Abstract
Respiratory virus infections contribute substantially both to hospitalizations of young children, and to morbidity in immunocompromised patients such as those with hematological malignancies. Their rapid and accurate diagnosis is essential to patient management. To evaluate the prospective utility of Seeplex® DPO technology in respiratory virus diagnosis, a panel of 99 respiratory samples positive by real‐time RT‐PCR for one or more viruses was assayed by the Seegene Seeplex® RV12 system. As well as being able to detect all 10 viruses in the real‐time RT‐PCR system with the exception of enteroviruses, RV12 can also distinguish between the two subgroups of RSV and detect two subgroups of coronaviruses. Seven of the nine viruses in common with the RT‐PCR were detected reliably by RV12. Eleven samples RT‐PCR‐positive for Metapneumovirus and five samples positive for influenza B were not detected by RV12. Seegene developed a second‐generation system, RV15, which not only allowed detection of three additional viruses, but also addressed the potential problems with RV12 specificity. To address these concerns, 84 respiratory samples positive for a range of viruses by real‐time PCR were assayed with RV15. The results of this evaluation improved significantly upon those seen with RV12. The high throughput capabilities and potential lower technical requirements afforded by the Seeplex® system may offer an alternative to real‐time RT‐PCR systems. J. Med. Virol. 83:1469–1475, 2011. © 2011 Wiley‐Liss, Inc.
Keywords: diagnosis, evaluation, novel
INTRODUCTION
Timely and accurate diagnosis of respiratory virus infection is essential to patient care in settings as diverse as hematopoietic stem cell transplant recipients [Jalal et al., 2007; Piralla et al., 2009], childhood bronchitis and pneumonia [Shay et al., 2001; Michelow et al., 2004] and the seasonal influenza epidemics implicated in substantial annual mortality [Mook et al., 2008]. As such infections often present with similar symptoms, laboratory analysis is essential.
Many laboratories rely still on non‐molecular approaches such as direct immunofluorescence and viral culture. However, in the UK, various real‐time PCR‐based systems have been employed for a number of years, their utility having been demonstrated largely by comparison with these more traditional methods [Templeton et al., 2004; Bustin and Mueller, 2005; Freymuth et al., 2006; Mahony et al., 2007; van de Pol et al., 2007; Kuypers et al., 2009; reviewed in Mahony, 2008]. Emerging technologies thus have to demonstrate significant benefit against these existing tools to achieve widespread adoption [Pabbaraju et al., 2008; Arens et al., 2010].
The novel Seeplex® system exploits Dual Priming Oligonucleotides (DPOs) as PCR primers, each comprising a standard primer‐length 5′ portion which initiates priming, separated by multiple inosine residues from a shorter, 6–12 nucleotide 3′ component which determines duplex extension. As well as facilitating melting temperature regulation during primer design, mispriming and PCR artifacts such as primer dimerization are almost eliminated, allowing simultaneous and efficient amplification of multiple complex targets in single reactions [Chun et al., 2007].
The system summary in Figure 1 shows a standard nucleic acid extraction, a cDNA step, and then a DPO PCR, in which the products of each target virus have different nucleotide lengths. Post‐PCR product analysis employs an automated bench‐top system with which target detection interpretations are inferred from PCR product sizes.
Figure 1.
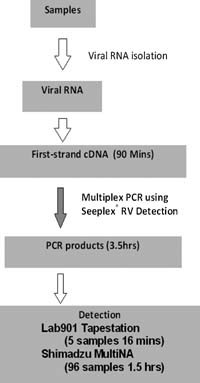
Workflow diagram of the Seeplex® diagnostic system.
The RV12 system enables simultaneous detection of 12 respiratory viruses in two reactions per sample. Its performance when compared to traditional diagnostic methods has been demonstrated frequently [Yoo et al., 2007; Drews et al., 2008; Roh et al., 2008; Kim et al., 2009], with potential time and resource savings when compared to more modern methods, particularly in high throughput settings where prompt diagnosis is important. More recently, the RV15 kit has been developed that targets 15 viruses in three reactions.
No studies evaluating either system against real‐time PCR‐based respiratory virus detection methods have yet been published. The results of a comparative evaluation of serial iterations of the Seeplex® system with the multiplex PCR‐based method that has been used for 5 years as the primary diagnostic assay for respiratory virus detection in a major London hospital are described.
MATERIALS AND METHODS
Samples were nasopharyngeal aspirates, bronchio‐alveolar lavages, or respiratory swabs (nose and/or throat) submitted to the laboratory for routine testing. Aspirates and lavages were diluted in saline and centrifuged to separate cellular material from mucoid and liquid components. Swabs were transported in viral transport medium and extracted directly. Two automated nucleic acid extractors were employed at different times during the course of this evaluation—the NucliSENS easyMAG platform (Biomérieux, Marcy l'Etoile, France) and the QIAamp One‐For‐All Nucleic Acid Kits on the Qiagen BioRobot MDx DSP (Qiagen, Crawley, UK). Immediately prior to extraction, samples were supplemented with a fixed quantity of phocine distemper virus (PDV), obtained from cell culture supernatant and stored in lots at −80°C.
Additionally, all samples that underwent easyMAG extraction were pre‐incubated at 56°C for 10 min with Buffer AL and 30 mAU/ml proteinase K (both Qiagen). Nucleic acid extracts were stored at −80°C between extraction and analysis.
Real‐Time RT‐PCR
Each sample extract was subjected to seven simultaneous real‐time RT‐PCRs, with each reaction detecting between one and three targets (Table I). The seventh reaction constituted a duplex control reaction, with primers and probes targeting both a human gene from the ras family (K‐Ras) [Hilbe et al., 2003] and Protein H (hemagglutinin) of the PDV internal control RNA.
Table I.
Distribution of Primer and Probe Combinations and Probe Dye‐Labels Within Each of the Seven Real‐Time RT‐PCR Reactions, That Is, Reaction 4 Had Adenovirus and Enterovirus Primers and Probes
| Reaction # | FAM channel | JOE channel | Cy5 channel |
|---|---|---|---|
| 1 | FLA | FLB | |
| 2 | RSV | ||
| 3 | PF2 | PF1 | PF3 |
| 4 | ADV | Enterovirus | |
| 5 | HRV | ||
| 6 | MPNV | ||
| 7 | PDV | K‐Ras |
Mastermixes and samples were dispensed robotically into the Rotorgene Gene‐Disc reaction vessels by a CAS1200 liquid handler (both Qiagen).
Quantitect Multiplex RT‐PCR Mix NoRox (Qiagen), specific primers and probe combinations (Table II, all obtained from Sigma‐Genosys, Gillingham, UK), and 10 µl of nucleic acid extract were mixed in 25 µl reactions. Real‐time cycling, detection, and post‐run analysis were performed on the 72‐well Rotorgene platform (Qiagen) with fixed fluorescence thresholds, allowing direct comparison both between samples and between runs. Positive controls (a panel of plasmids containing the specific PCR target as inserts) for each respiratory virus target were included in each run, and compared to expected threshold cycle (Ct) values. For each sample, Ct values for the PDV and K‐Ras control reactions were compared to expected values before parallel PCR results were considered valid.
Table II.
Primer and Probe Sequences for Each of the 10 Targets Detected by the Real‐Time Respiratory RT‐PCR System
| Virus | Sequence | Final conc. (nM) | |
|---|---|---|---|
| Influenza A (FLA) | Forward | CAAGACCAATCCTGTCACCTCTG | 900 |
| Reverse | TGCATTTTGGACAAAGCGTCTAC | 900 | |
| Probe | FAM‐AGTCCTCGCTCACTGGGCACGGT‐BHQ1 | 200 | |
| Influenza B (FLB) | Forward | GGCAACACCTGTTTCCATATTAAG | 900 |
| Reverse | GCCTGTTAATGCAGACAAAACTC | 900 | |
| Probe | JOE‐TCATAGGCAGCTCCGAAGCAAGACATGA‐BHQ1 | 200 | |
| Respiratory syncytial virus (RSV) | Forward | GAAGGGTCAAACATCTGTTTAACAAG | 900 |
| Reverse 1 | GTTTCAGCTWGTGGGAAGAAAGATA | 900 | |
| Reverse 2 | GTGTCAGCCTGTGGAAAGAAGGATA | 900 | |
| Probe | FAM‐TGATCCTGCATTRTCACARTACCATCCTCT‐BHQ1 | 200 | |
| Parainfluenza 1 (PF1) | Forward | ATTCAGACAGGATGGAACCGTTAA | 900 |
| Reverse | GATACTAAGCTTTGTTGTGACCTCAT | 900 | |
| Probe | JOE‐ACCAATGCCTTCAACTGTGTCTCCCGTG‐BHQ1 | 200 | |
| Parainfluenza 2 (PF2) | Forward | AGAGATAACAGGGTTTGAGAATAATTCAT | 900 |
| Reverse | CAAATGGAGTTTGGTGATTAAGGGTA | 900 | |
| Probe | FAM‐TCCAGATGCTCGATCAACTATGTCCAGAGG‐BHQ1 | 200 | |
| Parainfluenza 3 (PF3) | Forward | CGATTAGAGGCTTTCAGACAAGATG | 900 |
| Reverse | CTGTTGAGACCGCATGATTGAC | 900 | |
| Probe | Cy5‐CCACTGTGTCACCGCTCAATACCAGCC‐BHQ3 | 200 | |
| Enterovirus | Forward | CCCCTGAATGCGGCTAATCC | 900 |
| Reverse | GTCACCATAAGCAGCCAATATAAGAA | 900 | |
| Probe | AACACGGACACCCAAAGTAGTCGGTTCC | 200 | |
| Adenovirus (ADV) | Forward | GCCCCAGTGGTCTTACATGCACATC | 900 |
| Reverse | GCCACGGTGGGGTTTCTAAACTT | 900 | |
| Probe | FAM‐TGCACCAGACCCGGGCTCAGGTACTCCGA‐BHQ1 | 200 | |
| Rhinovirus (HRV) | Forward 1 | TGTGCTCRCTTTGAGTCCTC | 900 |
| Forward 2 | TGTGCTCAGTGTGCTTCCTC | 900 | |
| Reverse | TGAAACACGGACACCCAAAGTA | 900 | |
| Probe | Cy5‐CCCTGAATGYGGCTAACCTTAAMCCTGC‐BHQ3 | 200 | |
| Metapneumovirus (MPNV) | Forward | CATATAAGCATGCTATATTAAAAGAGTCTC | 500 |
| Reverse | CCTATTTCTGCAGCATATTTGTAATCAG | 250 | |
| Probe | FAM‐TGYAATGATGAGGGTGTCACTGCGGTTG‐BHQ1 | 500 | |
| K‐Ras | Forward | GCCTGCTGAAAATGACTGAATATAAAC | 600 |
| Reverse | TGATTCTGAATTAGCTGTATCGTCAAG | 600 | |
| Probe | JOE‐TGCCTACGCCACAAGCTCCAACTACCA‐BHQ1 | 100 | |
| PDV | Forward | GCGGGTGCCTTTTACAAGAAC | 600 |
| Reverse | CAGAATAAGCAAAATTGATAGGAACCAT | 600 | |
| Probe | FAM‐TCTTTCCTCAACCTCGTCCGTCACAAGT‐BHQ1 | 100 | |
Seeplex® RV12
Ninety‐nine respiratory extracts stored at −80°C were selected that previously had tested positive by RT‐PCR for one or more respiratory virus targets. With the exception of enterovirus and the internal control virus (PDV), which were not detected by RV12, all viral targets in the RT‐PCR were represented (Table III).
Table III.
Characteristics of the 100 Respiratory Extracts Tested by Seeplex RV12
| Single virus samples | n | Ct rangea |
|---|---|---|
| FLA | 9 | 18.5–31.4 |
| FLB | 9 | 22.7–37.6 |
| HRV | 9 | 21.2–38.3 |
| ADV | 9 | 23.4–38.1 |
| MPNV | 8 | 22.4–35.0 |
| RSV | 8 | 25.3–38.2 |
| PF1 | 13 | 23.8–37.9 |
| PF2 | 6 | 25.3–35.3 |
| PF3 | 11 | 25.3–37.5 |
| Dual virus samples | n |
|---|---|
| ADV and HRV | 2 |
| PF1 and ADV | 1 |
| RSV and ADV | 2 |
| RSV and HRV | 1 |
| HRV and MPNV | 4 |
| FLA and MPNV | 2 |
| FLA and RSV | 4 |
| PF3 and MPNV | 1 |
| PF2 and ADV | 1 |
Either single targets (left‐hand two columns of the top part) or two targets (two columns of the bottom part) were detected in each sample by in‐house multiplex RT‐PCR. The target range of RV12 comprises these nine viruses, together with two human coronavirus groups (229E/NL63 and OC43/HKU1). Also, RSV‐A and RSV‐B are detected separately.
Ct ranges are derived from both single and dual virus samples.
Random hexamer‐primed cDNA synthesis products were generated using the Revertaid system (Fermentas, York, UK), according to the manufacturer's instructions, and stored at −20°C until use.
Each cDNA preparation was subjected to the RV12 PCR procedure according to the manufacturer's instructions (Seegene, Seoul, South Korea). Briefly, parallel 20 µl reactions were set up, each containing RV12 mastermix, 8‐MOPS contamination control reagent, and 3 µl of cDNA. One of each pair was supplemented with 4 µl primer mix A, the other with 4 µl primer mix B. Thermal cycling conditions were as follows: 15 min at 94°C, followed by 40 cycles of 94°C for 30 sec, 60°C for 90 sec, and 72°C for 90 sec, followed by a single incubation of 10 min at 72°C. Completed reactions were analyzed using the MultiNA platform (Shimadzu Corporation, Kyoto, Japan), coupled to a dedicated Seegene software template. Analysis output comprised both a qualitative interpretation of presence or absence of each virus target, and a determination of peak intensity.
Seeplex® RV15
Eighty‐four respiratory extracts stored at −80°C testing positive for one or more RT‐PCR virus targets (Table IV) were subjected to cDNA synthesis using the Mu‐LV system (Invitrogen, Paisley, UK), according to the manufacturer's instructions. Reaction products were stored at −20°C until use. Eight samples of the 84 had been part of the previous RV12 analysis.
Table IV.
Characteristics of the 84 Respiratory Extracts Tested by Seeplex RV15
| Single virus samples | n | Ct rangea |
|---|---|---|
| FLA | 10 | 23.6–34.6 |
| FLB | 17* | 18.8–32.5 |
| HRV | 4 | 23.5–36.0 |
| ADV | 10 | 20.4–32.4 |
| MPNV | 7 | 20.6–31.5 |
| RSV | 9 | 23.4–36.4 |
| PF1 | 9 | 22.7–30.6 |
| PF2 | 5* | 20.7–32.7 |
| PF3 | 7 | 23.3–32.6 |
| Dual virus samples | n |
|---|---|
| ADV and PF3 | 1 |
| FLB and HRV | 1 |
| FLB and PF3 | 1 |
| FLA and PF2 | 1 |
| RSV and ADV | 2* |
Either single targets (left‐hand two columns of the top part) or two targets (two columns of the bottom part) were detected in each sample by in‐house multiplex RT‐PCR.
Four FLB, two PF2, and both RSV‐ADV dually infected samples had been tested previously in the RV12 evaluation. All viruses in these repeated samples had been previously detected by the RV12 system. In addition to the 12 targets detected by RV12, RV15 also detects human bocavirus, human parainfluenza 4, and enterovirus.
Ct ranges are derived from both single and dual virus samples.
Each cDNA preparation was subjected to the RV15 PCR procedure according to Seegene's instructions. Briefly, parallel 20 µl reactions were set up for each sample, as per the RV12 kit, but with three reactions per sample (primer mixes A, B, and C) rather than the two in the earlier kit. Thermal cycling conditions were as per the RV12 kit. Completed reactions were analyzed using the Tape Station platform (Lab901, Edinburgh, UK) linked to a specific Seeplex® RV15 interpretation software module. Again, results were presented both qualitatively and semi‐quantitatively, according to the migration rate and band intensity of each species present within completed reactions.
Redesigned Influenza B Primers
During the evaluation, a new panel of influenza B primers was developed by Seegene to replace those in the RV15 kit. The cDNA preparations of the 19 FLB‐positive (by RT‐PCR) samples analyzed by RV15 were subjected to amplification using these primers under the same conditions as for the primer mixes of the RV12 and RV15 kits, but in monoplex reactions. Analysis was performed as per RV15, above.
RESULTS
Seeplex® RV12
All targets detected by the RT‐PCR were also detected by Seeplex® RV12 in 88 of the 99 nucleic acid extracts tested with the Seeplex® RV12 system. In 7 of the remaining 11 samples, single infections were missed (4 × FLB, one each of HRV, PF1, and MPNV), and in four samples where two viruses had been detected by RT‐PCR, the virus target with the higher Ct was missed by RV12 (2 × MPNV, 1 × PF3, and 1 × ADV). Comparison of MultiNA peak heights and RT‐PCR Ct values for each virus target revealed a broad negative correlation between the two parameters, with the exception of HRV, where no correlation was observed (Fig. 2).
Figure 2.
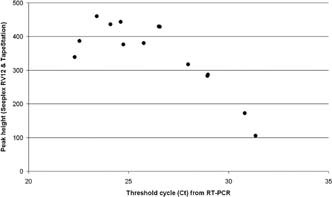
Plot showing how peak heights obtained using Seeplex® RV12 and the TapeStation microelectrophoresis platform display negative correlation with the threshold cycles obtained from the RT‐PCR method. Data shown are from influenza A‐positive samples.
In all instances where PF2 (7 samples), FLA (15), and/or RSV (15) were detected by RT‐PCR, these virus targets were detected with RV12 also. For each instance where PF1, PF3, HRV, and/or ADV were detected by RT‐PCR, all except one of each virus target were detected by Seeplex® RV12. Each instance of undetected virus represented a sample whose Ct was found in the higher part of the range for that virus. FLB and MPNV targets were detected less frequently. Four of the five FLB‐positive samples with the highest Ct values, and the three MPNV‐positive samples with the highest Ct values were not detected by RV12.
Conversely, ADV, RSV, HRV, and MPNV were detected by RV12 in extracts where the RT‐PCR system had failed to do so (in 3, 4, 4, and 1 sample, respectively), and with the exception of the RSV detections, had high peak heights, consistent with high target concentrations in the initial samples.
Seeplex® RV15
Eighty‐four extracts from respiratory samples RT‐PCR‐positive for at least one respiratory virus were tested by RV15. With the exception of ADV and FLB, all viruses detected by RT‐PCR were also detected by RV15, including all instances of MPNV (7/7). Inverse correlations between Ct and RV15 band intensity were less pronounced than that seen with RV12.
Three of nine ADV‐positive samples were not detected by RV15. All four FLB‐positive samples that had also been tested with RV12 remained FLB‐positive by RV15. However, a further five previously untested FLB‐positive samples failed to signal with RV15. Again, viruses were detected by RV15 that were not detected by the RT‐PCR: HRV, enterovirus, coronavirus, and bocavirus (1, 1, 2, and 1 sample, respectively). The RT‐PCR system did not target either coronavirus or bocavirus. Table V details the discrepant results between the RT‐PCR and Seeplex® systems.
Table V.
Summary of the Discrepant Results (Highlighted in Bold Type) Between the In‐House Real‐Time PCR and Seeplex® RV12 (a) and RV15 (b)
| Sample # | PCR result (Ct) | RV12 |
|---|---|---|
| (a) | ||
| 7 | FLB (35.77) | None |
| 29 | FLB (35.70) | None |
| 83 | FLB (32.01) | None |
| 94 | FLB (37.56) | None |
| 60 | HRV (33.54) | None |
| 43 | MPNV (35.32) | None |
| 32 | PF1 (35.21) | RSVb |
| 27 | HRV (29.34), MPNV (35.62) | HRV, ADV |
| 82 | FLA (30.81), MPNV (35.94) | FLA |
| 87 | MPNV (26.57), PF3 (33.92) | MPNV |
| 100 | PF2 (28.5), ADV (38.11) | PF2 |
| 17 | ADV (31.65) | ADV, 229E |
| 18 | ADV (26.85), RSV (32.64) | ADV, RSVa, HRV |
| 39 | RSV (30.24) | RSVa, OC43, ADV |
| 47 | ADV (33.44) | ADV, OC43 |
| 50 | FLA (28.95) | FLA, HRV |
| 55 | HRV (32.69) | HRV, OC43 |
| 66 | HRV (21.16) | HRV, RSVa |
| 70 | FLA (27.98) | FLA, RSVa |
| 71 | HRV (31.81) | HRV, RSVa |
| 90 | PF3 (31.81) | PF3, RSVb |
| 102 | PF2 (29.14) | PF2, HRV |
| Sample # | PCR result (Ct) | RV15 |
|---|---|---|
| (b) | ||
| 156 | FLB (26.50) | None |
| 158 | FLB (30.30) | None |
| 159 | FLB (30.41) | None |
| 160 | FLB (29.46) | None |
| 170 | FLB (31.64) | None |
| 142 | ADV (30.02) | None |
| 143 | ADV (29.76) | None |
| 150 | ADV (23.38) | None |
| 108 | PIV‐2 (26.98), FLA (28.70) | None |
| 182 | ADV (27.20) | HRV, RSV |
| 107 | MPNV (26.92) | MPNV, HRV |
| 134 | RSV (29.86) | RSVb, HEV |
| 165 | FLA (27.03) | FLA, OC43 |
| 176 | FLB (25.13) | FLB, OC43 |
| 183 | RSV (31.96) | RSVb, BocV |
Samples where Seeplex® missed targets detected by PCR are given first.
Redesigned Influenza B Primer Set
Using the new primer set designed to overcome the FLB‐detection limitations seen with RV12 and RV15, all FLB‐positive samples in which FLB had been detected by RV15 were FLB‐positive by the new primer set, as were four of the five samples FLB‐negative by RV15. The single sample where FLB was not detected by Seeplex® had given a Ct of 30 by RT‐PCR. The lack of amplification controls in this assay series precluded detection of PCR inhibition.
DISCUSSION
An in‐house multiplex RT‐PCR system and two iterations of a novel PCR‐based technology for the detection of respiratory viruses have been compared. In the first instance, 99 samples were evaluated, and in the second, 84. A further panel of 19 FLB‐positive samples was tested by a revised primer set within the novel Seeplex® system. Whilst direct comparison of the two systems is limited, due to the sample sets having been pre‐determined to be positive for one or more target viruses by one system, it can be stated that the overall performance of the Seeplex® RV12 and RV15 systems was very good, with several instances of viruses being detected where the RT‐PCR system had either failed or had been unable to do so.
Furthermore, with each virus, some negative correlation was observed between RT‐PCR threshold cycle and Seeplex® peak intensity, suggesting the latter could also be employed semi‐quantitatively.
The frequency of instances where targets detected by RT‐PCR were missed by Seeplex® was low, and may have been influenced by repeated freeze–thaw cycles of affected samples, nearly all of which were at the upper end of the relevant Ct range (i.e., with lower target concentrations). However, both MPNV and FLB targets were detected poorly by RV12, with concomitant low band intensities from samples where these targets were detected. Revision of the primer sets was a key component of the development of RV15. RV15 gave a 100% detection rate of all viruses detected by RT‐PCR, including seven MPNV‐positive samples, but no improvement in FLB detection. The latter primer set was modified further, and retesting the panel of known FLB‐positive samples gave substantially greater success. Although these new primers have since been included in the RV15, further evaluation within a standard Seeplex® multiplex PCR format is required.
A limitation to the Seeplex® system is its internal control facility. The artificial targets included in each PCR mastermix allows validation only of the PCR step, whereas the addition of PDV to each extraction allows the RT‐PCR system also to control for both the RNA extraction and the reverse transcription step. Inhibition of the RT‐PCR system, detected by poorly amplifying PDV target, occurs with a not insignificant frequency; it is essential to report that a lack of virus detection is not due to any technical deficiency within the system.
In the laboratory setting, the Seeplex® system looks to compare favorably with existing technologies where high throughput, ease of use, low technical requirements, and a fully kit‐based format are factors. Equally useful is the ability of the RV15 system to detect bocavirus and coronaviruses, whose clinical impact is becoming increasingly clear [Esposito et al., 2008; Brodzinski and Ruddy, 2009; Garcia‐Garcia et al., 2010].
Since the completion of this work, the Seeplex® RV15 kit now includes the redesigned influenza B primer set, human RNase P whole process control and now comprises a one‐step RT‐PCR system rather than the two‐step systems employed in these evaluations.
In summary, the Seeplex® RV15 produces results comparable to the in‐house system. The high throughput capabilities and low technical requirements afforded by Seeplex® recommend it as a practical alternative to real‐time RT‐PCR.
REFERENCES
- Arens MQ, Buller RS, Rankin A, Mason S, Whetsell A, Agapov E, Lee WM, Storch GA. 2010. Comparison of the Eragen Multi‐Code (R) Respiratory Virus Panel with conventional viral testing and real‐time multiplex PCR assays for respiratory viruses. J Clin Microbiol 48: 2387–2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodzinski H, Ruddy RM. 2009. Review of new and newly discovered respiratory tract viruses in children. Pediatr Emerg Care 25: 352–360. [DOI] [PubMed] [Google Scholar]
- Bustin SA, Mueller R. 2005. Real‐time reverse transcription PCR (qRT‐PCR) and its potential use in clinical diagnosis. Clin Sci (Lond) 109: 365–379. [DOI] [PubMed] [Google Scholar]
- Chun JY, Kim KJ, Hwang IT, Kim YJ, Lee DH, Lee IK, Kim JK. 2007. Dual priming oligonucleotide system for the multiplex detection of respiratory viruses and SNP genotyping of CYP2C19 gene. Nucleic Acids Res 35: e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drews SJ, Blair J, Lombos E, DeLima C, Burton L, Mazzulli T, Low DE. 2008. Use of the Seeplex RV Detection kit for surveillance of respiratory viral outbreaks in Toronto, Ontario, Canada. Ann Clin Lab Sci 38: 376–379. [PubMed] [Google Scholar]
- Esposito S, Bosis S, Niesters HG, Tremolati E, Sabatini C, Porta A, Fossali E, Osterhaus AD, Principi N. 2008. Impact of human bocavirus on children and their families. J Clin Microbiol 46: 1337–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freymuth F, Vabret A, Cuvillon‐Nimal D, Simon S, Dina J, Legrand L, Gouarin S, Petitjean J, Eckart P, Brouard J. 2006. Comparison of multiplex PCR assays and conventional techniques for the diagnostic of respiratory virus infections in children admitted to hospital with an acute respiratory illness. J Med Virol 78: 1498–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia‐Garcia ML, Calvo C, Falcon A, Pozo F, Perez‐Brena P, De Cea JM, Casas I. 2010. Role of emerging respiratory viruses in children with severe acute wheezing. Pediatr Pulmonol 45: 585–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim A, Ebnet C, Harste G, Pring‐Akerblom P. 2003. Rapid and quantitative detection of human adenovirus DNA by real‐time PCR. J Med Virol 70: 228–239. [DOI] [PubMed] [Google Scholar]
- Hilbe W, Dlaska M, Duba HC, Dirnhofer S, Eisterer W, Oberwasserlechner F, Mildner A, Schmid T, Kuhr T, Woll E. 2003. Automated real‐time PCR to determine K‐ras codon 12 mutations in non‐small cell lung cancer: Comparison with immunohistochemistry and clinico‐pathological features. Int J Oncol 23: 1121–1126. [PubMed] [Google Scholar]
- Jalal H, Bibby DF, Bennett J, Sampson RE, Brink NS, MacKinnon S, Tedder RS, Ward KN. 2007. Molecular investigations of an outbreak of parainfluenza virus type 3 and respiratory syncytial virus infections in a hematology unit. J Clin Microbiol 45: 1690–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SR, Ki CS, Lee NY. 2009. Rapid detection and identification of 12 respiratory viruses using a dual priming oligonucleotide system‐based multiplex PCR assay. J Virol Methods 156: 111–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuypers J, Campbell AP, Cent A, Corey L, Boeckh M. 2009. Comparison of conventional and molecular detection of respiratory viruses in hematopoietic cell transplant recipients. Transpl Infect Dis 11: 298–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maertzdorf J, Wang CK, Brown JB, Quinto JD, Chu M, de GM, van den Hoogen BG, Spaete R, Osterhaus AD, Fouchier RA. 2004. Real‐time reverse transcriptase PCR assay for detection of human metapneumoviruses from all known genetic lineages. J Clin Microbiol 42: 981–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahony JB. 2008. Detection of respiratory viruses by molecular methods. Clin Microbiol Rev 21: 716–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahony J, Chong S, Merante F, Yaghoubian S, Sinha T, Lisle C, Janeczko R. 2007. Development of a respiratory virus panel test for detection of twenty human respiratory viruses by use of multiplex PCR and a fluid microbead‐based assay. J Clin Microbiol 45: 2965–2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelow IC, Olsen K, Lozano J, Rollins NK, Duffy LB, Ziegler T, Kauppila J, Leinonen M, McCracken GH, Jr. 2004. Epidemiology and clinical characteristics of community‐acquired pneumonia in hospitalized children. Pediatrics 113: 701–707. [DOI] [PubMed] [Google Scholar]
- Mook P, Pebody R, Zhao H, Ellis J, Zambon M, Fleming DM, Watson JM. 2008. Surveillance of influenza and other respiratory viruses in the United Kingdom: October 2007 to May 2008. Available at http://www.hpa.org.uk/hpr/archives/Infections/2008/respiratory.htm#ann0708.
- Pabbaraju K, Tokaryk KL, Wong S, Fox JD. 2008. Comparison of the Luminex xTAG respiratory viral panel with in‐house nucleic acid amplification tests for diagnosis of respiratory virus infections. J Clin Microbiol 46: 3056–3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piralla A, Percivalle E, Di Cesare‐Merlone A, Locatelli F, Gerna G. 2009. Multicluster nosocomial outbreak of parainfluenza virus type 3 infection in a pediatric oncohematology unit: A phylogenetic study. Haematologica 94: 833–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh KH, Kim J, Nam MH, Yoon S, Lee CK, Lee K, Yoo Y, Kim MJ, Cho Y. 2008. Comparison of the Seeplex reverse transcription PCR assay with the R‐mix viral culture and immunofluorescence techniques for detection of eight respiratory viruses. Ann Clin Lab Sci 38: 41–46. [PubMed] [Google Scholar]
- Shay DK, Holman RC, Roosevelt GE, Clarke MJ, Anderson LJ. 2001. Bronchiolitis‐associated mortality and estimates of respiratory syncytial virus‐associated deaths among US children, 1979–1997. J Infect Dis 183: 16–22. [DOI] [PubMed] [Google Scholar]
- Templeton KE, Scheltinga SA, Beersma MF, Kroes AC, Claas EC. 2004. Rapid and sensitive method using multiplex real‐time PCR for diagnosis of infections by influenza a and influenza B viruses, respiratory syncytial virus, and parainfluenza viruses 1, 2 3, and 4. J Clin Microbiol 42: 1564–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Pol AC, van Loon AM, Wolfs TF, Jansen NJ, Nijhuis M, Breteler EK, Schuurman R, Rossen JW. 2007. Increased detection of respiratory syncytial virus, influenza viruses, parainfluenza viruses, and adenoviruses with real‐time PCR in samples from patients with respiratory symptoms. J Clin Microbiol 45: 2260–2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SJ, Kuak EY, Shin BM. 2007. Detection of 12 respiratory viruses with two‐set multiplex reverse transcriptase‐PCR assay using a dual priming oligonucleotide system. Korean J Lab Med 27: 420–427. [DOI] [PubMed] [Google Scholar]


