Abstract
Respiratory tract infection (RTI) involves a variety of viruses and bacteria, which can be conveniently detected by multiplex nucleic acid amplification testing (NAT). To compare the novel Luminex‐based NxTAG‐Respiratory Pathogen Panel (NxTAG‐RPP) with the routine multiplex‐ligation‐NAT based RespiFinder‐22® (RF‐22), 282 respiratory specimens including nasopharyngeal swabs (71%), broncho‐alveolar lavage (27%), throat swabs, tracheal secretions, and sputum (2%) from 116 children and 155 adults were extracted using a Corbett CAS1200 (Qiagen), and analyzed in parallel by the routine RF‐22 and NxTAG‐RPP. Concordant results were obtained in 263 (93.3%) cases consisting of concordant positives in 167 (59.2%) and concordant negatives in 96 (34%). Results were discordant in 19 (6.7%) consisting of 15 positive:negative, and 4 negative:positive results by NxTAG‐RPP versus RF‐22, respectively. Co‐infections were observed in 10.3% with NxTAG‐RPP and in 5.9% with RF‐22. Most additional viral pathogens identified by the NxTAG‐RPP involved dual infections with rhinovirus and RSV. Discordant samples were mainly due to low genome signals of Ct less than 36, when retested by QNAT suggesting a higher sensitivity of the NxTAG‐RPP, also when detecting multiple infections. Hands‐on time after extraction for 24 and 96 samples was 0.25 and <0.5 hr for the NxTAG‐RPP, and 2 and 4 hr for the RF‐22, respectively. The median turn‐around time was 6 hr (range 5–7 hr) for NxTAG‐RPP and 12 hr (range 8–16 hr) for RF‐22. The NxTAG‐RPP showed comparable detection rates for most respiratory pathogens, while hands‐on and turn‐around time were considerably shorter. The clinical significance of detecting multiple viruses needs further clinical evaluation. J. Med. Virol. 88:1319–1324, 2016 . © 2016 Wiley Periodicals, Inc.
Keywords: respiratory tract infection, respiratory tract infectious disease, nucleic acid testing, multiplex PCR, turn‐around‐time, hands‐on‐time
INTRODUCTION
Respiratory tract infection (RTI) involves a wide variety of different etiologic agents among viruses and bacteria. With increasing availability of specific and sensitive diagnostic assays using nucleic acid amplification testing (NAT), it has become clear that the etiological diagnosis based on signs and symptoms is unreliable, since the clinical presentation of different causative agents is often similar, especially in immunocompromised patients [Ison, 2012; Hirsch et al., 2013]. Of note, this limitation also applies to periods of higher pretest probability as during epidemics, as reported previously [Dumoulin et al., 2009; Grondahl et al., 2014]. Therefore, rapid and correct identification of RTI agents is important for infection control and treatment strategies. Although near‐patient testing is available and particularly important for treating influenza [Khanna et al., 2009; Beckmann and Hirsch, 2015], negative results are not satisfactory, especially for immunocompromised patients [Ison, 2012; Hirsch et al., 2013]. This challenge is most conveniently tackled by NAT for multiple pathogens in parallel. Multiplex NAT platforms are able to detect different viruses and bacteria in a single assay, but are often technically demanding in preparation, processing, and read‐outs, which increase the hands‐on‐time (HOT) and affect the turn‐around‐time (TAT) [Caliendo, 2011]. In 2009, we routinely introduced the RespiFinder platform [Reijans et al., 2008] to identify respiratory pathogens in symptomatic children and adults [Dumoulin et al., 2009; Nickel et al., 2009; Sidler et al., 2012], which detects 22 different pathogens using a multiplex ligation‐dependent probe amplification (MLPA) coupled to capillary electrophoresis (RF‐22). However, the procedure is demanding with considerable HOT and the current TAT of 8–16 hr is limiting its clinical utility, especially in immunocompromised and critically ill patients. Therefore, we compared the RF‐22 with the NxTAG‐respiratory pathogen panel (NxTAG‐RPP), which combines a multiplex RT‐PCR with bead hybridization for detection and identification of 18 viruses and 3 bacteria associated with virus‐like RTI.
MATERIALS AND METHODS
Clinical Specimens
The total of 282 respiratory specimens were obtained from 271 patients (119 females [43.9%] and 152 males [56.1%]; median age, 42 years; interquartile range [IQR] 2–63 years) for analysis by both: RespiFinder‐22® (RF‐22, PathoFinder, Maastricht, The Netherlands) and NxTAG‐RPP (Luminex, MV's‐Hertogenbosch, The Netherlands). Of the 116 pediatric patients (42.8%; median age, 1 year; IQR, 0–4 years), 44 (37.9%) were less than 1‐year‐old. Of the 155 adult patients (57.2%; median age, 61 years; IQR, 47–69 years), 77 (49.7%) were older than 60 years. The specimens analyzed are shown in Figure1A. A first set was tested by RF‐22 and then retrospectively by NxTAG‐RPP, consisting of 115 specimens obtained between September and December 2014. A second set of 167 consecutive samples submitted between March 2015 and April 2015 was analyzed prospectively in parallel by RF‐22 and NxTAG‐RPP. The specimens from adult patients were submitted from the University Hospital, the pediatric specimens from the University Children's Hospital in Basel, Switzerland.
Figure 1.
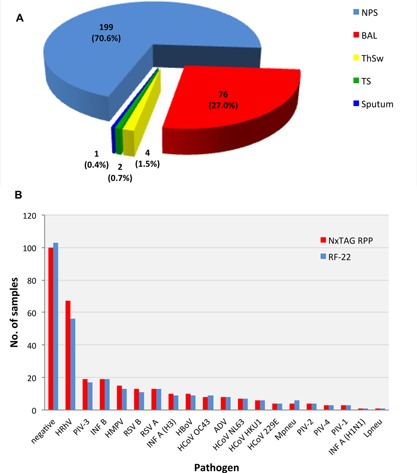
A: Specimen distribution. Distribution of 282 specimens from 116 pediatric and 155 adult patients. Blue, nasopharyngeal swabs (NPS); red, broncho‐alveolar lavage (BAL); yellow, throat swabs (ThSw); green, tracheal secretions (TS); dark blue, sputum. B: Detection rate by NxTAG‐RPP and by RespiFinder‐22. Number of pathogens detected in both cohorts of retrospective and prospective samples (n = 282). ADV, adenovirus; HBoV, human bocavirus; HCoV, human coronavirus; HMPV, human metapneumovirus; HRhV, human rhinovirus; INF, influenza; Lpneu, Legionella pneumophila; Mpneu, Mycoplasma pneumoniae; PIV, parainfluenza virus; RSV, respiratory syncytial virus. Red bars: detection by NxTAG‐RPP; blue bars: detection by RF‐22.
Nucleic Acid Extraction and Multiplex Pathogen Panels
Total nucleic acids were extracted for both assays from 200 µl of the respiratory specimen using the Corbett CAS‐1200 (Qiagen, Hilden, Germany) (TAT: 2 hr). RF‐22 was performed as described by the manufacturer with a HOT depending on the number of samples processed being 2 and 4 hr for 24 and 96 samples, before analysis by capillary electrophoresis yielding a TAT of 8–16 hr [Dumoulin et al., 2009; Beckmann and Hirsch, 2015]. The NxTAG‐RPP is a closed‐tube assay, consisting of a multiplex reverse transcription PCR and hybridization in a single step 96‐well format requiring a HOT of less than 15 min for both 24 or 96 samples, and a TAT of 5 hr. The NxTAG‐RPP results were obtained as mean fluorescence intensity (MFI) measured by the Luminex MAGPIX instrument and a research use only version of the TDAS software. MFI values above the threshold for a particular pathogen indicated a detectable target (positive result). The limit of detection was target dependent and indicated by the manufacturer. RF‐22 served as the primary reference test. Discordant results led to re‐testing using local in‐house quantitative PCRs (QNAT). Three specimens were sent to an external reference laboratory (Drs. C. Tapparell‐Vu, L. Kaiser, University Hospital Geneva).
Statistics
Sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV) were calculated for the NxTAG‐RPP in comparison to RF‐22 using 2 × 2 tables. Agreement between the two methods was assessed by the kappa value test. Kappa values from 0.21 to 0.4 represent fair agreement; from 0.41 to 0.6 moderate agreement; 0.61 to 0.8 and 0.81 to 0.99 indicate substantial and almost perfect agreement, respectively. To evaluate the statistical significance of the difference between NxTAG‐RPP and RF‐22 the two‐tailed McNemar's test was used.
RESULTS
Concordant results were obtained in 263 samples (93.3%) consisting of 167 (59.2%) concordant‐positives and 96 (34%) concordant‐negatives. Discordant results between both assays were seen in 19 (6.7%) consisting of 11 NxTAG‐RPP positive—RF‐22 negative pairs, and 8 NxTAG‐RPP negative—RF‐22 positive pairs. Single infections were found in 154 samples (54.6%). Most frequently detected were rhino‐/enterovirus (23.8%), parainfluenza‐3 (6.7%), and influenza B (6.7%). The clinical specimens consisted of nasopharyngeal swabs in 71% (82% obtained from children), and broncho‐alveolar lavage (BAL) in 27% (Fig. 1A), of which only 2 (0.7%) were obtained from children. The pathogen‐specific detection rate for the two multiplex assays is shown in Figure 1B.
Among the 115 retrospectively analyzed specimens, 86 pathogens were found in single or dual infections, resulting in a sensitivity of 97.7% and a specificity of 87.5% of the NxTAG‐RPP compared to the RF‐22. Among the 167 prospectively analyzed specimens, 117 pathogens were detected indicating a sensitivity of 95.1% and a specificity of 91.8% of the NxTAG‐RPP. When comparing the detection rates for the three most frequently found individual targets in the retrospective cohort 1 and in the prospective cohort 2, there was no statistically significant difference between both assays, with the exception of rhino‐/enterovirus detection showing with a higher positivity rate in the retrospective cohort (Table IIB).
Table II.
Comparing NxTAG‐RPP and RF‐22 for Individual Targets
| A. Overall | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Pathogen | TP | FP | FN | TN | Total | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | Κ |
| PIV‐1 | 3 | 0 | 0 | 279 | 282 | 100 | 100 | 100 | 100 | 1.00 |
| PIV‐2 | 4 | 0 | 0 | 278 | 282 | 100 | 100 | 100 | 100 | 1.00 |
| PIV‐4 | 3 | 0 | 0 | 279 | 282 | 100 | 100 | 100 | 100 | 1.00 |
| HMPV | 15 | 0 | 0 | 267 | 282 | 100 | 100 | 100 | 100 | 1.00 |
| RSV A | 13 | 0 | 0 | 269 | 282 | 100 | 100 | 100 | 100 | 1.00 |
| RSV B | 13 | 0 | 0 | 269 | 282 | 100 | 100 | 100 | 100 | 1.00 |
| INF A (H3) | 10 | 0 | 0 | 272 | 282 | 100 | 100 | 100 | 100 | 1.00 |
| INF A (H1N1) | 1 | 0 | 0 | 281 | 282 | 100 | 100 | 100 | 100 | 1.00 |
| INF B | 19 | 0 | 0 | 263 | 282 | 100 | 100 | 100 | 100 | 1.00 |
| HCoV 229E | 4 | 0 | 0 | 278 | 282 | 100 | 100 | 100 | 100 | 1.00 |
| HCoV HKU1 | 6 | 0 | 0 | 276 | 282 | 100 | 100 | 100 | 100 | 1.00 |
| Lpneu | 1 | 0 | 0 | 281 | 282 | 100 | 100 | 100 | 100 | 1.00 |
| ADV | 7 | 1 | 0 | 274 | 282 | 100 | 99.6 | 87.5 | 100 | 0.93 |
| HRhV | 60 | 7 | 2 | 213 | 282 | 96.8 | 96.8 | 89.6 | 99.1 | 0.91 |
| PIV‐3 | 18 | 1 | 1 | 262 | 282 | 94.7 | 99.6 | 94.7 | 99.6 | 0.94 |
| HBoV | 9 | 1 | 1 | 271 | 282 | 90.0 | 99.6 | 90.0 | 99.6 | 0.90 |
| HCoV OC43 | 8 | 0 | 1 | 273 | 282 | 88.9 | 100 | 100 | 99.6 | 0.94 |
| HCoV NL63 | 5 | 1 | 1 | 275 | 282 | 83.3 | 99.6 | 83.3 | 99.6 | 0.83 |
| Mpneu | 4 | 0 | 2 | 276 | 282 | 66.7 | 100 | 100 | 99.3 | 0.80 |
| B. According to cohort | ||||||||
|---|---|---|---|---|---|---|---|---|
| Total (%) | RF‐22 pos (%) | NxTAG‐RPP pos (%) | RF‐22 neg (%) | NxTAG‐RPP neg (%) | P | Sensitivity (%) | Specificity (%) | |
| Retrospective samples | 115 (100) | 80 (69.9) | 79 (68.7) | 35 (30.4) | 36 (31.3) | 1.0 | 97.7 | 87.5 |
| Prospective samples | 167 (100) | 100 (59.9) | 103 (61.7) | 67 (40.1) | 64 (38.3) | 0.25 | 95.1 | 91.8 |
| HRhV | ||||||||
| Retrospective samples | 115 (100) | 27 (23.5) | 37 (32.2) | 88 (76.5) | 78 (67.8) | 0.002 | 100 | 95.1 |
| Prospective samples | 167 (100) | 29 (17.4) | 30 (18.0) | 138 (82.6) | 137 (82.0) | 1.0 | 93.1 | 97.8 |
| PIV‐3 | ||||||||
| Retrospective samples | 115 (100) | 7 (6.1) | 8 (7.0) | 108 (93.9) | 107 (93.0) | 1.0 | 100 | 99.1 |
| Prospective samples | 167 (100) | 11 (6.6) | 10 (6.0) | 156 (93.4) | 157 (94.0) | 1.0 | 91.7 | 100 |
| INFB | ||||||||
| Retrospective samples | 115 (100) | 4 (3.5) | 3 (2.6) | 111 (96.5) | 112 (97.4) | 1.0 | 100 | 100 |
| Prospective samples | 167 (100) | 15 (9.0) | 16 (9.6) | 152 (91.0) | 151 (90.4) | 1.0 | 100 | 100 |
ADV, adenovirus; HBoV, human bocavirus; HCoV, human coronavirus; HMPV, human metapneumovirus; HRhV, human rhinovirus; INF, influenza; Lpneu, Legionella pneumophila; Mpneu, Mycoplasma pneumoniae; PIV, parainfluenza virus; RSV, respiratory syncytial virus; PPV, positive predicitve value; NPV, negative predictive value; κ (kappa), interobserver agreement.
Number and percentage of total positives and negatives detected by RF‐2 and NxTAG‐RPP in retrospective and prospective samples and the number and percentage of the most frequently found pathogens of the two cohorts, rhino‐/enterovirus (HRhV), influenza B (INFB), parainfluenza‐3 (PIV‐3).
Co‐infections were observed in 10.3% by NxTAG‐RPP and in 5.9% by RF‐22. Most additional viral pathogens identified by the NxTAG‐RPP involved dual infections with rhino‐/enterovirus and respiratory syncytial virus. Of the 31 multiple infections, four samples contained triple infections detected by both assays (Table I). In 15 cases, co‐infections were solely detected by NxTAG‐RPP (5.3%), in 14 cases by both NxTAG‐RPP and RF‐22 (5.0%), and in 2 cases only by RF‐22 (0.9%).
Table I.
Co‐Infection Rates by Assay Type
| NxTag‐RPP | n | RespiFinder‐22 | n |
|---|---|---|---|
| ADV + HCoV HKU1 + Inf B | 2 | ADV + HCoV HKU1 + Inf B | |
| HRhV + RSV B + HBoV | 1 | HRhV + RSV B + HBoV | |
| HMPV + PIV‐3 + HCoV HKU1 | 1 | HMPV + PIV‐3 + HCoV HKU1 | |
| HCoV NL63 + HCoV OC43 | 1 | HCoV NL63 + HCoV OC43 | |
| HRhV + HMPV | 2 | HRhV + HMPV | |
| HRhV + RSV A | 1 | HRhV + RSV A | |
| HRhV + RSV B | 1 | HRhV + RSV B | |
| HMPV + HBoV | 1 | HMPV + HBoV | |
| HMPV + Inf B | 1 | HMPV + Inf B | |
| HMPV + MYPN | 1 | HMPV + MYPN | |
| PIV‐4 + RSV A | 1 | PIV‐4 + RSV A | |
| ADV + HBoV | 1 | ADV + HBoV | |
| HCoV OC43 + HCoV NL63 | 1 | HCoV OC43 | |
| HCoV NL63 + hMPV | 1 | HCoV NL63 | |
| HRhV + PIV‐3 | 2 | HRhV | |
| HRhV + RSV B | 1 | HRhV | |
| HRhV + ADV | 1 | HRhV | |
| ADV + HRhV | 2 | ADV | |
| HBoV + HRhV | 1 | HBoV | |
| HCoV OC43 + HRhV | 1 | HCoV OC43 | |
| Inf A + HRhV | 1 | Inf A | |
| RSV A + HRhV | 1 | RSV A | |
| HCoV 229E + HRhV | 1 | HCoV 229E | |
| HCoV NL63 + HRhV | 1 | HCoV NL63 | |
| RSV A + HBoV | 1 | RSV A | |
| HRhV | HRhV + Mpneu | 2 |
Thirty‐one co‐infections detected by NxTAG‐RPP and RF‐22. Discordant results are shown in bold. ADV, adenovirus; HBoV, human bocavirus; HCoV, human coronavirus; HMPV, human metapneumovirus; HRhV, human rhinovirus; INF, influenza; Lpneu, Legionella pneumophila; Mpneu, Mycoplasma pneumoniae; PIV, parainfluenza virus; RSV, respiratory syncytial virusn, number.
Discordant samples were mainly due to low pathogen levels when retested by QNAT and due to a higher sensitivity of the NxTAG‐RPP assay resulting also in detecting multiple infections.
Using RF‐22 as a reference, NxTAG‐RPP had an overall higher positivity rate, a sensitivity of 96.2%, specificity of 90.3%, PPV of 94.9%, NPV of 92.7%, and a kappa value of 0.870 (Table IIA). Mycoplasma pneumoniae was detected in 2 cases (2.1%) during that time period.
DISCUSSION
RTI involve a variety of viruses and bacteria, most of which are conveniently detected by multiplex NAT. In this study, we present the direct comparison of our current routine assay, the RespiFinder‐22® (RF‐22) with the novel NxTAG‐RPP for RTI specimens from symptomatic adults and children. The overall concordance between both assay results was high with 263 (93.3%) samples and an overall positivity rate of more than 50%. The leading pathogens were rhino‐/enterovirus, parainfluenzavirus, influenzavirus, and respiratory syncytial virus (Fig. 1B). The pathogen‐specific results were highly concordant for parainfluenzavirus‐1, ‐2, ‐4, respiratory syncytial virus A and B, human metapneumovirus, influenza A (H3), influenza A (H1N1), influenza B, human coronaviruses 229E and HKU1, and Legionella pneumophila (κ‐value 1.0) (Table IIA), whereas adenovirus, rhino‐/enterovirus, human bocavirus, para‐influenzavirus‐3, and human coronaviruses OC43 had slightly lower κ‐values of ≥0.9. The most striking differences were observed for human coronaviruses NL63 (κ‐value 0.83) and M. pneumonia (κ‐value 0.80), which is still interpreted as an almost perfect agreement though the positivity rate was too low to draw conclusions. All discordant results that were verified by QNAT were associated with high cycle threshold (CT) values, indicating low genome loads (data not shown). The detection of additional rhino‐/enterovirus was the single most important cause for discordance and seems to result from a higher sensitivity of the NxTAG‐RPP compared to RF‐22. Clearly, the role of rhino‐/enterovirus detection, particularly in dual infections, requires additional studies as pointed out recently [Milano et al., 2010; Piralla et al., 2014]. NxTAG‐RPP suffered from occasionally invalid results due to a low bead count, which disappeared with more hands‐on experience of the technicians. Failure of the RF‐22 with invalid internal controls or other technical issues were seen in less than 2%, and required repeat testing. The specimen types consisted mostly of nasopharyngeal swabs, but BAL was submitted in one fourth of the cases, which requires an invasive procedure typically applied to patients with significant clinical problems. Of note, no difference was seen in the rates of discordant results in BAL (5/76; 6.6%) versus NPA (14/199; 7.0%).
Limitations of the current study are the low number of detections for some pathogens. M. pneumoniae was rare, and detected in two cases (2.1%) during that time period, suggesting that viruses are the main cause of RTIs in pediatric and adult patients [Ruuskanen et al., 2011]. L. pneumophila as not frequent with only one positive specimen, whereas there were no positive results for Chlamydophila pneumoniae. Here, particular attention should be paid to the performance of these assays in quality assurance programs. In view of the considerable number of different pathogens to be covered, quality assurance programs become a real financial and organizational challenge for the diagnostic laboratory given the goal of a yearly participation.
Given the overall high concordance of the RespiFinder‐22® and the novel NxTAG‐RPP observed in this study, other arguments such as the ease of handling, HOT, TAT, CE marking, and costs become critical issues. While CE marking has been obtained in the meantime, costs cannot be addressed at present. However, the ease of handling and HOT were clearly in favor of the novel NxTAG‐RPP. This is largely due to the lyophilized master mix in the sealed ready‐to‐use microtiter plates, and the bead technology, including PCR and hybridization in one step, which reduces the HOT to less than 30 min. Even for a large number of parallel samples, scalable depending on the seasonal demand to 96 test, the TAT is in the range of only 5–7 hr (median 6 hr). In contrast, the RespiFinder‐22® had a sample number‐dependent HOT being 2 and 4 hr for 24 and 96 samples, respectively (including PCR, hybridization, ligation, and denaturation), before analysis by capillary electrophoresis yielding a TAT ranging from 8 to 16 hr (median 12 hr).
Taken together, NxTAG‐RPP and RespiFinder‐22® provided highly concordant results. The sensitivity of the new assay is excellent and even seems to have superior sensitivity for some pathogens. Ease‐of‐handling, HOT, and TAT favor NxTAG‐RPP, but there remain general challenges in the interpretation of multiply positive NAT results in the clinics, even in symptomatic children and adults, that require additional clinical or laboratory qualifiers.
ETHICAL APPROVAL
Not required.
ACKNOWLEDGMENTS
We thank Sylvie Goepfert, Elisabeth Hohmann, and Sibylle Stauffer for excellent technical assistance, the technicians of the Division Infection Diagnostics in the Laboratory for Molecular Diagnostics and the Laboratory for virus isolation for general support with this project. We also wish to thank Prof L. Kaiser and Dr. C. Tapparel from the University Hospital Geneva (HUG) for independent confirmatory analysis of three samples, and Professors Ulrich Heininger and Urs Frey from the University Children's Hospital Basel (UKBB) for the excellent collaboration regarding pediatric infection diagnostics. The NxTAG‐RPP reagents and MagPIX instrument were provided by Dr. J. Daam from Luminex (Austin, Texas).
Conflicts of interest: none.
REFERENCES
- Beckmann C, Hirsch HH. 2015. Diagnostic performance of near‐patient testing for influenza. J Clin Virol 67:43–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caliendo AM. 2011. Multiplex PCR and emerging technologies for the detection of respiratory pathogens. Clin Infect Dis 52:S326–S330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumoulin A, Widmer AF, Hirsch HH. 2009. Comprehensive diagnostics for respiratory virus infections after transplantation or after potential exposure to swine flu A/H1N1: What else is out there? Transpl Infect Dis 11:287–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grondahl B, Ankermann T, von Bismarck P, Rockahr S, Kowalzik F, Gehring S, Meyer C, Knuf M, Puppe W. 2014. The 2009 pandemic influenza A(H1N1) coincides with changes in the epidemiology of other viral pathogens causing acute respiratory tract infections in children. Infection 42:303–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch HH, Martino R, Ward KN, Boeckh M, Einsele H, Ljungman P. 2013. Fourth European conference on infections in leukaemia (ECIL‐4): Guidelines for diagnosis and treatment of human respiratory syncytial virus, parainfluenza virus, metapneumovirus, rhinovirus, and coronavirus. Clin Infect Dis 56:258–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ison MG. 2012. Antiviral therapies for respiratory viral infections in lung transplant patients. Antivir Ther 17:193–200. [DOI] [PubMed] [Google Scholar]
- Khanna N, Steffen I, Studt JD, Schreiber A, Lehmann T, Weisser M, Fluckiger U, Gratwohl A, Halter J, Hirsch HH. 2009. Outcome of influenza infections in outpatients after allogeneic hematopoietic stem cell transplantation. Transpl Infect Dis 11:100–105. [DOI] [PubMed] [Google Scholar]
- Milano F, Campbell AP, Guthrie KA, Kuypers J, Englund JA, Corey L, Boeckh M. 2010. Human rhinovirus and coronavirus detection among allogeneic hematopoietic stem cell transplantation recipients. Blood 115:2088–2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickel CH, Stephan FP, Dangel M, Blume K, Gehrisch R, Dumoulin A, Tschudin S, Keller DI, Hirsch HH, Widmer AF, Bingisser R. 2009. First wave of the influenza A/H1N1v pandemic in Switzerland. Swiss Med Wkly 139:731–737. [DOI] [PubMed] [Google Scholar]
- Piralla A, Lunghi G, Percivalle E, Viganò C, Nasta T, Pugni L, Mosca F, Stronati M, Torresani E, Baldanti F. 2014. FilmArray® respiratory panel performance in respiratory samples from neonatal care units. Diagn Microbiol Infect Dis 79:183–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijans M, Dingemans G, Klaassen CH, Meis JF, Keijdener J, Mulders B, Eadie K, van Leeuwen W, van Belkum A, Horrevorts AM, Simons G. 2008. RespiFinder: A new multiparameter test to differentially identify fifteen respiratory viruses. J Clin Microbiol 46:1232–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruuskanen O, Lahti E, Jennings LC, Murdoch DR. 2011. Viral pneumonia. Lancet 377:1264–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidler JA, Haberthur C, Dumoulin A, Hirsch HH, Heininger U. 2012. A retrospective analysis of nosocomial viral gastrointestinal and respiratory tract infections. Pediatr Infect Dis J 31:1233–1238. [DOI] [PubMed] [Google Scholar]


