing, which leads to a reduction of HIV‐1 virions [ 71 ].
T CELL ACTIVATION
Activation of lymphocytes is fundamental to the immune response to antigens and requires two main signals. One signal is provided upon the engagement of TcR with the antigen presented with MHC on APCs. The other is delivered by costimulatory molecules, such as lymphocyte function‐associated protein 1 (LFA‐1), CD2, and CD28 on T cells, when they interact with cognate receptors on the APCs. With these two signals, the result is AP‐1 (c‐fos/Jun) induction and commitment production of IL‐2, the hallmark of T cell activation and differentiation [ 72 ]. Post‐transcriptional control plays an important regulatory role in almost all of the events that lead to transcriptional activation ( Fig. 2 ). A large proportion of genes is regulated by mRNA stability, as revealed with the large‐scale, gene‐expression profiling in activated T cells [ 73 , 74 ]. One of the most important signaling events in T cell activation that results in stabilization of cytokines, such as IL‐2, IFN‐γ, TNF‐α, and GM‐CSF, is induced by the costimulatory CD28 [ 75 ]. This signaling pathway does not only increase the promoter activity of the cytokines [ 76 ] but also stabilizes their mRNAs [ 75 ]. The increased IL‐2 mRNA stability is thought to be controlled differently and independent of the PI‐3K‐dependent induction of the IL‐2 promoter [ 77 , 78 ]. Unlike IL‐2 transcription, CD28‐induced stabilization of the IL‐2 mRNA and possibly other cytokine mRNAs does not require membrane colocalization with the TcR within CD28 in a multimolecular signaling complex, the central supramolecular activation cluster [ 72 , 77 ]. The role of TTP in IL‐2 production during T cell activation has been studied in detail [ 79 ]. The TcR/CD28‐engaged or ‐immobilized, anti‐CD3‐induced T cells derived from TTP knockout mice have elevated IL‐2 levels as a result of increased stabilization of the IL‐2 mRNA [ 79 ]. TTP is able to bind 32‐base AREs from the IL‐2 3′UTR and destabilizes reporter mRNA fused with a 3′UTR, which contains the IL‐2 ARE [ 79 ]. Like CD28, when LFA‐1 on T cells binds to ICAM‐1 on APCs, allowing strong adhesion between the two cells and promoting efficient T cell activation [ 80 ], it stabilizes certain cytokine mRNAs [ 81 ].
Figure 2.
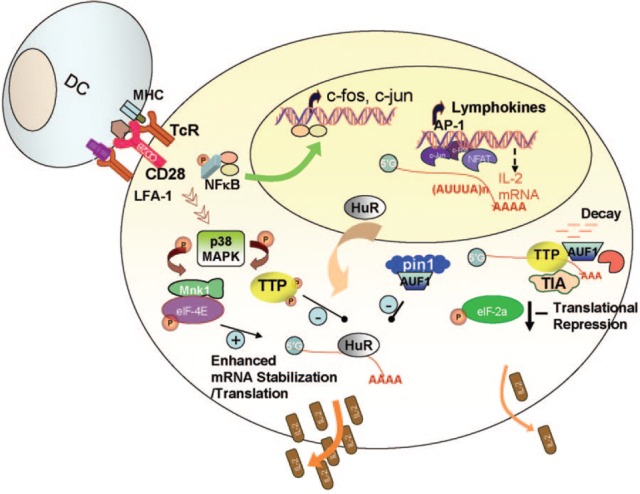
Enhanced mRNA stabilization and translation in T cell activation. APCs, like DC, present antigens efficiently within MHC to T cells through interaction with TcR. Facilitated by other costimulatory molecules, T cell activation occurs, resulting in induction of immediate early response gene products such as c‐fos and c‐jun. These, as AP‐1 complex, with NFAT activate the IL‐2 promoter. Costimulatory molecules, such as CD28 and LFA‐1 on T cells, transduce signals, which also trigger mRNA stabilization and other post‐transcriptional effects. For example, HuR translocates from the nucleus to the cytoplasm and stabilizes ARE‐mRNAs, and pin1 isomerizes another RNA‐binding protein, AUF1, leading to loss of its mRNA decay‐promoting function. These events contribute to early and rapid response of T cell activation and cytokine production. In later phase, shut‐off also can be facilitated by post‐transcriptional mechanisms including mRNA destabilization and translational arrest. Details are given in the text. pin1, Peptidyl‐prolyl isomerase; TIA, T cell intracellular antigen.
Although it has been reported that HuR translocation occurs in mouse T cells upon CD28‐induced signaling, without an effect on IL‐2 mRNA stabilization [ 82 ], LFA‐1 or CD28 engagement of human‐stimulated T cells promoted HuR translocation to the cytoplasm and stabilized TNF‐α and GM‐CSF [ 81 ]. This discrepancy may be a result of the species source of the lymphocytes used in the two studies. AREs are evolutionally conserved in mammalian, but the numbers of ARE pentamers may differ slightly among species, allowing differential responses [ 70 ].
A novel pathway in cytokine mRNA stabilization in T cell activation and differentiation, also occurring during eosinophils activation, has been described recently involving pin1 [ 83 , 84 ]. This protein isomerizes specific peptide bonds and thus, alters protein folding and activity; when it becomes activated in TcR/CD28‐stimulated CD4 T cells, it associates concurrently with AUF1 and HuR. This results in stabilization of GM‐CSF mRNA as a result of the loss of the mRNA decay‐promoting activity of AUF1, which is linked to the exosome (Fig. 2) . AUF1 may be isomerized by pin1, allowing the stabilizing action of HuR to predominate [ 83 ]. AUF1 is a nucleocytoplasmic‐shuttling protein and is associated with stabilization and destabilization activities; this appears to be dependent on the AUF1 isoform generated from four alternative‐spliced mRNAs and the cell type in question. For example, when AUF1 p37 and p42 isoforms are overexpressed, they bind to IL‐6 3′UTR, facilitated by a putative RNA stem‐loop structure; this process promotes the stabilization of IL‐6 mRNA [ 85 ].
TTP plays a significant role in regulating mRNA stability of cytokines such as IL‐2 during T cell activation. Thus, one can envision that for CD28‐ or LFA‐1‐induced stabilization to occur following T cell activation, TTP destabilization action must cease. This can happen by either or both of two scenarios: coordinated stabilization‐destabilization kinetics and TTP phosphorylation (Fig. 2) . With the coordinated kinetics model, stabilizing RNA‐binding proteins such as HuR can occur initially following immune cell activation, allowing rapid and early response of cytokine production. This is followed by the action of mRNA‐destabilizing proteins such as TTP, resulting in transient cytokine production. HuR translocation following TcR/CD28 engagement is seen, first within 1 h, and TTP is induced in activated T cells in a later time [ 86 ]. There is also the potential for interesting reciprocal regulation, which helps to amplify this “cycle”; HuR may stabilize TTP mRNA itself, thus helping to prime TTP for its destabilizing action on cytokine mRNAs. TTP has also been suggested to regulate the stability of its own mRNA in different cell types [ 20 , 21 ], although this has not been proved conclusively [ 4 ]; it is also possible that this regulation extends to activated T cells. The second scenario is that TTP becomes transiently phosphorylated by one of the kinases following CD28 or LFA‐1 engagement. The ultimate outcome is a transient immune response and controlled cytokine production.
During restimulation of Th2 cells, as opposed to priming of naïve T cells with APC, rapid dephosphorylation of eIF2α occurs and results in derepression of translational arrest of cytokine mRNAs such as IL‐4 mRNA [ 87 ]. This process involves translation repressor proteins, TIA‐1, and TIA‐1‐related protein, which operate during priming and restrict T cell function by recruiting untranslated mRNAs into stress granules, discrete cytoplasmic foci triggered by eIF2α phosphorylation [ 88 ]. Also, the p38 activity, which leads to IL‐4 mRNA stabilization, modulates differentiation of naive precursor T cells to Th2 direction [ 89 ].
NEGATIVE REGULATION OF THE IMMUNE SYSTEM AND ROLE OF microRNA (miRNA)
Post‐transcriptional control plays a significant part in specific pathways that regulate T cell activation and involves several types of modulators, including suppressive cytokines, miRNA, and adaptors. When cytokines such as IFN‐α/β, IL‐4, and IL‐10 dampen the immune response, they tend to destabilize mRNAs involved. IFN such as IFN‐β or ‐γ, when in combination with LPS, induces TTP transcription and thereby reduces proinflammatory cytokines, namely, TNF‐α and IL‐6 and the chemokines CCL2 (MCP‐1) and CCL3 (MIP‐1α) [ 90 ]. This happens in a p38 MAPK‐dependent manner, as revealed with TTP‐knockout mouse embryo fibroblasts [ 90 ]. The TTP transcription appears to require a functional, IFN‐γ‐activated sequence, which recruits STAT1 onto the TTP promoter. As TTP is considered an anti‐inflammatory mediator and suppresses proinflammatory cytokines, it is possible that at least some of the molecules with anti‐inflammatory action induce TTP or its activity. Indeed, glucocorticoids can induce the transcription of TTP in the A549 lung epithelial cell line; small interfering RNA to TTP prevented dexamethasone‐induced reduction of TNF‐α mRNA stability and activity of reporter fused with TNF‐α 3′UTR [ 91 ]. On the contrary, this action may be tissue‐type‐specific, as dexamethasone may inhibit rather than induce TTP in activated macrophages [ 92 ].
IL‐10 is a potent, suppressive cytokine, which can inhibit macrophage action and generation of Th2 lymphocytes. It is capable of inhibiting proinflammatory cytokines and Th1 cytokines, such as TNF‐α, IFN‐γ, IL‐2, IL‐3, and GM‐CSF. IL‐10 destabilizes TNF‐α mRNA and probably others by inhibiting the binding of HuR to the AREs in the 3′UTR; this effect appears also to be dependent on the p38 MAPK pathway [ 93 ]. Another anti‐inflammatory cytokine, IL‐4, can suppress FPR1 mRNA expression via a mechanism, which acts on primary transcripts prior to maturation and depends on the constitutive instability of pre‐existing mRNA [ 94 ].
A possible pathway, which may involve mRNA stability changes as a result of negative regulation of TcR/CD28‐induced T cell activation, is the association between adaptor in lymphocytes of unknown function (ALX) and membrane‐associated adaptor protein (LAX) adaptors. Work with ALX‐deficient knockout mice suggested that the ALX/LAX association may inhibit the p38 MAPK pathway and IL‐2 production [ 95 ]. Specifically, the upstream, regulatory MAPKs, MKK3/6, have been found to be constitutively activated in ALX‐deficient mice [ 95 ]. As p38 MAPK is involved in IL‐2 mRNA stabilization, it is possible that negative regulation by ALX occurs via a post‐transcriptional mechanism as well.
Negative regulation of T cell activation and cytokine production can also occur by a miRNA‐processing pathway. In general, miRNAs are considered post‐transcriptional, negative regulatory elements, which have the ability to dampen many biological processes including innate and adaptive immunity. A role in the regulation of T cell development has been proposed recently in a few studies [ 96 , 97 , 98 , 99 ]. Specific deletion of the dicer‐coding gene, dcr‐1, in mouse T cell lineage resulted in aberrant Th cell differentiation and failure to suppress IFN‐γ expression under Th2‐polarizing conditions [ 98 ]. Although no mechanism is discussed in this study, it is possible that miRNA targets the IFN‐γ mRNA, which harbors an ARE, which may in turn be a target for miR16 by a process in which TTP promotes mRNA decay. Involvement of miRNA in ARE‐mediated instability has been documented by demonstrating that dicer and miR16 are required in epithelial cells [ 100 ]. This TTP involvement would be an attractive option in explaining the IFN‐γ mRNA dysregulation in dicer‐deficient Th cells. Another important role of dicer in T cell development has been shown to occur in the development of regulatory T cells (Treg); absence of dicer leads to a reduced number of Treg and immunopathological disorders such as splenomegaly [ 97 ]. This study concludes that dicer facilitates the development of Treg in the thymus and the efficient induction of the necessary forkhead transcription factor, Foxp3, by TGF‐β in naive CD4 T cells [ 97 ]. Several miRNAs, miR146a/b, miR132, and miR155, are LPS‐responsive genes in human monocytes [ 99 , 101 ]. The miR155 is inducible in a human primary macrophage in response to different types of inflammatory mediators [ 101 ], but its effects on the immune cell function remain to be elucidated. The miR146a/b bind sequences in the 3′UTRs of two important adaptor molecules involved in LPS signaling, TRAF6 and IRAK1, and these UTRs inhibit expression of reporter constructs [ 99 ].
The role of miRNA silencing machinery in not limited to negative control of the immune system as described in the above examples but itself, has a direct innate immunity function. Drosha and dicer can act as intracellular, antiviral enzymes against HIV human monocytic cells, and the HIV Tat can act as a suppressor of the host RNA silencing [ 102 , 103 ].
OVERVIEW AND FINAL REMARKS
Although many recent studies emphasize protein–protein and the adaptor interactions on signaling events, which lead to transcriptional activation, it is expected that more studies will focus on post‐transcriptional mechanisms. The array of intracellular RNA‐protein interactions is manifested by the large number of ARE‐mRNAs and RNA‐binding proteins. Given that many mRNAs of the immune response contain AREs, and multiple signaling pathways are operative such as p38 MAPK and PI‐3K/AKT, one can imagine the vast probability of post‐transcriptional interactions in the immune system. Additional types of post‐transcriptional mechanisms, such as alternative splicing and post‐translational regulatory events, although not within the focus of this review, also contribute to the intricate and complex regulation of the immune system.
As cities cope with the rapid entry of people by using rapid transit systems, post‐transcriptional control of immune system helps in rapid activation of immune cells by mechanisms that involve rapid translocation of mRNA stabilization proteins such as HuR to effector cellular compartments and/or temporal inactivation, e.g., by phosphorylation or proteolysis of RNA decay proteins such as TTP or KSRP. Also, when people move out quickly from the transits at the point of exit, mechanisms in the immune system ensure a swift shut‐off and recovery. This happens by the reversal of events that led to mRNA stabilization, activation of decay‐promoting proteins, and translational arrest ( Fig. 3 ). These temporal and spatial events culminate in a transiently regulated immune system, which is able to cope with external and internal insults to the body without deleterious effects on the host. Specific abnormal situations may arise if the transient response of the immune system becomes prolonged, such as those that occur in chronic inflammatory and autoimmune diseases.
Figure 3.
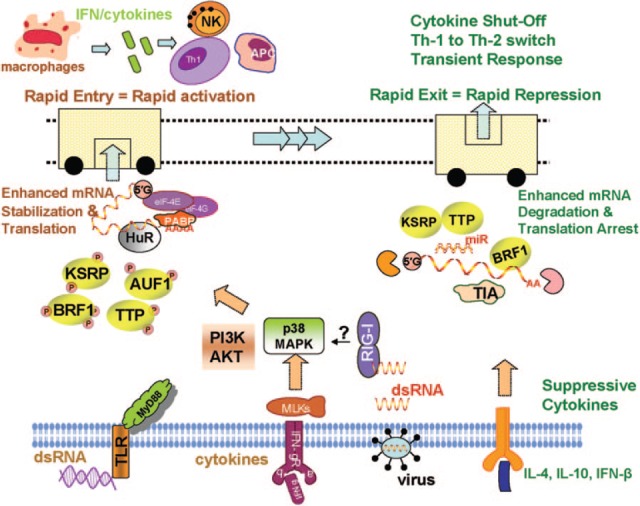
Rapid transit analogy with rapid activation and repression of gene expression. Receptor‐mediated signaling events following recognition of various types of foreign bodies or following interactions with positive and negative modulatory cytokines lead to transcriptional and post‐transcriptional activities. Rapid activation of immune cells and early response events such as cytokine production mimic rapid entry to transit systems, culminating in amplification of innate and adaptive responses, whereas rapid exit from the transit mimics the shut‐off mechanisms in the immune cells that lead to transient response. This is orchestrated by an array of RNA decay mechanisms and a vast number of instability‐prone mRNAs. PABP, Poly(A)‐binding protein; MLKs, mixed lineage kinases.
ACKNOWLEDGMENTS
The Program of BioMolecular Research is supported by the King Faisal Specialist Hospital and Research Center, King Abdelaziz City for Science and Technology, and U.S. National Institutes of Health. The author thanks Drs. Paul Bohjanen (University of Minnesota, Minneapolis, MN, USA) and Andy Clark (Imperial College, London, UK) for critical reading and comments about the manuscript.
REFERENCES
- 1. Bakheet, T. , Williams, B. R. , Khabar, K. S. (2006) ARED 3.0: the large and diverse AU‐rich transcriptome. Nucleic Acids Res. 34, D111–D114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barreau, C. , Paillard, L. , Osborne, H. B. (2005) AU‐rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 33, 7138–7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lopez de Silanes, I. , Galban, S. , Martindale, J. L. , Yang, X. , Mazan‐Mamczarz, K. , Indig, F. E. , Falco, G. , Zhan, M. , Gorospe, M. (2005) Identification and functional outcome of mRNAs associated with RNA‐binding protein TIA‐1. Mol. Cell. Biol. 25, 9520–9531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lai, W. S. , Parker, J. S. , Grissom, S. F. , Stumpo, D. J. , Blackshear, P. J. (2006) Novel mRNA targets for tristetraprolin (TTP) identified by global analysis of stabilized transcripts in TTP‐deficient fibroblasts. Mol. Cell. Biol. 26, 9196–9208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lal, A. , Mazan‐Mamczarz, K. , Kawai, T. , Yang, X. , Martindale, J. L. , Gorospe, M. (2004) Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs. EMBO J. 23, 3092–3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dean, J. L. , Wait, R. , Mahtani, K. R. , Sully, G. , Clark, A. R. , Saklatvala, J. (2001) The 3’ untranslated region of tumor necrosis factor α mRNA is a target of the mRNA‐stabilizing factor HuR. Mol. Cell. Biol. 21, 721–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Winzen, R. , Gowrishankar, G. , Bollig, F. , Redich, N. , Resch, K. , Holtmann, H. (2004) Distinct domains of AU‐rich elements exert different functions in mRNA destabilization and stabilization by p38 mitogen‐activated protein kinase or HuR. Mol. Cell. Biol. 24, 4835–4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tran, H. , Maurer, F. , Nagamine, Y. (2003) Stabilization of urokinase and urokinase receptor mRNAs by HuR is linked to its cytoplasmic accumulation induced by activated mitogen‐activated protein kinase‐activated protein kinase 2. Mol. Cell. Biol. 23, 7177–7188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chou, C. F. , Mulky, A. , Maitra, S. , Lin, W. J. , Gherzi, R. , Kappes, J. , Chen, C. Y. (2006) Tethering KSRP, a decay‐promoting AU‐rich element‐binding protein, to mRNAs elicits mRNA decay. Mol. Cell. Biol. 26, 3695–3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schmidlin, M. , Lu, M. , Leuenberger, S. A. , Stoecklin, G. , Mallaun, M. , Gross, B. , Gherzi, R. , Hess, D. , Hemmings, B. A. , Moroni, C. (2004) The ARE‐dependent mRNA‐destabilizing activity of BRF1 is regulated by protein kinase B. EMBO J. 23, 4760–4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Houseley, J. , LaCava, J. , Tollervey, D. (2006) RNA‐quality control by the exosome. Nat. Rev. Mol. Cell Biol. 7, 529–539. [DOI] [PubMed] [Google Scholar]
- 12. Fenger‐Gron, M. , Fillman, C. , Norrild, B. , Lykke‐Andersen, J. (2005) Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol. Cell 20, 905–915. [DOI] [PubMed] [Google Scholar]
- 13. Lykke‐Andersen, J. , Wagner, E. (2005) Recruitment and activation of mRNA decay enzymes by two ARE‐mediated decay activation domains in the proteins TTP and BRF‐1. Genes Dev. 19, 351–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Stoecklin, G. , Mayo, T. , Anderson, P. (2006) ARE‐mRNA degradation requires the 5′‐3’ decay pathway. EMBO Rep. 7, 72–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gaestel, M. (2006) MAPKAP kinases—MKs—two's company, three's a crowd. Nat. Rev. Mol. Cell Biol. 7, 120–130. [DOI] [PubMed] [Google Scholar]
- 16. Gherzi, R. , Trabucchi, M. , Ponassi, M. , Ruggiero, T. , Corte, G. , Moroni, C. , Chen, C. Y. , Khabar, K. S. , Andersen, J. S. , Briata, P. (2006) The RNA‐binding protein KSRP promotes decay of β‐catenin mRNA and is inactivated by PI3K‐AKT signaling. PLoS Biol. 5, e5. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17. Frevel, M. A. , Bakheet, T. , Silva, A. M. , Hissong, J. G. , Khabar, K. S. , Williams, B. R. (2003) p38 Mitogen‐activated protein kinase‐dependent and ‐independent signaling of mRNA stability of AU‐rich elementcontaining transcripts. Mol. Cell. Biol. 23, 425–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hitti, E. , Iakovleva, T. , Brook, M. , Deppenmeier, S. , Gruber, A. D. , Radzioch, D. , Clark, A. R. , Blackshear, P. J. , Kotlyarov, A. , Gaestel, M. (2006) Mitogen‐activated protein kinase‐activated protein kinase 2 regulates tumor necrosis factor mRNA stability and translation mainly by altering tristetraprolin expression, stability, and binding to adenine/uridine‐rich element. Mol. Cell. Biol. 26, 2399–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Brook, M. , Tchen, C. R. , Santalucia, T. , McIlrath, J. , Arthur, J. S. , Saklatvala, J. , Clark, A. R. (2006) Posttranslational regulation of tristetraprolin subcellular localization and protein stability by p38 mitogen‐activated protein kinase and extracellular signal‐regulated kinase pathways. Mol. Cell. Biol. 26, 2408–2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tchen, C. R. , Brook, M. , Saklatvala, J. , Clark, A. R. (2004) The stability of tristetraprolin mRNA is regulated by mitogen‐activated protein kinase p38 and by tristetraprolin itself. J. Biol. Chem. 279, 32393–32400. [DOI] [PubMed] [Google Scholar]
- 21. Brooks, S. A. , Connolly, J. E. , Rigby, W. F. (2004) The role of mRNA turnover in the regulation of tristetraprolin expression: evidence for an extracellular signal‐regulated kinase‐specific, AU‐rich element‐dependent, autoregulatory pathway. J. Immunol. 172, 7263–7271. [DOI] [PubMed] [Google Scholar]
- 22. Rigby, W. F. , Roy, K. , Collins, J. , Rigby, S. , Connolly, J. E. , Bloch, D. B. , Brooks, S. A. (2005) Structure/function analysis of tristetraprolin (TTP): p38 stress‐activated protein kinase and lipopolysaccharide stimulation do not alter TTP function. J. Immunol. 174, 7883–7893. [DOI] [PubMed] [Google Scholar]
- 23. Stoecklin, G. , Stubbs, T. , Kedersha, N. , Wax, S. , Rigby, W. F. , Blackwell, T. K. , Anderson, P. (2004) MK2‐induced tristetraprolin:14‐3‐3 complexes prevent stress granule association and ARE‐mRNA decay. EMBO J. 23, 1313–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ronkina, N. , Kotlyarov, A. , Dittrich‐Breiholz, O. , Kracht, M. , Hitti, E. , Milarski, K. , Askew, R. , Marusic, S. , Lin, L. L. , Gaestel, M. , Telliez, J. B. (2007) The mitogen‐activated protein kinase (MAPK)‐activated protein kinases MK2 and MK3 cooperate in stimulation of tumor necrosis factor biosynthesis and stabilization of p38 MAPK. Mol. Cell. Biol. 27, 170–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. He, C. , Schneider, R. (2006) 14‐3‐3σ is a p37 AUF1‐binding protein that facilitates AUF1 transport and AU‐rich mRNA decay. EMBO J. 25, 3823–3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Andersson, K. , Sundler, R. (2006) Posttranscriptional regulation of TNFα expression via eukaryotic initiation factor 4E (eIF4E) phosphorylation in mouse macrophages. Cytokine 33, 52–57. [DOI] [PubMed] [Google Scholar]
- 27. Buxade, M. , Parra, J. L. , Rousseau, S. , Shpiro, N. , Marquez, R. , Morrice, N. , Bain, J. , Espel, E. , Proud, C. G. (2005) The Mnks are novel components in the control of TNF α biosynthesis and phosphorylate and regulate hnRNP A1. Immunity 23, 177–189. [DOI] [PubMed] [Google Scholar]
- 28. Green, J. , Khabar, K. S. , Koo, B. C. , Williams, B. R. , Polyak, S. J. (2006) Stability of CXCL‐8 and related AU‐rich mRNAs in the context of hepatitis C virus replication in vitro. J. Infect. Dis. 193, 802–811. [DOI] [PubMed] [Google Scholar]
- 29. Datta, S. , Novotny, M. , Li, X. , Tebo, J. , Hamilton, T. A. (2004) Toll IL‐1 receptors differ in their ability to promote the stabilization of adenosine and uridine‐rich elements containing mRNA. J. Immunol. 173, 2755–2761. [DOI] [PubMed] [Google Scholar]
- 30. Novotny, M. , Datta, S. , Biswas, R. , Hamilton, T. (2005) Functionally independent AU‐rich sequence motifs regulate KC (CXCL1) mRNA. J. Biol. Chem. 280, 30166–30174. [DOI] [PubMed] [Google Scholar]
- 31. Sun, D. , Ding, A. (2006) MyD88‐mediated stabilization of interferon‐γ‐induced cytokine and chemokine mRNA. Nat. Immunol. 7, 375–381. [DOI] [PubMed] [Google Scholar]
- 32. Henness, S. , van Thoor, E. , Ge, Q. , Armour, C. L. , Hughes, J. M. , Ammit, A. J. (2006) IL‐17A acts via p38 MAPK to increase stability of TNF‐α‐induced IL‐8 mRNA in human ASM. Am. J. Physiol. Lung Cell. Mol. Physiol. 290, L1283–L1290. [DOI] [PubMed] [Google Scholar]
- 33. Dixon, D. A. , Tolley, N. D. , Bemis‐Standoli, K. , Martinez, M. L. , Weyrich, A. S. , Morrow, J. D. , Prescott, S. M. , Zimmerman, G. A. (2006) Expression of COX‐2 in platelet‐monocyte interactions occurs via combinatorial regulation involving adhesion and cytokine signaling. J. Clin. Invest. 116, 2727–2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Atasoy, U. , Watson, J. , Patel, D. , Keene, J. D. (1998) ELAV protein HuA (HuR) can redistribute between nucleus and cytoplasm and is upregulated during serum stimulation and T cell activation. J. Cell Sci. 111, 3145–3156. [DOI] [PubMed] [Google Scholar]
- 35. Beutler, B. (2005) The Toll‐like receptors: analysis by forward genetic methods. Immunogenetics 57, 385–392. [DOI] [PubMed] [Google Scholar]
- 36. Akira, S. , Uematsu, S. , Takeuchi, O. (2006) Pathogen recognition and innate immunity. Cell 124, 783–801. [DOI] [PubMed] [Google Scholar]
- 37. Han, J. (2006) MyD88 beyond Toll. Nat. Immunol. 7, 370–371. [DOI] [PubMed] [Google Scholar]
- 38. Lin, F. Y. , Chen, Y. H. , Lin, Y. W. , Tsai, J. S. , Chen, J. W. , Wang, H. J. , Chen, Y. L. , Li, C. Y. , Lin, S. J. (2006) The role of human antigen R, an RNA‐binding protein, in mediating the stabilization of Toll‐like receptor 4 mRNA induced by endotoxin: a novel mechanism involved in vascular inflammation. Arterioscler. Thromb. Vasc. Biol. 26, 2622–2629. [DOI] [PubMed] [Google Scholar]
- 39. Mandal, P. , Novotny, M. , Hamilton, T. A. (2005) Lipopolysaccharide induces formyl peptide receptor 1 gene expression in macrophages and neutrophils via transcriptional and posttranscriptional mechanisms. J. Immunol. 175, 6085–6091. [DOI] [PubMed] [Google Scholar]
- 40. Mandal, P. , Hamilton, T. (2007) Signaling in lipopolysaccharide‐induced stabilization of formyl peptide receptor 1 mRNA in mouse peritoneal macrophages. J. Immunol. 178, 2542–2548. [DOI] [PubMed] [Google Scholar]
- 41. Xu, Y. Z. , Di Marco, S. , Gallouzi, I. , Rola‐Pleszczynski, M. , Radzioch, D. (2005) RNA‐binding protein HuR is required for stabilization of SLC11A1 mRNA and SLC11A1 protein expression. Mol. Cell. Biol. 25, 8139–8149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chen, Y. L. , Huang, Y. L. , Lin, N. Y. , Chen, H. C. , Chiu, W. C. , Chang, C. J. (2006) Differential regulation of ARE‐mediated TNFα and IL‐1β mRNA stability by lipopolysaccharide in RAW264.7 cells. Biochem. Biophys. Res. Commun. 346, 160–168. [DOI] [PubMed] [Google Scholar]
- 43. Kim, Y. M. , Paik, S. G. (2005) Induction of expression of inducible nitric oxide synthase by Taxol in murine macrophage cells. Biochem. Biophys. Res. Commun. 326, 410–416. [DOI] [PubMed] [Google Scholar]
- 44. Du, M. , Roy, K. M. , Zhong, L. , Shen, Z. , Meyers, H. E. , Nichols, R. C. (2006) VEGF gene expression is regulated post‐transcriptionally in macrophages. FEBS J. 273, 732–745. [DOI] [PubMed] [Google Scholar]
- 45. Carrick, D. M. , Lai, W. S. , Blackshear, P. J. (2004) The tandem CCCH zinc finger protein tristetraprolin and its relevance to cytokine mRNA turnover and arthritis. Arthritis Res. Ther. 6, 248–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Taylor, G. A. , Carballo, E. , Lee, D. M. , Lai, W. S. , Thompson, M. J. , Patel, D. D. , Schenkman, D. I. , Gilkeson, G. S. , Broxmeyer, H. E. , Haynes, B. F. , Blackshear, P. J. (1996) A pathogenetic role for TNF α in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity 4, 445–454. [DOI] [PubMed] [Google Scholar]
- 47. Kontoyiannis, D. , Pasparakis, M. , Pizarro, T. T. , Cominelli, F. , Kollias, G. (1999) Impaired on/off regulation of TNF biosynthesis in mice lacking TNF AU‐rich elements: implications for joint and gut‐associated immunopathologies. Immunity 10, 387–398. [DOI] [PubMed] [Google Scholar]
- 48. Lu, J. Y. , Sadri, N. , Schneider, R. J. (2006) Endotoxic shock in AUF1 knockout mice mediated by failure to degrade proinflammatory cytokine mRNAs. Genes Dev. 20, 3174–3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Katsanou, V. , Papadaki, O. , Milatos, S. , Blackshear, P. J. , Anderson, P. , Kollias, G. , Kontoyiannis, D. L. (2005) HuR as a negative posttranscriptional modulator in inflammation. Mol. Cell 19, 777–789. [DOI] [PubMed] [Google Scholar]
- 50. Alvarez, M. E. , Bass, J. I. , Geffner, J. R. , Calotti, P. X. , Costas, M. , Coso, O. A. , Gamberale, R. , Vermeulen, M. E. , Salamone, G. , Martinez, D. , Tanos, T. , Trevani, A. S. (2006) Neutrophil signaling pathways activated by bacterial DNA stimulation. J. Immunol. 177, 4037–4046. [DOI] [PubMed] [Google Scholar]
- 51. Parilla, N. W. , Hughes, V. S. , Lierl, K. M. , Wong, H. R. , Page, K. (2006) CpG DNA modulates interleukin 1β‐induced interleukin‐8 expression in human bronchial epithelial (16HBE14o‐) cells. Respir. Res. 7, 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kuchtey, J. , Chefalo, P. J. , Gray, R. C. , Ramachandra, L. , Harding, C. V. (2005) Enhancement of dendritic cell antigen cross‐presentation by CpG DNA involves type I IFN and stabilization of class I MHC mRNA. J. Immunol. 175, 2244–2251. [DOI] [PubMed] [Google Scholar]
- 53. Prechtel, A. T. , Chemnitz, J. , Schirmer, S. , Ehlers, C. , Langbein‐Detsch, I. , Stulke, J. , Dabauvalle, M. C. , Kehlenbach, R. H. , Hauber, J. (2006) Expression of CD83 is regulated by HuR via a novel cis‐active coding region RNA element. J. Biol. Chem. 281, 10912–10925. [DOI] [PubMed] [Google Scholar]
- 54. Finlay, B. B. , McFadden, G. (2006) Anti‐immunology: evasion of the host immune system by bacterial and viral pathogens. Cell 124, 767–782. [DOI] [PubMed] [Google Scholar]
- 55. Hsu, W. L. , Saffran, H. A. , Smiley, J. R. (2005) Herpes simplex virus infection stabilizes cellular IEX‐1 mRNA. J. Virol. 79, 4090–4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Corcoran, J. A. , Hsu, W. L. , Smiley, J. R. (2006) Herpes simplex virus ICP27 is required for virus‐induced stabilization of the ARE‐containing IEX‐1 mRNA encoded by the human IER3 gene. J. Virol. 80, 9720–9729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Vockerodt, M. , Pinkert, D. , Smola‐Hess, S. , Michels, A. , Ransohoff, R. M. , Tesch, H. , Kube, D. (2005) The Epstein‐Barr virus oncoprotein latent membrane protein 1 induces expression of the chemokine IP‐10: importance of mRNA half‐life regulation. Int. J. Cancer 114, 598–605. [DOI] [PubMed] [Google Scholar]
- 58. McCormick, C. , Ganem, D. (2005) The kaposin B protein of KSHV activates the p38/MK2 pathway and stabilizes cytokine mRNAs. Science 307, 739–741. [DOI] [PubMed] [Google Scholar]
- 59. McCormick, C. , Ganem, D. (2006) Phosphorylation and function of the kaposin B direct repeats of Kaposi's sarcoma‐associated herpesvirus. J. Virol. 80, 6165–6170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Rezaee, S. A. , Cunningham, C. , Davison, A. J. , Blackbourn, D. J. (2006) Kaposi's sarcoma‐associated herpesvirus immune modulation: an overview. J. Gen. Virol. 87, 1781–1804. [DOI] [PubMed] [Google Scholar]
- 61. Kato, H. , Takeuchi, O. , Sato, S. , Yoneyama, M. , Yamamoto, M. , Matsui, K. , Uematsu, S. , Jung, A. , Kawai, T. , Ishii, K. J. , Yamaguchi, O. , Otsu, K. , Tsujimura, T. , Koh, C. S. , Reis e Sousa, C. , Matsuura, Y. , Fujita, T. , Akira, S. (2006) Differential roles of MDA5 and RIG‐I helicases in the recognition of RNA viruses. Nature 441, 101–105. [DOI] [PubMed] [Google Scholar]
- 62. Hornung, V. , Ellegast, J. , Kim, S. , Brzozka, K. , Jung, A. , Kato, H. , Poeck, H. , Akira, S. , Conzelmann, K. K. , Schlee, M. , Endres, S. , Hartmann, G. (2006) 5′‐Triphosphate RNA is the ligand for RIG‐I. Science 314, 994–997. [DOI] [PubMed] [Google Scholar]
- 63. Pichlmair, A. , Schulz, O. , Tan, C. P. , Naslund, T. I. , Liljestrom, P. , Weber, F. , Reis e Sousa, C. (2006) RIG‐I‐mediated antiviral responses to single‐stranded RNA bearing 5′‐phosphates. Science 314, 997–1001. [DOI] [PubMed] [Google Scholar]
- 64. Wagoner, J. , Austin, M. , Green, J. , Imaizumi, T. , Casola, A. , Brasier, A. , Khabar, K. S. , Wakita, T. , Gale Jr., M. , Polyak, S. J. (2007) Regulation of CXCL‐8 (interleukin‐8) induction by double‐stranded RNA signaling pathways during hepatitis C virus infection. J. Virol. 81, 309–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Polyak, S. J. , Khabar, K. S. , Rezeiq, M. , Gretch, D. R. (2001) Elevated levels of interleukin‐8 in serum are associated with hepatitis C virus infection and resistance to interferon therapy. J. Virol. 75, 6209–6211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kamitani, W. , Narayanan, K. , Huang, C. , Lokugamage, K. , Ikegami, T. , Ito, N. , Kubo, H. , Makino, S. (2006) Severe acute respiratory syndrome coronavirus nsp1 protein suppresses host gene expression by promoting host mRNA degradation. Proc. Natl. Acad. Sci. USA 103, 12885–12890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Gealy, C. , Denson, M. , Humphreys, C. , McSharry, B. , Wilkinson, G. , Caswell, R. (2005) Posttranscriptional suppression of interleukin‐6 production by human cytomegalovirus. J. Virol. 79, 472–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Fan, J. , Heller, N. M. , Gorospe, M. , Atasoy, U. , Stellato, C. (2005) The role of post‐transcriptional regulation in chemokine gene expression in inflammation and allergy. Eur. Respir. J. 26, 933–947. [DOI] [PubMed] [Google Scholar]
- 69. Taddeo, B. , Zhang, W. , Roizman, B. (2006) The U(L)41 protein of herpes simplex virus 1 degrades RNA by endonucleolytic cleavage in absence of other cellular or viral proteins. Proc. Natl. Acad. Sci. USA 103, 2827–2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Khabar, K. S. , Bakheet, T. , Williams, B. R. (2005) AU‐rich transient response transcripts in the human genome: expressed sequence tag clustering and gene discovery approach. Genomics 85, 165–175. [DOI] [PubMed] [Google Scholar]
- 71. Maeda, M. , Sawa, H. , Tobiume, M. , Tokunaga, K. , Hasegawa, H. , Ichinohe, T. , Sata, T. , Moriyama, M. , Hall, W. W. , Kurata, T. , Takahashi, H. (2006) Tristetraprolin inhibits HIV‐1 production by binding to genomic RNA. Microbes Infect. 8, 2647–2656. [DOI] [PubMed] [Google Scholar]
- 72. Jordan, M. S. , Singer, A. L. , Koretzky, G. A. (2003) Adaptors as central mediators of signal transduction in immune cells. Nat. Immunol. 4, 110–116. [DOI] [PubMed] [Google Scholar]
- 73. Raghavan, A. , Dhalla, M. , Bakheet, T. , Ogilvie, R. L. , Vlasova, I. A. , Khabar, K. S. , Williams, B. R. , Bohjanen, P. R. (2004) Patterns of coordinate down‐regulation of ARE‐containing transcripts following immune cell activation. Genomics 84, 1002–1013. [DOI] [PubMed] [Google Scholar]
- 74. Cheadle, C. , Fan, J. , Cho‐Chung, Y. S. , Werner, T. , Ray, J. , Do, L. , Gorospe, M. , Becker, K. G. (2005) Stability regulation of mRNA and the control of gene expression. Ann. N. Y. Acad. Sci. 1058, 196–204. [DOI] [PubMed] [Google Scholar]
- 75. Lindstein, T. , June, C. H. , Ledbetter, J. A. , Stella, G. , Thompson, C. B. (1989) Regulation of lymphokine messenger RNA stability by a surfacemediated T cell activation pathway. Science 244, 339–343. [DOI] [PubMed] [Google Scholar]
- 76. Fraser, J. D. , Irving, B. A. , Crabtree, G. R. , Weiss, A. (1991) Regulation of interleukin‐2 gene enhancer activity by the T cell accessory molecule CD28. Science 251, 313–316. [DOI] [PubMed] [Google Scholar]
- 77. Sanchez‐Lockhart, M. , Miller, J. (2006) Engagement of CD28 outside of the immunological synapse results in up‐regulation of IL‐2 mRNA stability but not IL‐2 transcription. J. Immunol. 176, 4778–4784. [DOI] [PubMed] [Google Scholar]
- 78. Sanchez‐Lockhart, M. , Marin, E. , Graf, B. , Abe, R. , Harada, Y. , Sedwick, C. E. , Miller, J. (2004) Cutting edge: CD28‐mediated transcriptional and posttranscriptional regulation of IL‐2 expression are controlled through different signaling pathways. J. Immunol. 173, 7120–7124. [DOI] [PubMed] [Google Scholar]
- 79. Ogilvie, R. L. , Abelson, M. , Hau, H. H. , Vlasova, I. , Blackshear, P. J. , Bohjanen, P. R. (2005) Tristetraprolin down‐regulates IL‐2 gene expression through AU‐rich element‐mediated mRNA decay. J. Immunol. 174, 953–961. [DOI] [PubMed] [Google Scholar]
- 80. Laudanna, C. (2005) Integrin activation under flow: a local affair. Nat. Immunol. 6, 429–430. [DOI] [PubMed] [Google Scholar]
- 81. Wang, J. G. , Collinge, M. , Ramgolam, V. , Ayalon, O. , Fan, X. C. , Pardi, R. , Bender, J. R. (2006) LFA‐1‐dependent HuR nuclear export and cytokine mRNA stabilization in T cell activation. J. Immunol. 176, 2105–2113. [DOI] [PubMed] [Google Scholar]
- 82. Seko, Y. , Azmi, H. , Fariss, R. , Ragheb, J. A. (2004) Selective cytoplasmic translocation of HuR and site‐specific binding to the interleukin‐2 mRNA are not sufficient for CD28‐mediated stabilization of the mRNA. J. Biol. Chem. 279, 33359–33367. [DOI] [PubMed] [Google Scholar]
- 83. Esnault, S. , Shen, Z. J. , Whitesel, E. , Malter, J. S. (2006) The peptidyl‐prolyl isomerase pin1 regulates granulocyte‐macrophage colony‐stimulating factor mRNA stability in T lymphocytes. J. Immunol. 177, 6999–7006. [DOI] [PubMed] [Google Scholar]
- 84. Shen, Z. J. , Esnault, S. , Malter, J. S. (2005) The peptidyl‐prolyl isomerase Pin1 regulates the stability of granulocyte‐macrophage colony‐stimulating factor mRNA in activated eosinophils. Nat. Immunol. 6, 1280–1287. [DOI] [PubMed] [Google Scholar]
- 85. Paschoud, S. , Dogar, A. M. , Kuntz, C. , Grisoni‐Neupert, B. , Richman, L. , Kuhn, L. C. (2006) Destabilization of interleukin‐6 mRNA requires a putative RNA stem‐loop structure, an AU‐rich element, and the RNA‐binding protein AUF1. Mol. Cell. Biol. 26, 8228–8241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Raghavan, A. , Robison, R. L. , McNabb, J. , Miller, C. R. , Williams, D. A. , Bohjanen, P. R. (2001) HuA and tristetraprolin are induced following T cell activation and display distinct but overlapping RNA binding specificities. J. Biol. Chem. 276, 47958–47965. [DOI] [PubMed] [Google Scholar]
- 87. Scheu, S. , Stetson, D. B. , Reinhardt, R. L. , Leber, J. H. , Mohrs, M. , Locksley, R. M. (2006) Activation of the integrated stress response during T helper cell differentiation. Nat. Immunol. 7, 644–651. [DOI] [PubMed] [Google Scholar]
- 88. Anderson, P. , Kedersha, N. (2006) RNA granules. J. Cell Biol. 172, 803–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Dodeller, F. , Skapenko, A. , Kalden, J. R. , Lipsky, P. E. , Schulze‐Koops, H. (2005) The p38 mitogen‐activated protein kinase regulates effector functions of primary human CD4 T cells. Eur. J. Immunol. 35, 3631–3642. [DOI] [PubMed] [Google Scholar]
- 90. Sauer, I. , Schaljo, B. , Vogl, C. , Gattermeier, I. , Kolbe, T. , Muller, M. , Blackshear, P. J. , Kovarik, P. (2006) Interferons limit inflammatory responses by induction of tristetraprolin. Blood 107, 4790–4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Smoak, K. , Cidlowski, J. A. (2006) Glucocorticoids regulate tristetraprolin synthesis and posttranscriptionally regulate tumor necrosis factor α inflammatory signaling. Mol. Cell. Biol. 26, 9126–9135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Jalonen, U. , Lahti, A. , Korhonen, R. , Kankaanranta, H. , Moilanen, E. (2005) Inhibition of tristetraprolin expression by dexamethasone in activated macrophages. Biochem. Pharmacol. 69, 733–740. [DOI] [PubMed] [Google Scholar]
- 93. Rajasingh, J. , Bord, E. , Luedemann, C. , Asai, J. , Hamada, H. , Thorne, T. , Qin, G. , Goukassian, D. , Zhu, Y. , Losordo, D. W. , Kishore, R. (2006) IL‐10‐induced TNF‐α mRNA destabilization is mediated via IL‐10 suppression of p38 MAP kinase activation and inhibition of HuR expression. FASEB J. 20, 2112–2114. [DOI] [PubMed] [Google Scholar]
- 94. Dai, Y. , Major, J. , Novotny, M. , Hamilton, T. A. (2005) IL‐4 inhibits expression of the formyl peptide receptor gene in mouse peritoneal macrophages. J. Interferon Cytokine Res. 25, 11–19. [DOI] [PubMed] [Google Scholar]
- 95. Perchonock, C. E. , Fernando, M. C. , Quinn III, W. J. , Nguyen, C. T. , Sun, J. , Shapiro, M. J. , Shapiro, V. S. (2006) Negative regulation of interleukin‐2 and p38 mitogen‐activated protein kinase during T‐cell activation by the adaptor ALX. Mol. Cell. Biol. 26, 6005–6015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Cobb, B. S. , Nesterova, T. B. , Thompson, E. , Hertweck, A. , O'Connor, E. , Godwin, J. , Wilson, C. B. , Brockdorff, N. , Fisher, A. G. , Smale, S. T. , Merkenschlager, M. (2005) T cell lineage choice and differentiation in the absence of the RNase III enzyme dicer. J. Exp. Med. 201, 1367–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Cobb, B. S. , Hertweck, A. , Smith, J. , O'Connor, E. , Graf, D. , Cook, T. , Smale, S. T. , Sakaguchi, S. , Livesey, F. J. , Fisher, A. G. , Merkenschlager, M. (2006) A role for dicer in immune regulation. J. Exp. Med. 203, 2519–2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Muljo, S. A. , Ansel, K. M. , Kanellopoulou, C. , Livingston, D. M. , Rao, A. , Rajewsky, K. (2005) Aberrant T cell differentiation in the absence of dicer. J. Exp. Med. 202, 261–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Taganov, K. D. , Boldin, M. P. , Chang, K‐J. , Baltimore, D. (2006) NF‐{κ}B‐dependent induction of microRNA miR‐146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. USA 103, 12481–12486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Jing, Q. , Huang, S. , Guth, S. , Zarubin, T. , Motoyama, A. , Chen, J. , Di Padova, F. , Lin, S. C. , Gram, H. , Han, J. (2005) Involvement of microRNA in AU‐rich element‐mediated mRNA instability. Cell 120, 623–634. [DOI] [PubMed] [Google Scholar]
- 101. O'Connell, R. M. , Taganov, K. D. , Boldin, M. P. , Cheng, G. , Baltimore, D. (2007) MicroRNA‐155 is induced during the macrophage inflammatory response. Proc. Natl. Acad. Sci. USA 104, 1604–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Triboulet, R. , Mari, B. , Lin, Y. L. , Chable‐Bessia, C. , Bennasser, Y. , Lebrigand, K. , Cardinaud, B. , Maurin, T. , Barbry, P. , Baillat, V. , Reynes, J. , Corbeau, B. , Jeang, K. T. , Benkirane, M. (2007) Suppression of microRNA‐silencing pathway by HIV‐1 during virus replication. Science 315, 1579–1582. [DOI] [PubMed] [Google Scholar]
- 103. Contreras, X. , Bennasser, Y. , Chazal, N. , Moreau, M. , Leclerc, C. , Tkaczuk, J. , Bahraoui, E. (2005) Human immunodeficiency virus type 1 Tat protein induces an intracellular calcium increase in human monocytes that requires DHP receptors: involvement in TNF‐α production. Virology 332, 316–328. [DOI] [PubMed] [Google Scholar]


