Abstract
The prominent feature of human cytomegalovirus (HCMV) is cell tropism specificity for human fetal nervous system, which leads to severe fetal nervous system damage especially in first‐trimester gestation. In this study, human astrocytes isolated from fetal brain were infected with HCMV AD169 and whole genome transcriptome profile was performed. The results showed that the gene expression of interferon stimulated genes (ISGs), chemokine and chemokine receptors were significantly up‐regulated (P < 0.01). The antiviral replication effects of IFIT1 (Interferon‐induced protein with tetratricopeptide repeats 1, Fc = 148.17) was investigated. Lentivirus with IFIT1 overexpression or knockdown was transduced into astrocytes, respectively. The viral mRNA, protein expression and HCMV titers were determined. The results showed that IE1, IE2, pp65, and viral titers were significantly decreased in IFIT1 overexpression group and enhanced in the knockdown group compared with control one (P < 0.01). Taken together, this study revealed IFIT1 played an important antiviral role in HCMV infected fetal astrocytes. The prominent feature of human cytomegalovirus (HCMV) is cellular tropism specificity for human fetal brain nervous system leading to severe fetal nervous damage especially in first‐trimester gestation. In this study, human astrocytes isolated from first‐trimester fetal brain were infected with HCMV AD169 and IFIT1 was studied for its antiviral replication effects. The results provided insights into the function of IFIT1 as a key factor in antiviral defense contributing to development of targeted therapeutics to fetal brain with HCMV infection. J. Med. Virol. 89:672–684, 2017 . © 2016 Wiley Periodicals, Inc.
Keywords: human cytomegalovirus, interferon, microarrays, nervous system, antiviral agents, disease control
INTRODUCTION
Human cytomegalovirus (HCMV; Human herpesvirus, a member of β‐herpes virus subfamily) is a ubiquitous pathogen and the most common infectious cause of congenital diseases [Stagno, 1985; Boppana et al., 2005; Colugnati et al., 2007]. Primary infection during pregnancy poses a 30–40% risk of intrauterine transmission, with severe adverse outcomes more likely if the infection occurs within the first trimester gestation. Due to strict viral host specificity, so far there is no animal model of HCMV infection available. Current state of knowledge of CMV pathogenesis and host defense against HCMV has come largely from clinical observation and in vitro studies with cell lines or primary human cells [Sinzger and Jahn, 1996].
Previous study in our laboratory has identified human fetal astrocytes permissive to HCMV infection [Zhang et al., 2013]. It was reported that astrocytes constitute important parts of host defense mechanism, and play important roles in preserving CNS (central nervous system) function and protecting CNS against infectious agents [Carson and Sutcliffe, 1999]. That HCMV or its proteins modulate gene expression in astrocytes has also been investigated. After being infected by HCMV, astrocytes produce as well as respond to a number of immune mediators, including interferon, ISG (interferon‐stimulated gene) proteins, cytokines, and chemokines [Grandvaux et al., 2002] to control the spread of invading HCMV.
Type I IFN and ISG proteins have been shown to possess antiviral properties in CNS. Early expression of cytokines in CNS has been demonstrated to correlate with the control of viral replication [Carson and Sutcliffe, 1999]. When virus contact with host cells, viral proteins and nucleic acids are detected by pathogen recognition receptors (PRRs) as pathogen‐associated molecular patterns (PAMPs). Subsequently, the signal transduction pathways are activated. Type I IFN and production of other cytokines are triggered. Type I IFN then augments the expression of several ISGs and subverts virus replication by a variety of mechanisms [Poland et al., 1994; Peisley and Hur, 2013]. However, not all ISGs are induced in IFN‐dependent manner. Some are induced in IRF3‐dependent manner [Grandvaux et al., 2002]. Moreover, not all ISG proteins confer direct antiviral effects, and the anti‐HCMV effects of ISG proteins have not been attributed to every ISG family members so far characterized. Inhibitory phenotypes directed at specific ISG proteins need to be studied for HCMV. Given the observed replication of the virus in the presence of multiple antiviral host proteins, it is probable that a number of ISG‐inhibitory phenotypes are to be uncovered for HCMV [Munir, 2013; Peisley and Hur, 2013]; studies on ISG protein against HCMV infection were carried out using either cell lines or mature astrocytes, which do not mimic the natural course of HCMV infection and there are many gaps to be filled in the understanding of host defense against HCMV infection in CNS. To overcome these limitations, we used human fetal astrocytes of the first trimester gestation to investigate the antiviral effects of ISG proteins, with expectation of developing a targeting therapy without therapy‐limiting side effects of IFNs.
IFIT1 (Interferon‐induced protein with tetratricopeptide repeats 1) is one of the identified ISGs [Wathelet et al., 1986; Levy et al., 2004]. All members in IFIT family contain multiple tetratricopeptide motifs that are known to mediate protein–protein interactions through scaffolds formed among tandem TPR repeats [Lamb et al., 1995]. IFIT1 is induced in response to type I IFNs stimulation, presence of double‐stranded RNAs, and virus infection [Guo et al., 2000; Terenzi et al., 2006]. IFIT1 interacts with the translation initiation factor eIF‐3, leading to the inhibition of protein synthesis. IFIT1 has been implicated to have similar antiviral effects as IFNs in fighting against West Nile virus and LCMV [Wacher et al., 2007]. It is also reported to inhibit human papillomavirus DNA replication by binding to the viral protein E1 [Terenzi et al., 2008]; studies demonstrated that the 2′‐O methylation of the 5′ cap of viral RNA functions to subvert innate host antiviral responses through escape of IFIT‐mediated suppression [Daffis et al., 2010]; It was discovered that IFIT proteins selectively recognize viral RNA via a 5′‐triphosphate group (PPP‐RNA), and lend insight into their downstream effector function [Pichlmair et al., 2011; Abbas et al., 2013]. Recently, it was reported that ISG54 and ISG56 may contribute to immune and inflammatory reactions elicited by TLR3/IFN‐β signaling pathway in astrocytes, and may play an important role both in antiviral immunity and in neuroinflammatory diseases [Imaizumi et al., 2014]. However, little has been reported to date on the protective mechanism of IFIT1 against HCMV infection.
In present study, human fetal astrocytes were infected with HCMV AD169 and whole genome transcriptome profile microarrays were performed. The results showed that HCMV infection regulated multiple gene expression, including up‐regulation of interferon‐stimulated genes, and genes of chemokine and chemokine receptors, indicating HCMV infection in astrocytes profoundly altered host cell biology. The expression of ISGs were up‐regulated dramatically in HCMV infected group with IFIT1 expression most significantly increased (change folds = 148.17). IFIT1 was further studied for its antiviral effects using overexpression and knockdown lentivirus transduction.
MATERIALS AND METHODS
Cell Culture and Virus Infection
Human astrocytes from three 65‐ to 75‐day‐old fetal brains and HELFs (human embryonic lung fibroblast cells) were obtained from aborted fetuses in the Department of Obstetrics and Gynecology of the Affiliated Hospital, Qingdao University Medical College. The use of aborted fetal brain was approved by Qingdao University Human Investigation Committee. The pregnant women who voluntarily underwent abortion showed negative peripheral blood HCMV antibody detection. Astrocytes were isolated and purified as previous studies [Luo et al., 2010; Zhang et al., 2013, 2014], cells were seeded onto different culture dishes respectively in DMEM/F12 medium supplemented with 10% FBS and 1% penicillin/streptomycin. After cells grew to about 70–80% convergence, the medium was replaced with DMEM/F12 medium supplemented with 2% FBS. Cells were infected with HCMV AD169 (m.o.i = 2.5) for 2 hr, then the medium was replaced with fresh DMEM/F12 medium supplemented with 2% FBS. Cells were maintained at 37°C in a humidified atmosphere containing 5% CO2 for 2, 4, 6, 72 hr.
Whole Genome Profile Microarrays
After 6 or 72 hr HCMV infection, total RNA was extracted using TRIzol Reagent (Invitrogen, Carlsbad, CA) and reversed to cDNA. Four micrograms of dscDNA were sent to the NimbleGen expression array platform. The NimbleGen platform proposes high‐density expression arrays in which every human gene is represented by several independent probes. These probes consist of 60‐mers isothermal oligonucleotides, which show more robust hybridization than shorter oligonucleotides. The NimbleGen human expression array (Format: 12 × 135k; Source: NCBI; Build: HG18, Build 36; Probe Length: 60 mer; Genes: 45033; Probes/Target: 3; Replicates: 1) involves 45,033 gene transcripts (i.e., different accession numbers corresponding to the same or different isoforms). DNA end‐labeling, hybridization, scanning, and data normalization were performed at NimbleGen, which provided the final data file. Array hybridization using the NimbleGen Hybridization System followed by washing with the NimbleGen wash buffer kit and Array scanning using the Axon GenePix 4000B microarray scanner (Molecular Devices Corporation, Sunnyvale, CA) were conducted. Scanned images were then imported into NimbleScan software (version 2.5) for grid alignment and expression data analysis. Expression data were normalized through quantile normalization and the Robust Multichip Average (RMA) algorithm included in the NimbleScan software. The Probe level files and Gene level files were generated after normalization. All gene level files were imported into Agilent GeneSpring GX software (version 11.5.1) for further analysis. Genes that at least nine out of nine samples greater than or equal to lower cut‐off: 50.0 were chosen for data analysis. Differentially expressed genes between two samples were identified through Fold Change filtering. Differentially expressed genes between two groups were identified through Volcano Plot filtering. A set of genes presenting an average of at least twofold induction/repression calculated from the different probes specific for each gene spotted on the NimbleGen chip were selected for promoter analysis. GO analysis were applied to determine the roles of these differentially expressed genes played in these biological GO terms.
qRT‐PCR
QRT‐PCR was performed in 96‐well optical plates (Sorenson Bioscience, Inc., Salt Lake City, UT) with cDNA using the MyiQ Single‐Color Real‐Time PCR Detection System (Bio‐Rad Laboratories, Hercules, CA) in 20 µl reaction volumes. A master mix was made according to manufacturer's instructions using SYBR green supermix (Bio‐Rad Laboratories). Forward and reverse primers were used at a concentration of 250 nM per well, made in RNase/DNase‐free H2O. Primer sequences for qPCR were as follows: IFIT1: Forward 5′‐TTGCCTGGATGTATTACCAC‐3′, reverse 5′‐GCTTCTTGCAAATGTTCTCC‐3′; MXA: 5′‐AGGACCATCGGAATCTTGAC‐3′, reverse 5′‐TCAGGTGGAACACGAGGTT‐3′; CCL5: Forward 5′‐ATCCCTCACCGTCATCCTCGTTG‐3′, reverse 5′‐TTGGCACACACTTGGCGGTTC‐3′; CXCL2: Forward 5′‐GAAGTCATAGCCACTCTCAAGG‐3′, reverse 5′‐TTCCGTTGAGGGACAGCA‐3′; IL‐8: Forward 5′‐GGCACAAACTTTCAGAGACAG‐3′, reverse 5′‐ACACAGAGCTGCAGAAATAGG‐3′; IL‐11: Forward5′‐CCCAGTTACCCAAGCATCCA‐3′, reverse 5′‐AGACAGAGAACAGGGAATTAAATGTGT‐3′; OAS1: Forward 5′‐AGGTGGTAAAGGGTGGCT‐3′, reverse 5′‐TGCTTGACTAGGCGGATG‐3′; Vimentin: Forward 5′‐CGTACGTCAGCAATATGAAAGTGTG‐3′; TCAGAGAGGTCAGCAAACTTGGA‐3′; GFAP: Forward 5′‐ACTACATCGCCCTCCACATC‐3′, reverse 5′‐CAAAGGCACAGTTCCCAGAT‐3′; IE2: Forward 5′‐ATGAACCACCCTCCTCTTCC‐3′, reverse 5′‐GATATTGCGCACCTTCTCGT‐3′; IE1: Forward 5′‐GAGTCCTCTGCCAAGAGAAA‐3′, reverse 5′‐GAGTTCTGCCAGGACATCTTT‐3′; β‐Actin: Forward 5′‐TGGAACGGTGAAGGTGACAG‐3′, reverse 5′‐GGCTTTTAGGATGGCAAGGG. β‐actin and GAPDH primer sets were included for normalization. Data analysis were performed using Bio‐Rad iQ5 optical system software version 2.1.
Western Blot Analysis
Astrocytes were harvested at different time points. Cells were trypsinized, pelleted, and extracted for cell protein. Protein lysates were electrophoresed using SDS–PAGE. Proteins were transferred to PVDF membrane (Millipore, Billerica, MA), and the membrane with transferred proteins were incubated with anti‐IFIT1, 2, 3, 5 (Santa Cluz Biotechnology, Santa Cruz, CA), anti‐IE1 (Santa Cruz Biotechnology), anti‐IE2 (Beijing Biosynthesis Biotechnology Co. Ltd, Beijing, China), and anti‐pp65 antibodies (Beijing Biosynthesis Biotechnology Co. Ltd), respectively, (all antibodies diluted at 1:100 in TBST) for 60 min at 37°C and then with horseradish peroxidase‐labeled secondary antibody for 45 min at 37°C. After washing in TBST, the membrane was developed using ECL reagents and photos were taken. Protein bands were quantified using the Quantity One software (Bio‐Rad Laboratories). Paired mean comparisons were performed to assess concentration dependent change in the intensity of native protein bands. All western blot analyses were performed three times, with representative images shown.
Lentivirus Package and Lentivirus‐Mediated Transduction of Astrocytes and HELFs
Twenty microgram pGC‐LV‐IFIT1 vector (IFIT1 empty pGC‐LV was for negative ctontrol vector), 15 µg pHelper 1.0 vector and 10 µg pHelper 2.0 vector DNA solution were transfected to 393T cells with lipofectamine 2000 (11668–500, Invitrogen). After 48 hr transfection, cell supernatant was collected, centrifuged, and filtered with 0.45 μm filter for crude extract, then purified for lentivirus concentration with Plus‐20 centrifugal ultrafiltration device (UFC910024, Millipore) at 4000 rpm centrifugation.
Human fetal astrocytes and HELFs were seeded in 6‐well tissue culture plates at 3 × 105 cells/well and incubated at 37°C. The cells were transduced with lv/IFIT1/EGFP (accession number of IFIT1: NM_001548), lv/ctl/EGFP (GV208, Genechem, Shanghai, China), lv/shIFIT1/EGFP (accession number of IFIT1: NM_001548) or lv/shIFIT1/EGFP (GV115, Shanghai Genechem) at m.o.i of 5.0 in a fresh complete culture medium. The medium containing lentivirus was discarded and replaced with fresh complete medium at 6 hr post transfection. The cells were observed under flow cytometry (FCM) and confocal microscope at 48 hr post transfection to evaluate the efficiency of EGFP‐expressing cells and then infected or mock infected with HCMV AD169 (m.o.i = 2.5) for 24 and 48 hr.
Immunocytochemistry
Astrocytes were immersed in 4% paraformaldehyde for 0.5 hr at room temperature, rinsed in PBS, permeabilized in 0.1% Triton X‐100 for 30 min at room temperature. Ten percent normal goat serum was used for blocking for 0.5 hr at room temperature. The cells were incubated with primary antibody against IE1 (Santa Cruz Biotechnology), IFIT1 (Santa Cruz Biotechnology), IE2 (Beijing Biosynthesis Biotechnology Co. Ltd), and pp65 antibody (Beijing Biosynthesis Biotechnology Co. Ltd), (all antibodies diluted 1:100 in PBS), for 60 min at 37°C. Then the cells were rinsed and incubated with the FITC‐conjugated goat anti‐mouse or FITC/Cy3‐conjugated goat anti‐rabbit IgG (diluted 1:100) for 30 min at 37°C. DAPI or PI was used to stain nuclear. Cells were viewed under laser confocal microscopy.
Virus Plaque Titration
Various dilutions (10−3, 10−4, 10−5, 10−6, 10−7, 10−8, 10−9) of virus with DMEM/F12 medium were added to 75% confluent astrocytes or HELFs and incubated at 37°C for 2 hr. After virus adsorption, HCMV AD169 was tittered by plaque titration and expressed as the number of plaque‐forming units per milliliter. The culture medium was removed from infected cultures and centrifuged for determining virus titer of supernatant, while attached cells were washed with PBS and scraped into culture medium for intracellular virus titer determination at 24, 48 hr p.i.
Viability, Proliferation, and Apoptosis Detected by Colorimetric MTT and FCM
Cell viability and proliferation assay was performed by the colorimetric [3(4,5‐dimethylthiazol‐2‐yl)‐2,5‐diphenyltetrazolium bromide] (MTT) assay according to the kit instruction (Sigma–Aldrich). Cell morphism was observed under an inverted Olympus microscope. Yellow MTT is reduced to purple formazan, due to the metabolic activity of living cells. MTT (5 mg/ml) was added to astrocytes allowing to metabolize for 30 min at 37°C. Blue crystals were dissolved in DMSO. Absorbance values were measured at 492 nm (DG3022) at 0, 48, 72, 96, 120 hr p.i. The experiment was repeated in triplicate. Infected and mock infected astrocytes were collected, centrifuged, and 100 μl DMEM/F12 containing 2% FBS was added to resuspend cells and transferred to 1.5 ml Eppendorf tubes; 100 μl of Guava Nexin Reagent (annexin V‐PE vs. 7‐AAD; EMD Millipore) was added to each Eppendorf tube, incubated for 20 min at room temperature in the dark, and then analyzed using FCM (0500–1430, Guava Easy‐Cyte Mini; Guava Technologies Inc., Hayward, CA) as soon as possible. The experiment was repeated in triplicate.
Statistical Analysis
Student's t‐test using the two‐tailed distribution was used to determine significant differences in virus replication and gene expression. For comparison the difference between multiple groups, statistical significance was analyzed using a one‐way analysis of variance (ANOVA) followed by post Newman–Keul's test. P values of <0.05 were considered statistically significant.
RESULTS
Whole Genome Transcriptome Profile Analysis and Verification
Human fetal astrocytes were purified and identified by immunocytochemistry (A1–A3). When infected with HCMV AD169 and obtained at 6 hr post infection (p.i), astrocytes were examined for mRNA expression with transcriptome microarray using the platform developed by NimbleGen (Madison, WI). As one gene is often represented by several transcripts, we focused only on those genes represented by two or more transcripts and displaying a mean fold difference ≥2.0 and t‐test P‐value <0.05. With these criteria, we found that about 154 genes, including 138 up‐regulated and 16 down‐regulated, were modulated at 6 hr p.i. Gene expressions with more than 10‐fold change were shown in Table I. To verify the results, the mRNA expression of up‐regulated genes IFIT1, MXA, MXB, CCL5, CXCL2, BST2, IL8, IL11 at 6 hr p.i vs con and astrocyte genes GFAP and Vim were confirmed by qRT‐PCR (Fig. 1B). A good correlation was found between the two analyses, although almost all genes had higher FC values in qRT‐PCR than in microarray analysis. Gene Ontology (GO) analysis showed that the top five categories in biological process all belonged to cell response to stimuli, indicating the role of astrocytes against virus infection at 6 hr p.i (Table II).
Table I.
Genes (Fold Change More Than 10) in HCMV AD169 Infected Astrocytes
| SEQ_ID | P‐value | Fold change | Up/down | Gene name | Description |
|---|---|---|---|---|---|
| NM_001001887 | 4.61E‐04 | 148.17294 | Up | IFIT1 | Interferon‐induced protein with tetratricopeptide repeats 1 |
| NM_001547 | 6.57E‐05 | 91.12055 | Up | IFIT2 | Interferon‐induced protein with tetratricopeptide repeats 2 |
| NM_001031683 | 0.001913484 | 69.62959 | Up | IFIT3 | Interferon‐induced protein with tetratricopeptide repeats 3 |
| NM_002462 | 0.006052201 | 52.563366 | Up | MXA | MXA myxovirus (influenza virus) resistance 1, interferon inducible protein p78 |
| NM_016816 | 0.002228843 | 46.65933 | Up | OAS1 | 2′,5′‐oligoadenylate synthetase 1 |
| NM_198213 | 0.009020009 | 37.92200 | Up | OASL | 2′‐5′‐oligoadenylate synthetase‐like |
| NM_002463 | 0.012798179 | 32.80558 | Up | MXB | Myxovirus (influenza virus) resistance 2 |
| NM_002985 | 0.016673766 | 31.12011 | Up | CCL5 | Chemokine (C‐C motif) ligand 5 |
| NM_017414 | 0.008110123 | 26.03400 | Up | USP18 | Ubiquitin specific peptidase 18 |
| NM_002535 | 0.008042933 | 24.61983 | Up | OAS2 | 2′‐5′‐oligoadenylate synthetase 2 |
| NM_003641 | 0.003658616 | 18.35694 | Up | IFITM1 | Interferon induced transmembrane protein 1 |
| NM_006417 | 0.011669416 | 17.90868 | Up | IFI44 | Interferon‐induced protein 44 |
| AJ634912 | 0.019668868 | 16.89670 | Up | PTGS2 | Prostaglandin‐endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) |
| NM_006187 | 0.013115374 | 15.88929 | Up | OAS3 | 2′‐5′‐oligoadenylate synthetase 3 |
| XM_036729 | 0.006409143 | 14.64420 | Up | USP41 | Ubiquitin specific peptidase 41 |
| NM_004335 | 0.003946278 | 14.48724 | Up | BST2 | Bone marrow stromal cell antigen 2 |
| NM_017654 | 0.009343174 | 13.81734 | Up | SAMD9 | Sterile alpha motif domain containing 9 |
| NM_002089 | 0.04555019 | 12.10514 | Up | CXCL2 | Chemokine (C‐X‐C motif) ligand 2 |
| NM_004848 | 0.005747211 | 12.00902 | Up | C1orf38 | Chromosome 1 open reading frame 38 |
| NM_001037335 | 0.006599061 | 11.18215 | Up | PRIC285 | Peroxisomal proliferator‐activated receptor A interacting complex 285 |
| NM_003745 | 0.006988464 | 10.714319 | Up | SOCS1 | Suppressor of cytokine signaling 1 |
| NM_014314 | 0.010410038 | 10.233637 | Up | DDX58 | DEAD (Asp‐Glu‐Ala‐Asp) box polypeptide 58 |
Fold Change cut‐off, 2.0; Condition pairs, 6 hr p.i vs con; hr p.i, hour post infection; con, mock infected control.
Figure 1.
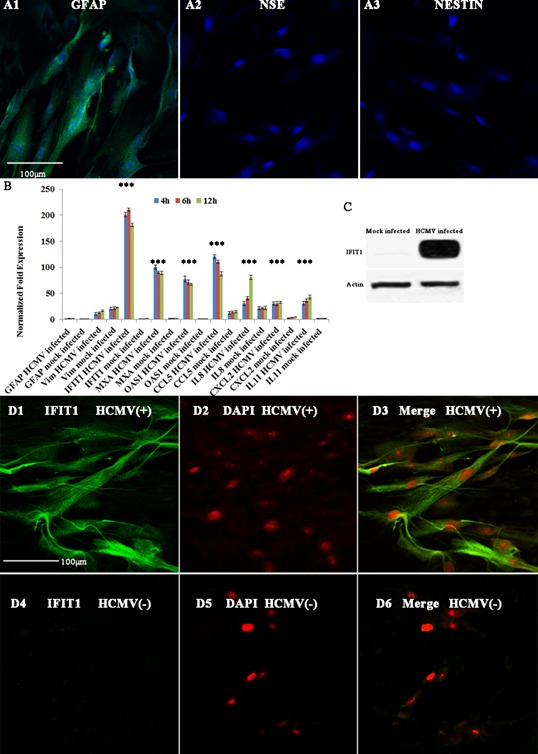
Identification of purified astrocytes and expression of IFIT1 upon HCMV infection by RT‐qPCR, immunocytochemistry and western blot. A, Identification of purified astrocytes; A1, GFAP; A2, NSE positive neurons; A3, NSTIN positive neural stem cells. B, RT‐qPCR confirmation of microarray analysis. The β‐actin and GAPDH primer sets were included for normalization. GFAP, glial fibrillary acidic protein; Vim, vimentin; IFIT1, Interferon‐induced protein with tetratricopeptide repeats 1; MxA, myxovirus (influenza virus) resistance A; OAS1, 2′,5′‐oligoadenylate synthetase 1; CCL5, chemokine (C‐C motif) ligand 5; IL8, interleukin 8; CXCL2, chemokine (C‐X‐C motif) ligand 2; IL11, interleukin 11. D1–D6, IFIT1 protein expression at 24 hr p.i by immunocytochemistry. D1, IFIT1 stained astrocytes; D2, DAPI stained astrocytes; D3, merge of IFIT1 and DAPI; D4, IFIT1 stained astrocytes; D5, DAPI stained astrocytes; D6, merge of IFIT1 and DAPI. C, IFIT1 protein expression by western blot. HCMV(+), HCMV infected (m.o.i = 2.5); HCMV(−), mock infected; H p.i, hours post infection; *, statistical significance P < 0.05.
Table II.
Categories of Up‐Regulated Genes in HCMV Infected Astrocytes
| Gene ontology term | P‐value | Genes |
|---|---|---|
| GO:0009615—response to virus | 6.8E‐16 | DDX58//PML//SAMHD1//BST2//ISGF3G//IFI44//IRF7//MX1//MX2// |
| PLSCR1//EIF2AK2//ZC3HAV1//CCL5//TRIM25//IFITM1//G1P2 | ||
| GO:0051707—response to other organism | 3.7E‐13 | DDX58//PML//CARD4//ISGF3G//IFI44//IRF7//MX1//MX2//PLSCR1//EIF2AK2 |
| //ZC3HAV1//CCL5//TRIM25//IFITM1//G1P2//PTGS2//SOCS1//SAMHD1//BST2 | ||
| GO:0051704—multi‐organism process | 1.9E‐12 | DDX58//PML//CARD4//ISGF3G//IFI44//IRF7//MX1//MX2//PLSCR1// |
| EIF2AK2//ZC3HAV1//CCL5//TRIM25//IFITM1//G1P2//STAT5A | ||
| //PTGS2//KLF9//TAP1//SOCS1//ICAM1//SP110//TFRC//SAMHD1//BST2 | ||
| GO:0009607—response to biotic stimulus | 1.6E‐11 | DDX58//PML//CARD4//ISGF3G//IFI44//IRF7//MX1//MX2//PLSCR1/ |
| /EIF2AK2//ZC3HAV1//CCL5//TRIM25//IFITM1//G1P2 | ||
| //PTGS2//SOCS1//SAMHD1//BST2 | ||
| GO:0006952—defense response | 4E‐10 | DDX58//PML//CCL5//TFRC//CARD4//CFH//TMEM23// |
| CXCL2//NFKBIZ//BMP2//C1ORF38//PTGS2//TAP1//IL1R1// | ||
| TRIM25//IFITM1//SAMHD1//STAT5A//BST2//MX1//MX2 | ||
| GO:0002376— immune system process | 1.1E‐09 | ICAM1//IL7R//DDX58//PML//SFRP1//CFH//G1P3//SAMHD1//CD274// |
| CXCL2//IL1R1//OAS1//OAS2//OAS3//CCL5//OASL//BST2//TAP1 | ||
| //CSF1//SOD2//IL11//STAT5A//LMO2//TRIM25//IFITM1 | ||
| GO:0006955—immune response | 4.5E‐09 | ICAM1//IL7R//DDX58//PML//CFH//BST2//TAP1//IL1R1//TRIM25// |
| IFITM1//SAMHD1//STAT5A//G1P3//CD274//CXCL2//OAS1// | ||
| OAS2//OAS3//CCL5//OASL | ||
| GO:0001816—cytokine production | 0.01009 | PTGS2//DDX58//CARD4//STAT5A//ISGF3G |
Fold Change cut‐off, 2.0; Condition pairs, 6 hr p.i vs con; hr p.i, hour post infection; con, mock infected control.
The expression levels of ISGs were up‐regulated dramatically (Table I), including IFIT1 (fold change, Fc = 148.17), IFIT2 (Fc = 91.12), IFIT3 (Fc = 69.62), IFITM1 (Fc = 18.35), MXA (Fc = 52.56), MXB (Fc = 32.80), OAS1 (Fc = 46.65), OAS2 (Fc = 24.61), OASL (Fc = 37.92), USP18 (Fc = 26.03), USP41 (Fc = 14.64), IFI44 (Fc = 17.90), and BST2 (Fc = 14.48). Chemokines were also up‐regulated, including IL‐11 (Fc = 3.50), CXCL2 (Fc = 12.10) and CCL5 (Fc = 31.12). IFIT1, showing most significant changes in gene expressions, was chosen for further investigation of its antiviral effect. IFIT1 protein expression was determined by immunocytochemistry (Fig. 1D1–D6) and western blot (Fig. 1C). The results of both assays showed significant expression of IFIT1 in HCMV infected astrocytes compared with mock infected control.
Morphological Observation of Astrocytes and HELFs Transduced With IFIT1 Overexpression Lentivirus
IFIT1 overexpression lentivirus (lv/IFIT1/EGFP) was transduced into human fetal astrocytes and HELFs. After 48 hr transfection, the efficiency of transfection was observed under laser confocal microscope and analyzed by FCM, which showed more than 98.5% cells were positive (Fig. 2A1–A4). These cells were infected with HCMV AD169 (m.o.i = 2.5), and cell morphology changes were monitored under laser confocal microscope at different time points. There were no morphological changes in lv/IFIT1/EGFP transduced astrocytes at 72 hr p.i (Fig. 2B1 and B2) and mock infected groups (Fig. 2B5 and B6) compared with lv/ctl/EGFP transduced control group, the later exhibited classical morphological changes at 72 hr p.i (Fig. 2B3 and B4). On the contrary, HELFs exhibited classical pathogenic effects at 72 hr p.i in both lv/IFIT1/EGFP (Fig. 2B7 and B8) and lv/ctl/EGFP transduced groups (Fig. 2B9 and B10), while the mock infected group had no morphological changes (Fig. 2B11 and B12). The results indicated a different antiviral role of IFIT1 in astrocytes and HELFs.
Figure 2.
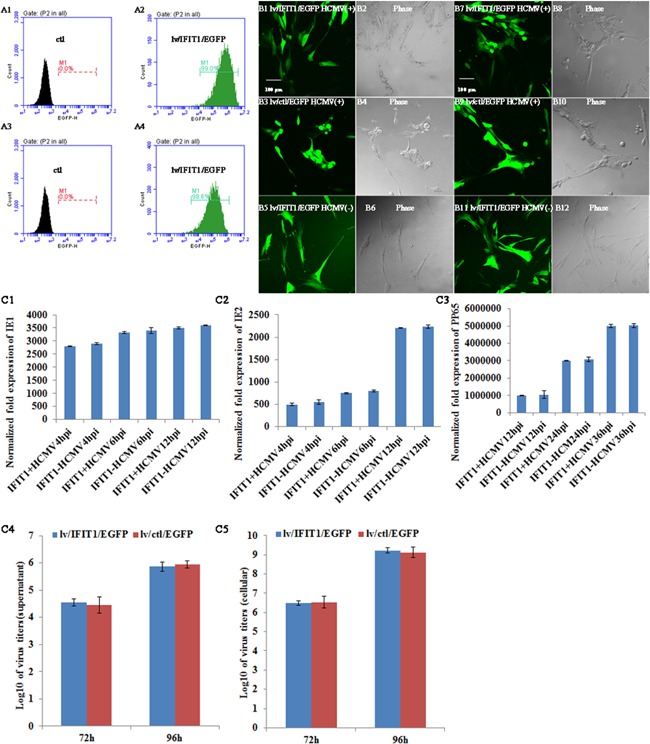
Overexpression of IFIT1 did not regulate HCMV AD169 replication in HELFs. A1 and A2, transduction efficiency by FCM. A1, non‐transduced astrocytes; A2, lv/IFIT1/EGFP transduced astrocytes after 48 hr; A3, non‐transduced HELFs; A4, lv/IFIT1/EGFP transduced HELFs after 48 hr. B1–B12, cell morphology observation under con‐focal microscope. B1, B2, B5, and B6, lv/IFIT1/EGFP transduced astrocytes; B3 and B4, lv/ctl/EGFP transduced astrocytes; B7, B8, B11, and B12, lv/IFIT1/EGFP transduced HELFs; B9 and B10, lv/ctl/EGFP transduced HELFs. C1–C3, IE1, IE2, and pp65 by RT‐qPCR C4, Virus titer of supernatant at 72 and 96 hr p.i; C5, intracellular virus titer at 72 and 96 hr p.i. Lv/IFIT1/EGFP, IFIT1 lentivirus; lv/ctl/EGFP, lentivirus negative control; HCMV(+), HCMV infected; HCMV(−), mock infected; H p.i, hours post infection.
Overexpression of IFIT1 and Its Effect on Viral Replication
When lv/IFIT1/EGFP was transduced into HELFs, viral mRNA expression (immediate‐early [IE1, IE2] and late [pp65] stages) and virus titer were detected in HCMV infected HELFs. The results showed that there was no statistical difference in both viral mRNA expression and virus titer between lv/IFIT1/EGFP transduced group and control one (Fig. 2C1–C5). When lv/IFIT1/EGFP was transduced into astrocytes, the expression levels of viral mRNA and protein at different stages (IE1, IE2, and pp65) were detected with qRT‐PCR or western blot. The results showed significant reduction in the expression of viral mRNA (Fig. 3A1–A3) and protein (Fig. 3B) at all stages (IE1, IE2, and pp65) compared with their corresponding control group lv/ctl/EGFP (P < 0.01).
Figure 3.
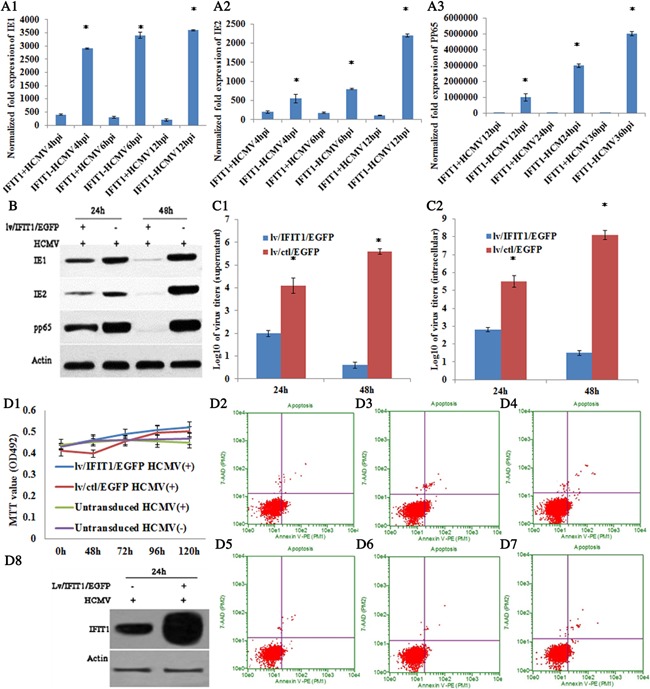
Overexpression of IFIT1 restricted HCMV AD169 replication. A1–A3, mRNA expression of IE1, IE2, and pp65 by RT‐qPCR. B, protein expression of IE1, IE2, and pp65 by western blot at 24 and 48 hr p.i. C1, Virus titer of supernatant; C2, intracellular virus titer. D1, MTT analysis; D2–D4, Cell apoptosis for lv/IFIT1/EGFP transduced astrocytes; D2, 0 hr p.i; D3, 72 hr p.i; D4, 120 hr p.i; D5–D7, lv/ctl/EGFP transduced astrocytes; D5, 0 hr p.i; D6, 72 hr p.i; D7, 120 hr p.i. D8, Protein expression of IFIT1 at 24 hr p.i. HCMV−, mock infected, HCMV+, HCMV infected; *, statistical significance P < 0.05.
To further identify the effect of IFIT1 on HCMV infected human fetal astrocytes, the release of infectious HCMV particles in cell culture medium and virus titer of intracellular astrocytes were determined with plaque titration method on a monolayer astrocytes at 72 hr p.i. The results showed that HCMV titer both inside and outside cells were significantly reduced in lv/IFIT1/EGFP transduced astrocytes compared with lv/ctl/EGFP‐transduced control (P < 0.001) (Fig. 3C1 and C2). Additionally, MTT and FCM for cell proliferation and apoptosis were analyzed, which showed no significant difference at different time points in lv/IFIT1/EGFP or lv/ctl/EGFP transduced group (P < 0.05) (Fig. 3D1–D7), suggesting that it was IFIT1 that mediated antiviral effects in human fetal astrocytes. Additionally, transduced astrocytes over‐expressed IFIT1 protein was determined by western blot analysis after infected with HCMV AD169 for 24 hr. Very high level increase in IFIT1 expression was observed in astrocytes transduced with lv/IFIT1/EGFP, while medium level increase was detected in non‐transduced astrocytes (Fig. 3D8)
Knockdown of IFIT1 and Its Effect on Viral Replication
To investigate whether knockdown of IFIT1 expression modulates HCMV replication, three small interference RNAs (shRNAs) for knocking down the expression of IFIT1 were synthesized. Astrocytes were transduced with control (scrambled) shRNA (lv/ctl/EGFP) or IFIT1 shRNA lentivirus (lv/shIFIT1/EGFP) for 48 hr, and then the cells were infected with HCMV AD169 at m.o.i = 2.5 for an additional 24 or 48 hr to determine antiviral replication effects of IFIT1. The results suggested that neither shRNA‐1 nor shRNA‐2 affected the expression of other ifit (ifit2, 3, 5) genes (Fig. 4A1). The inhibitory rate of shRNA‐1 was higher than that of shRNA‐2.Therefore, shRNA‐1 was chosen as an interference tool for all subsequent studies and the inhibitory property of shRNA‐1 was determined by western blot. The results showed significant inhibition in lv/shRNA/EGFP transduced group compared with lv/ctl/EGFP transduced group (Fig. 4A2).
Figure 4.
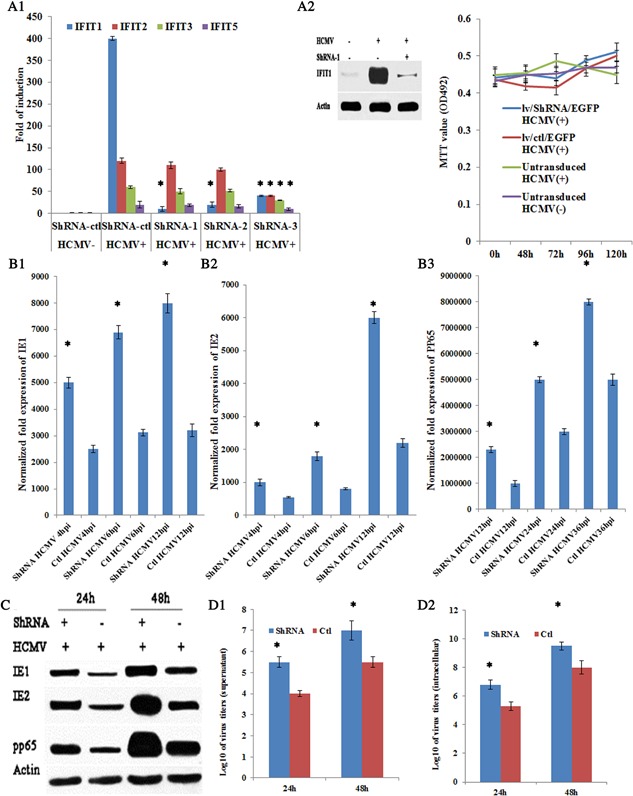
Knockdown of IFIT1 increased HCMV replication. A1, mRNA of ifit genes was determined by RT‐qPCR with treatment of shIFIT1‐1, shIFIT1‐2, or shIFIT1‐3; A2, protein expression of IFIT1 after treatment with lv/shRNA‐1/EGFP; B1–B3, mRNA expression of IE1, IE2, and pp65 by RT‐qPCR. C, protein expression of IE1, IE2, and pp65 after treatment with lv/shIFIT‐1/EGFP; D1, virus titer of supernatant; D2, intracellular virus titer. E, MTT analysis. HCMV−, mock infected astrocytes, HCMV+, HCMV infected astrocytes; *, statistical significance P < 0.05.
Total RNA and protein were isolated at 6 or 72 hr p.i and analyzed for HCMV replication by qRT‐PCR or western blot. The results showed that mRNA and protein expression of IE1, IE2, and pp65 were all markedly enhanced in lv/shRNA/EGFP transduced cells compared with control group (Fig. 4B1–B3 and C). Then HCMV infectivity of viral titers was detected. Results showed titer of inside and outside of cells increased in cells deficient in IFIT1 compared with control groups (Fig. 4D1 and D2). Finally, MTT analysis for cell viability and proliferation was also performed, which showed no significant difference either in lv/shRNA/EGFP transduced or lv/ctl/EGFP transduced groups at 0, 48, 72, 96, 120 hr p.i (P < 0.05) (Fig. 4E), which evidenced it was lv/shRNA/EGFP that mediated inhibition of IFIT1 antiviral effects. Taken together, the results demonstrated that HCMV replication increased in IFIT1 knockdown astrocytes compared with control.
DISCUSSION
HCMV is a widespread human pathogen. The symptoms of primary HCMV infection in healthy adults are generally mild, but infection during first trimester gestation can lead to serious neurodevelopment sequelea in infants. Fetal CNS is especially vulnerable to CMV‐induced injury [Poland et al., 1994; McCarthy et al., 2000]. Astrocytes are the most abundant cell type found in the CNS, outnumbering neurons 10‐1. These glial cells have the capacity to react to a variety of CNS disturbances such as viral infection through the production of immune mediators and it is well established that primary human astrocytes are fully permissive for CMV replication [Ho et al., 1991; Lokensgard et al., 1999]. Host antiviral responses to HCMV begin quickly after virus‐cell contact involving interaction with a growing list of pattern recognition receptors that initiate intracellular signaling and consequent expression and secretion of numerous immunologically active cytokines.
In this study, HCMV infected human fetal astrocytes were studied for the effects HCMV on genome transcription of astrocytes and antiviral activity of astrocytes. Firstly, we performed genome wide gene expression of HCMV infected human fetal astrocytes in vitro. With a 2.0‐fold change as a cutoff, it showed HCMV infection led to extensive dysregulation of cellular gene expression in astrocytes. One hundred and fifty‐four gene expressions (138 up‐regulated, 16 down‐regulated) were modulated in HCMV infected astrocytes compared with mock infected group at 6 hr p.i. GO analysis showed the top five categories in biological process and all belonged to cell response to stimuli (Table II). ISGs involved in antiviral response process in early infection time [Ho and van den Pol, 2007] exhibited the highest fold changes in HCMV infected astrocytes compared with mock infected group at 6 hr p.i. However, this result was not consistent with the finding that microglia not astrocytes could produce antiviral cytokines in response to CMV infection [Lokensgard et al., 2002]. The reason could be that the response to HCMV infection in fetal astrocytes was different from mature astrocytes or cell lines. These up‐regulated ISGs included IFIT1, IFIT2, IFIT3, IFITM1, MXA, MXB, OAS1, OAS2, OASL, USP18, USP41, IFI44, and BST2. It also showed that gene expression levels of chemokine and chemokine receptors (IL‐11, MCP‐1, IL‐8, RANTES, IP‐10, and MIP‐1α, Table III) were up‐regulated compared to mock infected group at 6 hr p.i. This result was consistent with the report that CMV‐infected human brain vascular pericytes and astrocytes released substantial amounts of the chemokines MCP‐1, MIP‐1α, and IL‐8 [Lokensgard et al., 2002; Alcendor et al., 2012]. Several studies have demonstrated that chemokine play important roles in regulating brain inflammation during viral encephalitis because of their chemotactic properties, and the importance of CCL5, CCL3, CCL4, IL‐11, IL‐8, and MCP‐1 in regulating immune responses in the CNS have been emphasized [Weiss et al., 1998; Kutsch et al., 2000; Zlotnik, 2000]. Our results showed different fold changes of these cytokines and chemokines in HCMV infected astrocytes (Table III). Additionally, it has also been reported that early local expression of cytokines with antiviral properties in the CNS such as TNFα, IFNα, IFNβ, and IFNɣ correlates with control of viral replication. IFN treatment induces the expression of a large set of IFN‐stimulated genes (ISGs), whose protein products prevent infection with many pathogens, including HCMV [Samuel, 2001]. In present study, the results showed different fold changes with IL‐1β, IL‐8, IL‐6 in HCMV infected astrocytes compared with mock infected group at 6 hr p.i, while TNFα, IFNα, IFNβ, or IFNɣ exhibited little gene expression level in HCMV or mock infected astrocytes and fold changes were no statistical significance in HCMV infected astrocytes compared with mock infected control. The results suggested that HCMV infected astrocytes defended themselves against HCMV infection through expressing of ISGs, cytokines, and chemokines which either exert direct antiviral activities, or recruit antiviral immune cells such as microglia to the sites of infection [Peterson et al., 1997] and ISGs exerted important role in antiviral action. However, it is also plausible that only a subset of ISGs is essential for protection against a specific pathogen or groups of related pathogens. Thus, identification of pathogen‐specific ISGs might lead to development of targeted therapeutics.
Table III.
Astrocytes Responsiveness With HCMV AD169 Infection
| Gene name | Up/down regulation | Fold change | P‐value |
|---|---|---|---|
| RIG‐I | Up | 10.2 | <0.05 |
| IPS‐1 | Up | 3.2 | <0.05 |
| MyD88 | Up | 3.2 | <0.05 |
| TRIF | Up | 2.2 | <0.05 |
| IRF3 | Up | 2.3 | <0.05 |
| IRF7 | Up | 11.3 | <0.05 |
| IRF9 | Up | 5.2 | <0.05 |
| STAT1 | Up | 2.2 | <0.05 |
| STAT2 | Up | 3.2 | <0.05 |
| NF‐κB | Up | 2.1 | <0.05 |
| IFNβ | — | — | >0.05 |
| IFNα1 | — | — | >0.05 |
| IFNλ3 | — | — | >0.05 |
| IFNɣ | — | — | >0.05 |
| TNFα | — | — | >0.05 |
| IL‐1β | Up | 4.3 | <0.05 |
| IL‐6 | Up | 10.1 | <0.05 |
| IL‐10 | — | — | — |
| RANTES | Up | 31.1 | <0.05 |
| IL‐8 | Up | 7.1 | <0.05 |
| MIP‐1α | Up | 7.1 | <0.05 |
| MCP‐1 | Up | 5.3 | <0.05 |
| CXCL2 | Up | 12.1 | <0.05 |
| IL‐11 | Up | 3.5 | <0.05 |
| IL‐4 | — | — | — |
Fold Change cut‐off, 2.0; Condition pairs, 6 hr p.i vs con; IFN, interferon; IL, interleukin; IP, IFN‐inducible protein; MCP, monocyte chemotactic protein; MIP, macrophage inflammatory protein; —, no/very low expression; TNF, tumor necrosis factor. hr p.i, hour post infection; con, mock infected control.
Secondly, IFIT1 with the highest Fc among all up‐regulated genes (6 hr p.i vs con) was studied for its antiviral effects. Overexpression Lentivirus lv/IFIT1/EGFP gene was transduced into astrocytes or HELFs, respectively. Cells were infected with HCMV AD169 for 72 hr. Cell appearance showed no change in lv/IFIT1/EGFP transduced astrocytes compared with lv/ctl/EGFP transduced control group at 72 hr p.i, while morphological changes were observed in lv/IFIT1/EGFP and lv/ctl/EGFP transduced HELFs. The different antiviral effects of IFIT1 on astrocytes and HELFs might be caused different expression of cellular factors, which may be responsible for the modulation of viral gene expression [Towler et al., 2012], and reinforce the reason to investigate the specific antiviral replication role of IFIT1 in fetal astrocytes. Furthermore, mRNA expression of IE1, IE2, and pp65 were detected by QRT‐PCR at 4, 6, 12 hr p.i and protein expression were detected by western blot at 24, 48 hr p.i, due to the delayed expression of protein compared to mRNA. The results showed that both mRNA and protein expressions were significantly reduced compared with lv/ctl/EGFP transduced control (P < 0.001). HCMV replication cycle in fetal astrocytes was 48–72 hr (showed in our previous study). Therefore, the release of HCMV infectious particles in cell culture medium and HCMV titer within astrocytes were determined at 48 and 72 hr p.i. The results showed significant decreases in the release of HCMV infectious particles in cell culture medium and in HCMV titer within astrocytes in lv/IFIT1/EGFP transduced astrocytes compared with the control (P < 0.001).
Finally, to further verify the antiviral effects of IFIT1, shIFIT1‐1 was used for knocking down IFIT1 gene. After shIFIT1‐1 lentivirus was transduced into astrocytes for 48 hr, cells were infected with HCMV AD169. Viral mRNA, protein expression levels and HCMV titers were determined, which showed significant increases in mRNA expressions of IE1, IE2, pp65, and viral titers in knockdown group compared with the control (P < 0.01), suggesting the antiviral replication effects of IFIT1 in fetal astrocytes. This is the first report on antiviral effects of IFIT1 in human fetal astrocytes to date and these findings also pointed to the potential importance of studying human primary first‐trimester gestation astrocytes because the antiviral effects of particular cytokines depend on specific cell surface receptors that may not be present or functional on established glial cell lines.
Functional studies have shown that IFITs inhibit the translation of viral proteins via different mechanisms. In early studies, IFITs were found to bind eukaryotic initiation factor 3 (eIF3) and interfere with viral protein translation [Hui et al., 2003]. Recent studies showed that IFITs can distinguish the different features between host and viral RNAs, block West Nile virus, Japanese encephalitis virus, and coronavirus mutants that lack 2′‐O‐methyltransferase activity. Inhibition by IFIT1 occurs by preferentially sequestering capped RNA lacking a 2′‐O‐methyl group and preventing eukaryotic translation initiation factors from binding to the RNA template specifically [Daffis et al., 2010; Habjan et al., 2013; Kimura et al., 2013]. IFIT1 was also identified as an eIF3‐associated protein fractionating with hepatitis C virus (HCV) translation complexes in IFN‐treated cells [Wang et al., 2003]. IFIT1 blocks translation driven by the HCV internal ribosome entry site (IRES) and the inhibition depends on eIF3 binding by IFIT1. However, another study suggested IFIT1 preferentially inhibits cap‐dependent translation but not translation of the EMCV IRES [Hui et al., 2003], since the EMCV IRES is different from that of HCV and functions by a different mechanism. IFIT1 also inhibits human papillomavirus DNA replication by binding to the viral protein E1 [Terenzi et al., 2008]. Nevertheless, several important questions still remain to be addressed. Our data verified that IFIT1 possessed anti‐HCMV function by inhibiting virus replication in human fetal astrocytes. However, the underlying mechanism remains unclear. There are possibilities that IFIT1 inhibits DNA replication by binding to the viral protein, or by suppressing HCMV protein translation by binding eIF3. It will be interesting to ponder new information on the uncovered the processes described here and on how to exploit for novel clinical antiviral or therapeutic strategies. Further studies are necessary to look into the precise role of this effector molecule.
In conclusion, this study provided insights into the function of IFIT1 as a key factor in antiviral defensing, and regulation of IFIT1 expression level is effective for modulating viral replication in HCMV infected human fetal astrocytes. The findings of present study might contribute to the development of targeted therapeutics towards fetal brain HCMV infection.
ETHICAL STANDARD
The authors ensured that all procedures were performed in compliance with relevant laws and institutional guidelines and that the Qingdao University Human Investigation Committee has approved them. There was informed consent prior to obtaining the fetus.
AUTHORS' CONTRIBUTIONS
All authors had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. Study concept and design: Bin Wang. Acquisition of data: Li Zhang. Analysis and interpretation of data: Li Zhang, Ling Li, Dong‐meng Qian. Drafting of the manuscript: Li Zhang, Hong Yu, Mei‐lan Xue. Critical revision of the manuscript for important intellectual content: Ming Hu, Xu‐xia Song. Statistical analysis: Mei‐lan Xue, Dong‐meng Qian. Obtained funding: Bin Wang, Li Zhang. Administrative, technical, and material support: Xu‐xia Song. Study supervision: Bin Wang.
ACKNOWLEDGMENTS
The authors thank the Pasteur Institute in France for kindly providing us the HCMV AD169. They also thank the Department of Gynecology and Obstetrics of the Affiliated Hospital of Qingdao University School of Medicine for providing human fetus preparation. Useful suggestions and critical revision given by Dr. Guo‐ying Wang of Qingdao University are also acknowledged.
This article was published online on 9 September 2016. Subsequently an error was found in affiliation and corresponding field, and the correction was published on 14 October 2016.
Conflicts of interest: None.
REFERENCES
- Abbas YM, Pichlmair A, Gorna MW, Superti‐Furga G, Nagar B. 2013. Structural basis for viral 5'‐PPP‐RNA recognition by human IFIT proteins. Nature 494:60–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcendor DJ, Charest AM, Zhu WQ, Vigil HE, Knobel SM. 2012. Infection and upregulation of proinflammatory cytokines in human brain vascular pericytes by human cytomegalovirus. J Neuroinflammation 9:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boppana SB, Fowler KB, Pass RF, Rivera LB, Bradford RD, Lakeman FD, Britt WJ. 2005. Congenital cytomegalovirus infection: Association between virus burden in infancy and hearing loss. J Pediatr 146:817–823. [DOI] [PubMed] [Google Scholar]
- Carson MJ, Sutcliffe JG. 1999. Balancing function vs. self defense the CNS as an active regulator of immune responses. J Neurosci Res 55:1–8. [DOI] [PubMed] [Google Scholar]
- Colugnati FA, Staras SA, Dollard SC, Cannon MJ. 2007. Incidence of cytomegalovirus infection among the general population and pregnant women in the United States. BMC Infect Dis 7:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daffis S, Szretter KJ, Schriewer J, Li Youn JS, Errett J, Lin TY, Schneller S, Zust R, Dong H, Thiel V, Sen GC, Fensterl V, Klimstra WB, Pierson TC, Buller RM, Gale M, Jr. , Shi PY, Diamond MS. 2010. 2'‐O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature 468:452–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandvaux N, Servant MJ, tenOever B, Sen GC, Balachandran S, Barber GN, Lin R, Hiscott J. 2002. Transcriptional profiling of interferon regulatory factor 3 target genes: Direct involvement in the regulation of interferon‐stimulated genes. J Virol 76:5532–5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Peters KL, Sen GC. 2000. Induction of the human protein P56 by interferon, double‐stranded RNA, or virus infection. Virology 267:209–219. [DOI] [PubMed] [Google Scholar]
- Habjan M, Hubel P, Lacerda L, Benda C, Holze C, Eberl CH, Mann A, Kindler E, Gil‐Cruz C, Ziebuhr J, Thiel Pichlmair VA. 2013. Sequestration by IFIT1 impairs translation of 2'O‐unmethylated capped RNA. PLoS Pathog 9:e1003663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho WS, van den Pol AN. 2007. Bystander attenuation of neuronal and astrocyte intercellular communication by murine cytomegalovirus infection of glia. J Virol 81:7286–7292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho WZ, Song L, Douglas SD. 1991. Human cytomegalovirus infection and trans‐activation of HIV‐1 LTR in human brain‐derived cells. J Acquir Immune Defic Syndr 4:1098–1106. [PubMed] [Google Scholar]
- Hui DJ, Bhasker CR, Merrick WC, Sen GC. 2003. Viral stress‐inducible protein p56 inhibits translation by blocking the interaction of eIF3 with the ternary complex eIF2. GTP. Met‐tRNAi. J Biol Chem 278:39477–39482. [DOI] [PubMed] [Google Scholar]
- Imaizumi T, Numata A, Yano C, Yoshida H, Meng P, Hayakari R, Xing F, Wang L, Matsumiya T, Tanji K, Tatsuta T, Murakami M, Tanaka H. 2014. ISG54 and ISG56 are induced by TLR3 signaling in U373MG human astrocytoma cells: Possible involvement in CXCL10 expression. Neurosci Res 84:34–42. [DOI] [PubMed] [Google Scholar]
- Kimura T, Katoh H, Kayama H, Saiga H, Okuyama M, Okamoto T, Umemoto E, Matsuura Y, Yamamoto M, Takeda K. 2013. Ifit1 inhibits Japanese encephalitis virus replication through binding to 5' capped 2'‐O unmethylated RNA. J Virol 87:9997–10003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutsch O, Oh J, Nath A, Benveniste EN. 2000. Induction of the chemokines interleukin‐8 and IP‐10 by human immunodeficiency virus type 1 tat in astrocytes. J Virol 74:9214–9221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb JR, Tugendreich S, Hieter P. 1995. Tetratrico peptide repeat interactions: To TPR or not to TPR? Trends Biochem Sci 20:257–259. [DOI] [PubMed] [Google Scholar]
- Levy DN, Aldrovandi GM, Kutsch O, Shaw GM. 2004. Dynamics of HIV‐1 recombination in its natural target cells. Proc Natl Acad Sci USA 101:4204–4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokensgard JR, Cheeran MC, Gekker G, Hu S, Chao CC, Peterson PK. 1999. Human cytomegalovirus replication and modulation of apoptosis in astrocytes. J Hum Virol 2:91–101. [PubMed] [Google Scholar]
- Lokensgard JR, Cheeran MC, Hu S, Gekker G, Peterson PK. 2002. Glial cell responses to herpesvirus infections: Role in defense and immunopathogenesis. J Infect Dis 186:S171–S179. [DOI] [PubMed] [Google Scholar]
- Luo MH, Hannemann H, Kulkarni AS, Schwartz PH, O'Dowd JM, Fortunato EA. 2010. Human cytomegalovirus infection causes premature and abnormal differentiation of human neural progenitor cells. J Virol 84:3528–3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy M, Auger D, Whittemore SR. 2000. Human cytomegalovirus causes productive infection and neuronal injury in differentiating fetal human central nervous system neuroepithelial precursor cells. J Hum Virol 3:215–228. [PubMed] [Google Scholar]
- Munir M, Berg M. 2013. The multiple faces of proteinkinase R in antiviral defense. Virulence 4:85–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peisley A, Hur S. 2013. Multi‐level regulation of cellular recognition of viral dsRNA. Cell Mol Life Sci 70:1949–1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson PK, Hu S, Salak‐Johnson J, Molitor TW, Chao CC. 1997. Differential production of and migratory response to beta chemokines by human microglia and astrocytes. J Infect Dis 175:478–481. [DOI] [PubMed] [Google Scholar]
- Pichlmair A, Lassnig C, Eberle CA, Gorna MW, Baumann CL, Burkard TR, Burckstummer T, Stefanovic A, Krieger S, Bennett KL, Rulicke T, Weber F, Colinge J, Muller M, Superti‐Furga G. 2011. IFIT1 is an antiviral protein that recognizes 5'‐triphosphate RNA. Nat Immunol 12:624–630. [DOI] [PubMed] [Google Scholar]
- Poland SD, Bambrick LL, Dekaban GA, Rice GP. 1994. The extent of human cytomegalovirus replication in primary neurons is dependent on host cell differentiation. J Infect Dis 170:1267–1271. [DOI] [PubMed] [Google Scholar]
- Samuel CE. 2001. Antiviral actions of interferons. Clin Microbiol Rev 14:778–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinzger C, Jahn G. 1996. Human cytomegalovirus cell tropism and pathogenesis. Intervirology 39:302–319. [DOI] [PubMed] [Google Scholar]
- Stagno S, Whitley RJ. 1985. Herpesvirus infections of pregnancy. Part I: Cytomegalovirus and Epstein‐Barr virus infections. N Engl J Med 313:1270–1274. [DOI] [PubMed] [Google Scholar]
- Terenzi F, Hui DJ, Merrick WC, Sen GC. 2006. Distinct induction patterns and functions of two closely related interferon‐inducible human genes, ISG54 and ISG56. J Biol Chem 281:34064–34071. [DOI] [PubMed] [Google Scholar]
- Terenzi F, Saikia P, Sen GC. 2008. Interferon‐inducible protein, P56, inhibits HPV DNA replication by binding to the viral protein E1. EMBO J 27:3311–3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towler JC, Ebrahimi B, Lane B, Davison AJ, Dargan DJ. 2012. Human cytomegalovirus transcriptome activity differs during replication in human fibroblast, epithelial and astrocyte cell lines. J Gen Virol 93:1046–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacher C, Muller M, Hofer MJ, Getts DR, Zabaras R, Ousman SS, Terenzi F, Sen GC, King NJ, Campbell IL. 2007. Coordinated regulation and widespread cellular expression of interferon‐stimulated genes (ISG) ISG‐49, ISG‐54, and ISG‐56 in the central nervous system after infection with distinct viruses. J Virol 81:860–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Pflugheber J, Sumpter R, Jr. , Sodora DL, Hui D, Sen GC, Gale M Jr. 2003. Alpha interferon induces distinct translational control programs to suppress hepatitis C virus RNA replication. J Virol 77:3898–3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wathelet M, Moutschen S, Defilippi P, Cravador A, Collet M, Huez G, Content J. 1986. Molecular cloning, full‐length sequence and preliminary characterization of a 56‐kDa protein induced by human interferons. Eur J Biochem 155:11–17. [DOI] [PubMed] [Google Scholar]
- Weiss JM, Downie SA, Lyman WD, Berman JW. 1998. Astrocyte‐derived monocyte‐chemoattractant protein‐1 directs the transmigration of leukocytes across a model of the human blood‐brain barrier. J Immunol 161:6896–6903. [PubMed] [Google Scholar]
- Zhang L, Li L, Wang B, Qian DM, Song XM, Hu M. 2013. Human cytomegalovirus infection modulates thrombospondins 1 and 2 in primary fetal astrocytes. Neuroreport 24:526–535. [DOI] [PubMed] [Google Scholar]
- Zhang L, Li L, Wang B, Qian DM, Song XX, Hu M. 2014. HCMV induces dysregulation of glutamate uptake and transporter expression in human fetal astrocytes. Neurochem Res 39:2407–2418. [DOI] [PubMed] [Google Scholar]
- Zlotnik A, Yoshie O. 2000. Chemokines: A new classification system and their role in immunity. Immunity 12:121–127. [DOI] [PubMed] [Google Scholar]


