Short abstract
First cross‐sectional expression profile of SAMHD1 in human tissue provides insight into its regulation on HIV target cells and effects its expression or phosphorylation state by proinflammatory cytokines.
Keywords: innate immunity, restriction factor, HIV, HSV
Abstract
The deoxynucleoside triphosphate triphosphohydrolase and 3′ → 5′ exonuclease SAMHD1 restricts HIV‐1 infection in noncycling hematopoietic cells in vitro, and SAMHD1 mutations are associated with AGS. Little is known about the in vivo expression and functional regulation of this cellular factor. Here, we first assessed the SAMHD1 protein expression profile on a microarray of 25 human tissues from >210 donors and in purified primary cell populations. In vivo, SAMHD1 was expressed in the majority of nucleated cells of hematopoietic origin, including tissue‐resident macrophages, DCs, pDCs, all developmental stages of thymic T cells, monocytes, NK cells, as well as at lower levels in B cells. Of note, SAMHD1 was abundantly expressed in HIV target cells residing in the anogenital mucosa, providing a basis for its evaluation as a cellular factor that may impact the efficiency of HIV transmission. Next, we examined the effect of the activation status and proinflammatory cytokine treatment of cells on expression and phosphorylation of SAMHD1. Activated, HIV‐susceptible CD4+ T cells carried pSAMHD1(T592), whereas resting CD4+ T cells and macrophages expressed the unphosphorylated protein with HIV‐restrictive activity. Surprisingly, stimulation of these primary cells with IFN‐α, IFN‐γ, IL‐4, IL‐6, IL‐12, IL‐18, IL‐27, or TNF‐α affected neither SAMHD1 expression levels nor threonine 592 phosphorylation. Only IL‐1β moderately down‐regulated SAMHD1 in activated CD4+ T cells. Taken together, this study establishes the first cross‐sectional protein expression profile of SAMHD1 in human tissues and provides insight into its cell cycle‐dependent phosphorylation and unresponsiveness to multiple proinflammatory cytokines.
Abbreviations
- AGS
Aicardi‐Goutières syndrome
- BDCA
blood dendritic cell antigen
- DC
dendritic cell
- DC‐SIGN
dendritic cell‐specific ICAM
- IFI16
IFN‐γ‐inducible protein 16
- MDDC
monocyte‐derived dendritic cell
- MDM
monocyte‐derived macrophage
- MLC
myosin light chain
- MxA
myxovirus resistance A
- NP‐40
Nonidet P‐40
- pDC
plasmacytoid dendritic cell
- pSAMHD1(T592)
phosphorylation of sterile alpha motif and histidine/aspartic acid domain‐containing protein 1 at threonine 592
- SAMHD1
sterile alpha motif and histidine/aspartic acid domain‐containing protein 1
- TAg
SV40 large T antigen
- TMA
tissue microarray
Introduction
Mutations within the SAMHD1 gene lead to a rare genetic disorder, the AGS, a primary immunodeficiency disorder that resembles an immune system's response pattern to certain chronic viral infections, displaying a massive production of the antiviral cytokine IFN‐α and a detrimental activation of the immune system [1]. The deficiency in SAMHD1 expression results in a complex disease characterized by hepatosplenomegaly, a chronic cerebrospinal fluid lymphocytosis, and an early‐onset encephalopathy that leads to severe intellectual and physical handicap [2]. The potential involvement of SAMHD1 in innate immunity had been suggested initially by studies of its mouse ortholog Mg11, which is IFN inducible in macrophages and DCs [3]. Subsequent studies showed an increased SAMHD1 expression upon stimulatory DNA treatment of macrophages [4, 5] and its up‐regulation in the context of viral infections [6, 7–8]. Whereas the SAM domain mediates protein–protein interactions, the HD domain possesses dNTP triphosphohydrolase activity [9, 10], through which SAMHD1 hydrolyzes dNTPs to deoxynucleosides.
SAMHD1 was identified as a potent restriction factor for HIV [11, 12], other nonhuman retroviruses [13], and herpesviruses, including HSV‐1 [14, 15]. For HIV, the current mode‐of‐action model suggests that SAMHD1, through its enzymatic function as a dNTP triphosphohydrolase, diminishes intracellular dNTP pools in noncycling cells, decreasing their concentrations below a threshold required for efficient HIV‐1 cDNA synthesis by the viral RT [13, 16, 17–18]. Furthermore, a metal‐dependent 3′ → 5′ exonuclease activity of SAMHD1 against ssDNAs and ssRNAs was demonstrated in vitro [19]. The RNAse activity was suggested recently to play a pivotal role for the HIV‐restrictive activity of SAMHD1 by directly degrading the HIV‐1 RNA of incoming particles [20].
A few studies in cultured HIV target cells have documented SAMHD1 expression in monocytes, MDMs, MDDCs, pDCs, and CD4+ T cells [12, 16, 21, 22, 23–24]. However, sites of SAMHD1 expression in vivo have not been systematically defined.
The primary aim of our study was thus to establish the in vivo expression profile of SAMHD1 in human tissues, with a focus on defining cells that harbor this nucleotide metabolism‐modulating enzyme and that are also target cells of HIV‐1 and HSV. In the context of its HIV‐restrictive activity, we also sought to assess the effect of T cell activation and stimulation with a number of proinflammatory cytokines on the overall expression level, as well as pSAMHD1(T592). In addition, we aimed at identifying SAMHD1‐expressing cells that may play a role in AGS pathophysiology.
MATERIALS AND METHODS
Paraffinized tissues
All paraffinized tissue specimens were provided by the tissue bank of the National Center for Tumor Diseases (Heidelberg, Germany) and their use approved by Heidelberg University's Ethics Committee (Germany; Approval No. 206/2005). Based on these specimens, an in‐house, whole‐body TMA containing a total of 468 paraffinized tissue sections from 25 different healthy organs from over 210 donors in total was assembled [25]. This TMA consists of a total of 5 coverslips, each containing 90–100 round sections (diameter of 2 mm) from paraffinized tissue blocks (see Fig. 1A and B, for examples). In more detail, histopathological tissue blocks, without evidence of tumor and/or inflammation, were selected from donors that had undergone surgery for non‐neoplastic reasons, i.e. tonsillitis, diverticulosis, and phimosis, or for neoplastic reasons, i.e. colorectal carcinoma or lung carcinoma. Other tissues, i.e., heart and brain, were derived from autopsies. In any case, the tissues used for the construction of the TMA were tumor and inflammation free, as assessed by histopathology. Donors were 60% men/boys and 40% women/girls, and ages were between 5 and 82 yr. Underlying conditions other than the indication for surgery have not been evaluated. Healthy tissues represented on the TMA analyses were salivary gland (n = 15), thyroid (n = 16), thymus (n = 10), lung (n = 20), heart (n = 21), esophagus (n = 21), stomach (n = 20), small intestine (n = 19), large intestine (n = 24), liver (n = 18), gallbladder (n = 14), spleen (n = 27), pancreas (n = 7), kidney (n = 24), adrenal gland (n = 5), bladder (n = 7), prostate (n = 25), testis (n = 22), uterus (n = 5), skin (n = 5), fat (n = 14), skeletal muscle (n = 11), ovary (n = 7), tonsil (n = 8), and appendix (n = 8). After immunohistochemical staining for SAMHD1, TMA slides were scanned for image acquisition by use of the Aperio ScanScope CS (Leica Microsystems, Buffalo Grove, IL, USA), analyzed with Aperio ImageScope software (Leica Microsystems), and rated, in principle, based on the immunoreactive scoring system of Allred et al. [26] and as recently reported [25]. Each section was assigned 2 scores: 1 score for the fraction of cells that stained positive (0, <1% positive cells; 1, 1–10% positive cells; 2, 11–33% positive cells; 3, 34–66% positive cells; 4, >66% positive cells) and another score for the intensity of staining (0, no staining; 1, weak staining; 2, intermediate staining; 3, strong staining). The overall amount of SAMHD1 was then calculated as the sum of the scores for the portion of positive, tissue‐specific cells and for staining intensity. A total score of 7 was categorized as high staining, scores of 5 and 6 as medium staining, and scores of 2–4 as low staining (see Fig. 1C).
Figure 1.
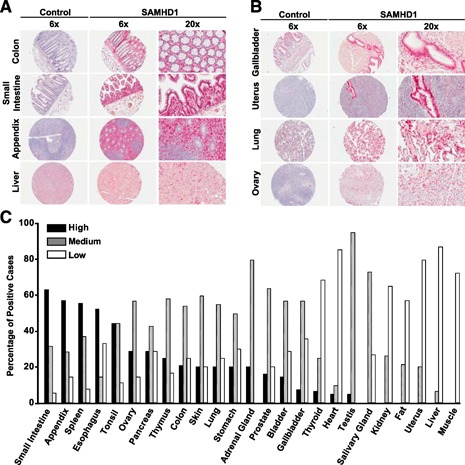
SAMHD1 is expressed in the majority of human tissues. (A and B) Images of immunohistochemical stainings of TMA slides. Stainings were performed with a specific polyclonal anti‐pan‐SAMHD1 antibody [21] (right) or a rabbit control serum (left), followed by biotinylated donkey anti‐rabbit secondary antibodies. Subsequently, sections were exposed to the avidin‐containing ABC‐AP kit, followed by substrate development with New Fuchsin. Nuclei were counterstained with hematoxylin. (A) Selected tissues, staining positive, mainly for resident or infiltrating mononuclear hematopoietic cells. (B) Selected tissues, displaying organ‐specific or specialized SAMHD1‐positive cells. (C) Rating of SAMHD1 expression in human tissues. The expression of SAMHD1 was quantified with a proportion and intensity scoring system [25, 26]. Histogram bars depict the percentage of cases with high (black), medium (gray), or low (open) expression ratings. The percentage of samples with negative ratings is not shown.
Immunoenzyme and double‐immunofluorescence staining
Antigen retrieval and immunoenzyme staining were performed as described [25]. Preimmune rabbit IgG served as a negative control. Immunoenzyme stainings of SAMHD1 were performed on 2 μm paraffin sections of formalin‐fixed tissues by use of standard avidin‐biotin antialkaline phosphatase techniques (Vectastain; Vector Laboratories, Burlingame, CA, USA; Super Sensitive Link‐Label IHC Detection System; BioGenex, Fremont, CA, USA). Slides were viewed with an Olympus BX45 microscope (Olympus, Center Valley, PA, USA), and pictures were taken with an Olympus U‐CMAD3 F‐View camera (Olympus).
Double‐immunofluorescence stainings were performed by use of pairwise combinations of primary antibodies. As a secondary reagent, biotinylated goat anti‐mouse antibodies were used for 30 min at room temperature, followed by Cy3‐conjugated streptavidin antibodies. The SAMHD1 rabbit primary reagent was directly detected by use of an anti‐rabbit antibody coupled to Alexa Fluor 488 (Invitrogen, Carlsbad, CA, USA). Pictures were taken at an Olympus IX81 microscope with an Olympus U‐CMAD3 F‐View camera and imported into cell^M software or with a Nikon Eclipse Ti microscope (Nikon Instruments, Melville, NY, USA) by use of an Orca‐Flash 2.8 camera (Hamamatsu, Hamamatsu City, Japan) and imported into NIS elements Basic Research software (Nikon Instruments). All primary and secondary antibodies used in this study are listed in Supplemental Table 1.
Immunoblotting
Cells and organ biopsies were resuspended directly in SDS lysis buffer, containing 4% SDS, or lysed in NP‐40 buffer (1% NP‐40, 150 mM NaCl, 40 mM Tris, pH 8) for 30 min on ice. Proteins were applied on a 10% or 12.5% SDS‐PAGE and subsequently transferred onto a nitrocellulose membrane. Blocked membranes were consecutively probed with rabbit (Proteintech Europe, Manchester, United Kingdom) or mouse (Abcam, Cambridge, United Kingdom) polyclonal anti‐SAMHD1 antiserum for detection of pan‐SAMHD1 expression and an anti‐MAPK and anti‐GAPDH (Santa Cruz Biotechnology, Santa Cruz, CA, USA) or anti‐MLC (Sigma‐Aldrich, St. Louis, MO, USA) antibody (Supplemental Table 1). For detection of pSAMHD1(T592), rabbits were immunized with peptide IAPLI(pT)PQKKE, and immunoreactive sera were affinity purified [22]. Cells were lysed and applied on a 4–20% gradient SDS‐PAGE. After transfer, polyvinylidene difluoride membranes were probed consecutively with primary antibodies/antisera for detection of the indicated proteins. Secondary antibodies were conjugated to HRP for ECL‐based detection.
Intracellular SAMHD1 staining
For fixation and antibody staining, PBMCs, freshly isolated tonsil cells, or thymocytes were fixed for 10 min at room temperature with BD Cytofix Fixation Buffer (BD Biosciences, San Jose, CA, USA) and permeabilized in BD Phosflow Perm Buffer III (BD Biosciences) for 2 min (PBMCs, tonsil) or 30 min (thymocytes) at 4°C in the dark. After 2 washes (PBS/1% FCS/0.09% sodium azide), cells were stained with the rabbit anti‐pan‐SAMHD1 antiserum, followed by Alexa 660‐conjugated goat anti‐rabbit antibodies and the respective directly labeled markers. A control rabbit antiserum as well as SAMHD1‐negative Jurkat‐TAg cells were always included as informative references (see also Fig. 2A). A FACSCalibur flow cytometer and BD CellQuest Pro 4.0.2 software (BD PharMingen, San Diego, CA, USA) were used for analyses. For flow cytometric analysis of thymocytes, cells were first stained in staining buffer (PBS, 1% BSA, and 0.09% sodium azide) for surface markers, followed by fixation and permeabilization for pan‐SAMHD1 staining, as described above. Flow cytometric analysis was carried out on an LSR II cytometer by use of FACSDiva software (both BD Biosciences). The live cell population was gated based on forward‐ and side‐scatter.
Figure 2.
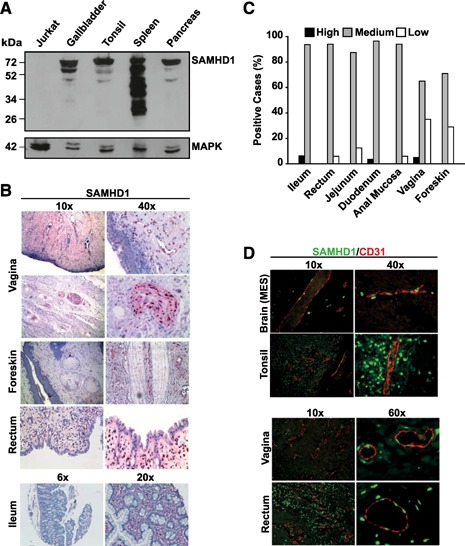
SAMHD1 is abundantly expressed in solid organs, gut‐associated lymphoid tissue, at mucosal sites of anogenital HIV transmission, and in CD31+ vascular endothelial cells in vivo. (A) Western blot analysis of SAMHD1 in homogenates from selected human tissues (gallbladder, tonsil, spleen, and pancreas) and SAMHD1‐negative Jurkat‐TAg cells. Cell lysates were separated by SDS‐PAGE, and nitrocellulose membranes were probed with a polyclonal rabbit anti‐SAMHD1 antiserum. MAPK: loading control. (B) Stainings of ileum, rectal, foreskin, and vaginal tissues were performed with a polyclonal anti‐SAMHD1 antibody, followed by incubation with secondary antibody solutions and substrate development with Permanent Red. Nuclei were counterstained with hematoxylin. (C) Rating of SAMHD1 expression in GALT and HIV transmission‐site tissues, according to the scoring system used in Fig. 1C. Histogram bars depict the percentage of cases with high (black), medium (gray), or low (open) expression ratings. The percentage of samples with negative ratings is not shown. (D) Immunofluorescence coexpression analysis of SAMHD1 (green nuclear staining) and vessel endothelium (CD31+, red membranous, and cytoplasmic staining) in selected human tissues. Shown are 2‐channel merged images. MES, Mesencephalon.
Cell lines and primary cell isolation
Human cell lines THP‐1 and Jurkat‐TAg were cultivated under standard conditions, as reported previously [27, 28]. PBMCs from healthy blood donors [approved by Frankfurt University's Ethics Committee (Approval No. 329/10), Germany] were purified by Ficoll‐Hypaque gradient centrifugation, in principle, as reported [29] Subsequently, cells were isolated by magnetic labeling of the respective lineage marker (MicroBeads; Miltenyi Biotec, San Diego, CA, USA), followed by magnetic separation (either positive or negative) via an AutoMACS (Miltenyi Biotec).
The following cell purities were obtained for the positively selected cells (see Fig. 3C): CD4+ T cells (99%), CD14+ monocytes (90–92%), CD20+ CD19‐selected B cells (66–72%), CD8+ T cells (97%), CD56+ NK cells (97%), CD11c+ MDDCs (92%). To obtain MDM, negatively separated CD14+ monocytes were differentiated in DMEM containing 10% FCS, 100 U/ml penicillin, 100 mg/ml streptomycin, and 10% human AB serum for 10 days. CD4+ T cells were isolated with the RosetteSep Human CD4+ T Cells Enrichment Cocktail (Stemcell Technologies, Vancouver, BC, Canada) and cultivated in the absence (resting) or presence (activated) of PHA‐P (5 µg/ml; Sigma‐Aldrich) and IL‐2 (100 IU/ml; Biomol, Enzo Life Sciences, Farmingdale, NY, USA) or anti‐CD3/anti‐CD28 antibodies (each 1 µg/ml; BD Biosciences) and IL‐2 (100 IU/ml; Biomol, Enzo Life Sciences), as described previously [21].
Figure 3.
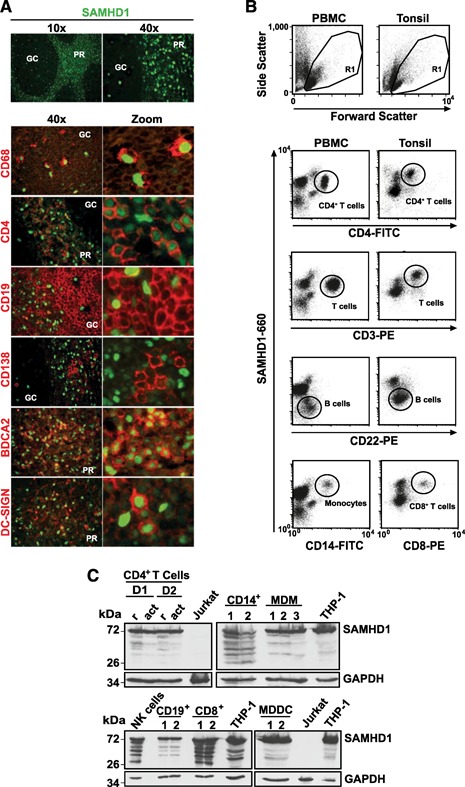
SAMHD1 is expressed in all cells of the hematopoietic compartment in vivo. (A) Expression analysis of SAMHD1 (green nuclear staining) alone (A, upper) and coexpression analysis in macrophages (red cytoplasmic CD68+ staining), CD4+ T cells (red membrane staining), B cells (surface CD19+ staining), plasma cells (CD138+), pDC (red surface BDCA2+ staining), or DC (red surface DC‐SIGN+ staining) in human tonsil tissue sections. Shown are 2‐channel merged images. GC, Germinal center; PR, perifollicular region. Expression analyses of SAMHD1 by (B) flow cytometry and (C) immunoblotting in different cell lineages circulating in peripheral blood (B, left, and C) or residing in tonsil (B, right). (B) Freshly isolated PBMCs or tonsil cells were fixed, permeabilized, and stained for SAMHD1, together with the respective lineage markers, and analyzed by flow cytometry. Shown are the forward‐/side‐scatter plots, with the R1 live gate indicated, and dot plots showing SAMHD1/lineage marker costainings. Subpopulations are circled. The data are representative of 3 (PBMC) and 2 (tonsil) donors analyzed. (C) Lysates of purified T cells, CD4+ (r, resting; act, activated) and CD8+, monocytes (CD14+), MDM, NK cells (CD56+), B cells (CD19+, CD22+), MDDC, and reference cell lines that express no (Jurkat‐TAg) or high (THP‐1) endogenous SAMHD1 levels were separated by SDS‐PAGE. D1, D2 indicate different donors; numbers on the left indicate the apparent molecular weight in kDa. Nitrocellulose membranes were probed with a polyclonal rabbit anti‐pan‐SAMHD1 antibody. GAPDH, Loading control.
Child thymus tissue, removed during cardiac surgery, was obtained and used following the guidelines of the Medical Ethical Commission of Ghent University Hospital (Belgium; B670201215078). Informed consent was obtained and samples used according to the Declaration of Helsinki. Thymus suspension was obtained after mechanical homogenization of the tissue, and mononuclear cells were isolated by density‐layer centrifugation (Lymphoprep; Axis‐Shield, Oslo, Norway). Tonsil tissue was removed during routine tonsillectomy from HIV‐, hepatitis B virus‐, and hepatitis C virus‐negative patients with informed consent. The use of anonymous surgical waste was approved by Heidelberg University's Ethics Committee (Approval No. 077/2005). Fresh tonsil cultures were prepared by collagenase digestion (collagenase type I; Worthington Biochemical, Lakewood, NJ, USA) at 37°C for 15 min, followed by mechanical homogenization and PBS washing. The resulting single‐cell suspension was directly stained and applied to flow cytometric analysis.
Cytokine treatment
For cytokine treatment, MDM or CD4+ T cells were incubated with IFN‐α (Roche, Basel, Switzerland); IFN‐γ, IL‐4, IL‐12, IL‐27, and TNF‐α (PeproTech, Rocky Hill, NJ, USA); and IL‐6 and IL‐1β (R&D Systems) or IL‐18 (MBL International, Woburn, MA, USA) at concentrations ranging between 50 and 10.000 IU/ml (or ng/ml) as indicated. After 24 h or 48 h, cells were lysed and applied to immunoblot analysis or fixed and stained for intracellular SAMHD1 and subsequent flow cytometric analysis.
Statistical analysis
To assess statistical significance, the unpaired Student's t‐test was used. P > 0.05 was considered not significant.
RESULTS
TMA‐based protein expression profiling of SAMHD1
A cross‐sectional SAMHD1 expression profiling was performed on a TMA containing 468 individual sections derived from 25 paraffinized nontransformed tissues from over 210 patients. First, we surveyed SAMHD1 protein expression on the TMA, based on a combined proportion and intensity scoring system, in principle, as reported [25, 26]. SAMHD1‐expressing cells were detected in all organs analyzed ( Fig. 1 and Supplemental Fig. 1A and B), with expression scores varying considerably among tissues: the scores of small intestine, appendix, spleen, esophagus, tonsil, ovary, pancreas, and thymus were highest (>80% medium or high‐expression score). Generally low or negative expression scores were determined for fatty tissue, uterus, liver, and muscle (Fig. 1 and Supplemental Fig. 1A and B).
In a complementary approach, strong SAMHD1 expression in homogenates of human tissue samples was confirmed by immunoblot analyses of snap‐frozen tissue samples of gallbladder, tonsil, spleen, and pancreas ( Fig. 2A ). The SAMHD1‐negative human T cell line Jurkat‐TAg served as a specificity control. Besides the major protein band at 72 kDa, seen in all tissues and reported previously in cell lines [11, 12], additional, prominent, lower molecular weight bands were noted, particularly in a spleen lysate (Fig. 2A) and some of the purified cell populations from peripheral blood ( Fig. 3C ), which may be a result of splice variants, post‐translational modifications, or partial proteolytic cleavage. Collectively, although expression levels were variable, SAMHD1 was detected in all organs and tissues analyzed.
SAMHD1 is abundantly expressed at primary HIV entry sites
Recently, SAMHD1, expressed in cultured monocytes, MDDCs, MDMs, and resting CD4+ T cells, was demonstrated to function as a restriction factor for HIV‐1 infection [11, 21, 23, 27, 30]. Given the potential role of SAMHD1 in limiting transmission of HIV‐1, we explored its expression in anogenital mucosa and gut‐associated lymphoid tissue. Immunohistochemical staining of vagina, foreskin, anal mucosa and rectum tissue sections revealed abundant, constitutive expression of the restriction factor in infiltrating and resident‐nucleated hematopoietic cells in mucosa and submucosa of these transmission site tissues (Fig. 2B–D and Supplemental Fig. 2 for IgG control staining). This expression analysis provides a basis for exploring the role of SAMHD1 in modulating the efficiency of establishment of an HIV‐1 infection in a new host.
SAMHD1 expression in organ‐specific and specialized cell types
By histomorphological criteria, we identified SAMHD1 expression in a number of specialized or organ‐specific cells in the TMA: pneumocytes in the lung, exocrine secretory ducts in salivary glands and pancreas, epithelium of the gallbladder and esophagus (stratum basale), endometrium of the uterus, stroma of the ovary, seminiferous tubules of the testis, cells in the adrenal gland with a stronger expression in the zona glomerulosa and zona reticularis compared with the zona fasciculata, keratinocytes in the basal layer of the skin, and gastric glands (Fig. 1A and B and Supplemental Fig. 1A and B). In addition, neurocytes of peripheral nerves in vagina stained highly positive for SAMHD1 (Fig. 2B). Furthermore, coimmunofluorescence analyses of SAMHD1 (primarily nuclear staining) and the endothelial marker CD31 (membranous‐cytoplasmic staining) revealed marked coexpression of the triphosphohydrolase/exonuclease in vascular endothelial cells in brain, tonsil, vagina, and rectum (Fig. 2D), as well as in liver and pancreas (data not shown).
Expression of SAMHD1 in hematopoietic cells
Next, we performed a systematic, in‐depth analysis of SAMHD1 expression in primary cells of hematopoietic origin. Based on flow cytometric codetection with lineage‐specific markers, histomorphological criteria, costainings in tissue sections, and immunoblotting of purified cell populations, SAMHD1 expression could be demonstrated in all hematopoietic lineages. In tissue, SAMHD1 expression was documented in CD68+ macrophages (Fig. 3A and Supplemental Fig. 3), DC‐SIGN+ DC, BDCA2+ pDC (Fig. 3A), and thymic and tonsillar CD4+ and CD8+ T cells ( Figs. 3 and 4 ). Isolated primary cell populations showed expression of SAMHD1 in CD14+ monocytes (Fig. 3B and C), MDM (Fig. 3C), MDDC (Fig. 3C), CD56+ NK cells (Fig. 3C), and peripheral CD4+ and CD8+ T cells ( Figs. 3B and C and 5A and B ). In CD19+ and CD22+ B cells, as well as CD138+ plasma cells, low‐level expression (Fig. 3B and C; by immunoblot and flow cytometry, see also Supplemental Fig. 4A) or even undetectable expression (Fig. 3A; by in situ immunofluorescence microscopy) of SAMHD1 was documented.
Figure 4.
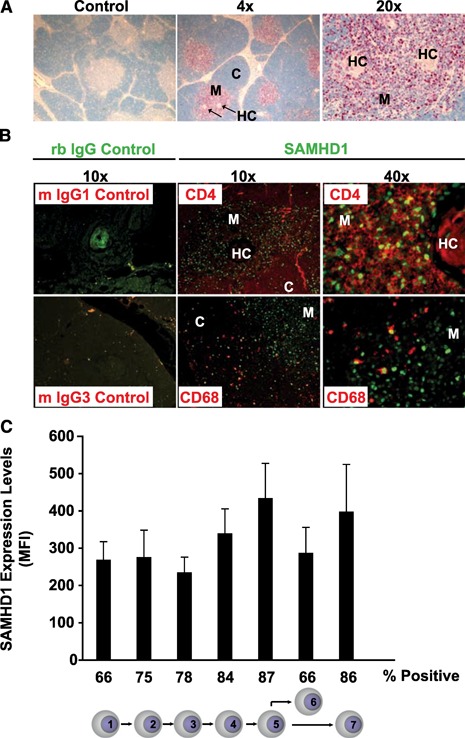
SAMHD1 expression in the human thymus. (A) Immunohistochemical analysis of pan‐SAMHD1 expression in thymic tissue sections. Control, Sections stained with preimmune rabbit serum (low power field); HC, Hassallˈs corpuscle; M, medulla; C, cortex. (B) Coexpression analysis of SAMHD1 (green nuclear staining) in CD4+ T cells (top, red CD4+ staining) and macrophages (lower, red CD68+ staining) by immunofluorescence microscopy. Control (left panel), Preimmune rabbit (rb) serum‐stained sections at low power field combined with murine (m) isotype controls IgG1 or IgG3, respectively. Two‐channel merged pictures are shown. (C) SAMHD1 expression analysis in developing human thymocytes ex vivo. The intracellular SAMHD1 expression levels in developing thymocyte populations were determined by flow cytometry (see Supplemental Fig. 5 for details). Histogram bars represent the arithmetic mean + sd of the mean fluorescence intensity (MFI) of SAMHD1 expression in the respective thymocyte subsets of 4 donors. Thymocyte differentiation was categorized into the 7 stages indicated: 1) CD34+CD1−, recent thymic immigrants; 2) CD3+CD4+CD8−, immature, single‐positive cells; 3) CD3−CD4+CD8+, double‐positive thymocytes undergoing β‐selection; 4) CD3+CD4+CD8+, double‐positive thymocytes that have undergone β‐selection; 5) CD3+CD27+CD69+, positively selected thymocytes; 6) CD3+CD4−CD8+, mature CD8+ thymic T cells; and 7) CD3+CD4+CD8−, mature CD4+ thymic T cells. The numbers indicated below the histogram represent the percentage of SAMHD1+ cells within the respective subset.
Figure 5.
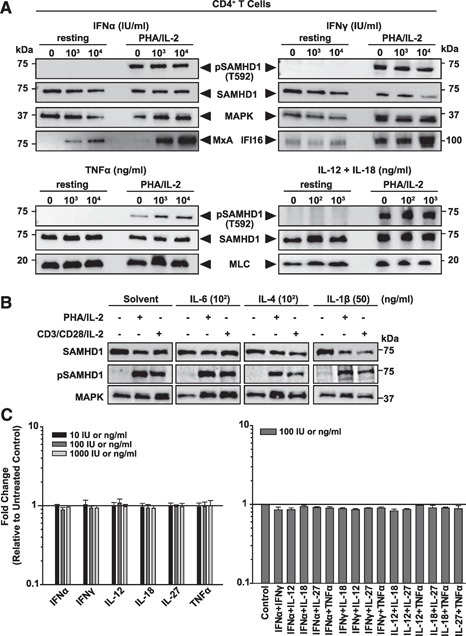
Impact of T cell activation and cytokine stimulation on SAMHD1 expression and phosphorylation. (A and B) Immunoblot analyses of CD4+ T cells after 24 h or 48 h of cytokine treatment, respectively. Cells were incubated with indicated concentrations of IFN‐α, IFN‐γ, TNF‐α, or IL‐12/IL‐18 (A), IL‐6, IL‐4, and IL‐1β (B), lysed and analyzed with anti‐pSAMHD1(T592) and anti‐pan‐SAMHD1 antibodies. MxA and IFI16 served as positive controls for IFN‐induced gene expression. MAPK and MLC: loading controls. Shown is 1 representative donor out of 3 donors (A) or 2 donors (B) for CD4+ T cells. (C) PBMC‐derived CD4+ T cells (left panel) and tonsil‐derived lymphoid aggregate cultures (right panel) were cultivated in the presence of the indicated cytokines or cytokine combinations for 24 h and subsequently analyzed for SAMHD1 expression by flow cytometry. Shown are arithmetic means + sem of the factor of change of the mean fluorescence intensity of cytokine‐treated cells relative to untreated control cells of a total of 3 donors (CD4+ T cells) and 2 donors (tonsil derived cells).
In the thymus, a lymphoid organ, particularly relevant for HIV pathogenesis [31, 32], SAMHD1, was expressed in various lymphoid and nonlymphoid cell types (Fig. 4 and Supplemental Fig. 5). The triphosphohydrolase/exonuclease was present during all stages of thymocyte development in 66–86% of cells starting from CD34+ CD1− thymic immigrants up to CD4+CD8+ and single‐positive thymocytes (Fig. 4C and Supplemental Fig. 5). There was a slight trend toward increased SAMHD1 expression with increasing thymocyte maturation. In addition, SAMHD1 was found in CD68+ macrophages in the thymic cortex and in mature and immature medullary epithelial cells (Fig. 4A and B, and data not shown).
Based on the identification of SAMHD1 expression in a large number of nucleated cell types of the hematopoietic lineage, the high SAMHD1 staining scores in the TMA in all lymphatic organs (e.g., thymus, spleen, tonsil; Fig. 1) and intestinal‐tract samples with associated lymphatic tissue (e.g., small intestine, appendix, colon; Figs. 1 and 2C), as well as low to medium scores in organs with a significant fraction of resident, transient, or infiltrating mononuclear hematopoietic cells (e.g., liver, thymus, gallbladder, kidney), are readily explained. Thus, this cross‐sectional analysis reveals that constitutive SAMHD1 expression can be found in the majority of nucleated cells of hematopoietic origin.
Impact of T cell activation and cytokine stimulation on SAMHD1 expression and phosphorylation
We next wanted to explore whether major proinflammatory cytokines can affect overall expression levels of SAMHD1. To this end, PBMC‐derived CD4+ T cells and tonsil‐derived lymphoid aggregate cultures (Fig. 5), as well as MDMs ( Fig. 6 ), were cultivated in the presence or absence of different concentrations of human IFN‐α, IFN‐γ, IL‐4, IL‐6, IL‐12, IL‐18, IL‐27, IL‐1β, or TNF‐α, alone or in combination, for 24 or 48 h. Subsequent analyses by immunoblotting (Figs. 5A and B and 6) and intracellular flow cytometry (Fig. 5C) revealed no significant cytokine‐mediated alterations of SAMHD1 expression levels. As a notable exception, the stimulation of activated CD4+ T cells by IL‐1β induced a moderate decrease of SAMHD1 expression (Fig. 5B). As controls of cytokine activity, treatment with IFN‐α or IFN‐γ stimulated MxA or IFI16 expression, respectively, in these HIV target cells (Figs. 5A and 6). We next explored whether the activation stimulus affects SAMHD1 expression levels in CD4+ T cells. To this end, CD4+ T cells were exposed to PHA‐P/IL‐2 or anti‐CD3/anti‐CD28 antibodies, together with IL‐2 (Fig. 5B). Neither of these T cell activation protocols significantly affected SAMHD1 levels, as reported previously [21, 24].
Figure 6.
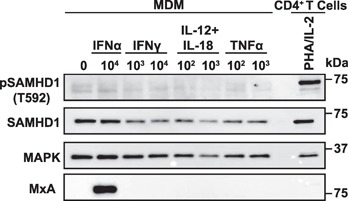
Impact of cytokine stimulation on SAMHD1 expression and phosphorylation in MDMs. Immunoblot analysis of MDM after 24 h of treatment with indicated concentrations of different cytokines (ng/ml for IL‐12/IL‐18/TNF‐α and IU/ml for IFN‐α/IFN‐γ). Cells were lysed and analyzed with anti‐pSAMHD1(T592)) and anti‐pan‐SAMHD1 antibodies. Activated CD4+ T cells served as positive control for pSAMHD1(T592) detection. MxA served as a positive control for IFN‐induced gene expression and MAPK as a loading control. Shown is 1 representative donor out of 7.
As pSAMHD1(T592) has been reported to correlate with a loss of its HIV‐restrictive capacity [23, 33], we also tested whether cytokine stimulation altered the phosphorylation status of SAMHD1 in these primary target cells. To this end, we generated an affinity‐purified, T592 phosphosite‐specific polyclonal rabbit antiserum. Nitrocellulose membranes with separated lysates from cytokine‐treated cells were reprobed with this antiserum. pSAMHD1(T592) was detectable in primary CD4+ T cells only following T cell activation but not in quiescent cells from the same donor (Fig. 5A and B). Remarkably, pSAMHD1(T592) expression was unaltered by cytokine treatment in resting and activated CD4+ T cells (Fig. 5A and B). In contrast to cycling CD4+ T cells, pSAMHD1(T592) was undetectable in differentiated MDM, and also for these myeloid cells, neither IFN‐α, IFN‐γ, IL‐12/IL‐18, nor TNF‐α stimulation had an impact on this status (Fig. 6). Collectively, neither T cell activation nor the addition of major proinflammatory cytokines, with the notable exception of IL‐1β, modulated SAMHD1ˈs expression levels or phosphorylation at threonine 592 in primary lymphoid and myeloid HIV target cells.
DISCUSSION
Our cross‐sectional in vivo profiling of the dNTP triphosphohydrolase/3′ → 5′ exonuclease SAMHD1 identified its widespread expression in nontransformed tissues, including a number of highly specialized and organ‐specific cell types throughout the human body, as well as in the majority of nucleated cell types of hematopoietic origin. Our whole‐body TMA expression approach allowed us to examine cellular in vivo targets of pathogenic viruses that have been shown to be inhibited by the nucleotide‐modulating antiviral factor SAMHD1 in tissue culture. For cells that can, in principle, be infected by HIV‐1 in vivo, SAMHD1 in situ expression was demonstrated in monocytes, macrophages, DCs, pDCs, and mature CD4+ T cells, as well as in all developmental stages of thymic T cells. Thus, virtually all major HIV target cells in an infected individual potentially carry the SAMHD1 protein. For cell types infected by HSV in vivo, vascular endothelial cells, keratinocytes, and neurocytes were found to express SAMHD1, also suggesting that this viral pathogen may frequently encounter the restriction factor in the context of its natural infection. In the lung, alveolar pneumocytes, which express the restriction factors SAMHD1 (present study), as well as CD317/tetherin [25], are targeted by various respiratory pathogens, including influenza viruses [34], severe acute respiratory syndrome coronavirus [35], and respiratory syncytial virus [36]. At least for CD317, influenza viruses seem to have evolved mechanisms of viral antagonism [37, 38]. For these respiratory viruses, it is currently unclear whether SAMHD1 impacts their replication and/or whether they encode a SAMHD1 antagonist similar to the viral protein X proteins of HIV‐2 and certain SIV strains [39].
Giventhe dNTP triphosphohydrolase and 3′ → 5′ exonuclease activities of SAMHD1 and the impact that these nucleotide‐modulating activities could have on cell‐cycle progression and cellular proliferation, we wondered whether its in vivo expression pattern was restricted to noncycling cells. This was clearly not the case: SAMHD1 was expressed in cycling, highly proliferative primary cells (e.g., endometrium and activated T cells), as well as noncycling cells (e.g., vascular endothelium, resting T cells, and macrophages). This broad expression pattern suggests that the regulation of the physiologic functions of SAMHD1 may occur by direct modulation of its enzymatic and antiviral activities rather than at the transcriptional/translational level. In line with this notion, recent studies reported that cyclin‐dependent kinase 1 mediated phosphorylation of SAMHD1 at residue T592 regulates its capacity to restrict HIV‐1 [22, 23]. Interestingly, pSAMHD1(T592) does not seem to regulate the protein's catalytic dNTPase activity [23, 33, 40], suggesting, at least in part, a regulatory uncoupling of its antiviral and enzymatic functions.
It is important to define which cytokines and stimuli affect expression and function of SAMHD1 in primary HIV target cells. Recently, Cribier et al. [33] reported the loss of pSAMHD1(T592) in activated CD4+ T cells and MDM following exposure to IFN‐α. Whereas in our study, pSAMHD1(T592) was also readily detected in CD4+ T cells upon TCR stimulation, in line with other reports [23, 33], we did not observe a reduction in SAMHD1 phosphorylation by treatment with IFN‐α or IFN‐γ, despite an apparent induction of type I and type II IFN‐positive controls MxA and IFI16, respectively. A recent study reported an ∼2‐fold induction of SAMHD1 expression in MDM by treatment with a combination of IL‐12 and IL‐18, each at 100 ng/ml [41]. We were neither able to confirm this cytokine‐mediated up‐regulation of the restriction factor at 100 or 1000 ng/ml nor did we observe changes in the pSAMHD1(T592) expression of cells. Likewise, this cytokine combination neither affected overall levels nor phosphorylation of SAMHD1 in CD4+ T cells. In line with our observation, St Gelais et al. [24] did not observe an increase in SAMHD1 levels in CD4+ T cells or MDDCs following exposure to type I IFN. Of note, IL‐1β was the only cytokine that down‐regulated SAMHD1 significantly in activated CD4+ T cells. However, it is unclear whether this translates into any change in the HIV susceptibility of cells, as SAMHD1 is not believed to pose a restriction for HIV in proliferating T cells. For assessment of the SAMHD1 status and cytokine responses in MDM, the monocyte isolation and differentiation protocol may be of particular relevance: the M‐CSF‐based rapid differentiation protocol (3 days) used in 1 study [22] resulted in strong pSAMHD1(T592) expression. In contrast, our results, in agreement with Allouch et al. [42], showed a low‐level or absent pSAMHD1(T592) expression in MDM differentiated with human AB serum for 7–10 days. Besides differentiation regimes, the purity of the initial monocyte preparation from PBMC may also affect this analysis, as residual T cells can become activated during the monocyte differentiation process and thus, contribute to an appreciable pSAMHD1(T592) signal in an immunoblot analysis of bulk cell populations. Collectively, the reported discrepancies of SAMHD1 expression, phosphorylation, and regulation in primary CD4+ T cells and MDM may, in part, stem from differences in methodology and will have to be addressed carefully by additional studies.
The whole‐body TMA approach identified constitutive SAMHD1 protein expression in vascular endothelium in a number of tissues, including the CNS, gastrointestinal tract, lymphatic tissues, genital tract, liver, and pancreas. This is of particular interest, as cerebral and cutaneous symptoms in AGS patients have been linked to vasculature [43]. Tubuloreticular inclusions in endothelial cells and cerebral arterial stenoses are found in the context of stroke in these patients. Moreover, half of the patients with AGS have painful, itchy skin lesions [44] that are associated with an inflammation of small blood vessels. Together, one can speculate that under conditions of SAMHD1 deficiency, an inappropriate accumulation of nucleic acids triggers autoimmune responses [1, 45, 46–47] that underlie the development of the observed inflammatory processes.
The broad expression profile of SAMHD1 may, in part, allow SAMHD1 to act as a ubiquitous guard that protects cells from retroviral insult. Based on the current understanding of its antiviral mode of action, the effectiveness of this cellular barrier depends on the cycling and possibly phosphorylation status of target cells. Taken together, the cross‐sectional SAMHD1 expression profile established in the current report will instruct studies into the biologic role of this nucleic acid‐modulating enzyme in physiology and infectious diseases.
AUTHORSHIP
S. Schmidt and O.T.K. conceived of and designed the experiments and wrote the paper. S. Schmidt, K.S., T.A., and E.E. performed experiments. J.L.K., S. Sertel, and F.L. contributed critical reagents. B.V. and O.T.F. provided expertise and discussion.
DISCLOSURES
The authors declare no conflict of interest.
Supporting information
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
ACKNOWLEDGMENTS
This work was, in part, funded by grants from BioBanking (CRC938/TP Z2 and DZIF/TI; to F.L.), and Deutsche Forschungsgemeinshaft Grants KE742/5‐1 (to O.T.K.) and FA 398/13/1 (to O.T.F.). The authors thank the Robert Koch‐Institute for financial support of the National Reference Center for Retroviruses. The authors are grateful for support from Goethe University. The authors thank Hanna‐Mari Baldauf for support and Bruno Kyewski for discussion. The authors thank Jutta Scheuerer for technical assistance and Pieter Meuwissen for supervised experimental contributions.
REFERENCES
- 1. Crow, Y.J. , Rehwinkel, J. (2009) Aicardi‐Goutieres syndrome and related phenotypes: linking nucleic acid metabolism with autoimmunity. Hum. Mol. Genet. 18, R130–R136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Goutieres, F. (2005) Aicardi‐Goutieres syndrome. Brain Dev. 27, 201–206. [DOI] [PubMed] [Google Scholar]
- 3. Li, N. , Zhang, W. , Cao, X. (2000) Identification of human homologue of mouse IFN‐gamma induced protein from human dendritic cells. Immunol. Lett. 74, 221–224. [DOI] [PubMed] [Google Scholar]
- 4. Rice, G.I. , Bond, J. , Asipu, A. , Brunette, R.L. , Manfield, I.W. , Carr, I.M. , Fuller, J.C. , Jackson, R.M. , Lamb, T. , Briggs, T.A. , Ali, M. , Gornall, H. , Couthard, L.R. , Aeby, A. , Attard‐Montalto, S.P. , Bertini, E. , Bodemer, C. , Brockmann, K. , Brueton, L.A. , Corry, P.C. , Desguerre, I. , Fazzi, E. , Cazorla, A.G. , Gener, B. , Hamel, B.C. , Heiberg, A. , Hunter, M. , van der Knaap, M.S. , Kumar, R. , Lagae, L. , Landrieu, P.G. , Lourenco, C.M. , Marom, D. , McDermott, M.F. , van der Merwe, W. , Orcesi, S. , Prendiville, J.S. , Rasmussen, M. , Shalev, S.A. , Soler, D.M. , Shinawi, M. , Spiegel, R. , Tan, T.Y. , Vanderver, A. , Wakeling, E.L. , Wassmer, E. , Whittaker, E. , Lebon, P. , Stetson, D.B. , Bonthron, D.T. , Crow, Y.J. (2009) Mutations involved in Aicardi‐Goutièeres syndrome implicate SAMHD1 as regulator of the innate immune response. Nat. Genet. 41, 829–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Liao, W. , Bao, Z. , Cheng, C. , Mok, Y.K. , Wong, W.S. (2008) Dendritic cell‐derived interferon‐gamma‐induced protein mediates tumor necrosis factor‐alpha stimulation of human lung fibroblasts. Proteomics 8, 2640–2650. [DOI] [PubMed] [Google Scholar]
- 6. Hartman, Z.C. , Kiang, A. , Everett, R.S. , Serra, D. , Yang, X.Y. , Clay, T.M. , Amalfitano, A. (2007) Adenovirus infection triggers a rapid, MyD88‐regulated transcriptome response critical to acute‐phase and adaptive immune responses in vivo. J. Virol. 81, 1796–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Préhaud, C. , Mégret, F. , Lafage, M. , Lafon, M. (2005) Virus infection switches TLR‐3‐positive human neurons to become strong producers of beta interferon. J. Virol. 79, 12893–12904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhao, D. , Peng, D. , Li, L. , Zhang, Q. , Zhang, C. (2008) Inhibition of G1P3 expression found in the differential display study on respiratory syncytial virus infection. Virol. J. 5, 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Goldstone, D.C. , Ennis‐Adeniran, V. , Hedden, J.J. , Groom, H.C. , Rice, G.I. , Christodoulou, E. , Walker, P.A. , Kelly, G. , Haire, L.F. , Yap, M.W. , de Carvalho, L.P. , Stoye, J.P. , Crow, Y.J. , Taylor, I.A. , Webb, M. (2011) HIV‐1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature 480, 379–382. [DOI] [PubMed] [Google Scholar]
- 10. Powell, R.D. , Holland, P.J. , Hollis, T. , Perrino, F.W. (2011) Aicardi‐Goutieres syndrome gene and HIV‐1 restriction factor SAMHD1 is a dGTP‐regulated deoxynucleotide triphosphohydrolase. J. Biol. Chem. 286, 43596–43600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hrecka, K. , Hao, C. , Gierszewska, M. , Swanson, S.K. , Kesik‐Brodacka, M. , Srivastava, S. , Florens, L. , Washburn, M.P. , Skowronski, J. (2011) Vpx relieves inhibition of HIV‐1 infection of macrophages mediated by the SAMHD1 protein. Nature 474, 658–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Laguette, N. , Sobhian, B. , Casartelli, N. , Ringeard, M. , Chable‐Bessia, C. , Ségéral, E. , Yatim, A. , Emiliani, S. , Schwartz, O. , Benkirane, M. (2011) SAMHD1 is the dendritic‐ and myeloid‐cell‐specific HIV‐1 restriction factor counteracted by Vpx. Nature 474, 654–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gramberg, T. , Kahle, T. , Bloch, N. , Wittmann, S. , Müllers, E. , Daddacha, W. , Hofmann, H. , Kim, B. , Lindemann, D. , Landau, N.R. (2013) Restriction of diverse retroviruses by SAMHD1. Retrovirology 10, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hollenbaugh, J.A. , Gee, P. , Baker, J. , Daly, M.B. , Amie, S.M. , Tate, J. , Kasai, N. , Kanemura, Y. , Kim, D.H. , Ward, B.M. , Koyanagi, Y. , Kim, B. (2013) Host factor SAMHD1 restricts DNA viruses in non‐dividing myeloid cells. PLoS Pathog. 9, e1003481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kim, E.T. , White, T.E. , Brandariz‐Nuñez, A. , Diaz‐Griffero, F. , Weitzman, M.D. (2013) SAMHD1 restricts herpes simplex virus 1 in macrophages by limiting DNA replication. J. Virol. 87, 12949–12956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kim, B. , Nguyen, L.A. , Daddacha, W. , Hollenbaugh, J.A. (2012) Tight interplay among SAMHD1 protein level, cellular dNTP levels, and HIV‐1 proviral DNA synthesis kinetics in human primary monocyte‐derived macrophages. J. Biol. Chem. 287, 21570–21574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lahouassa, H. , Daddacha, W. , Hofmann, H. , Ayinde, D. , Logue, E.C. , Dragin, L. , Bloch, N. , Maudet, C. , Bertrand, M. , Gramberg, T. , Pancino, G. , Priet, S. , Canard, B. , Laguette, N. , Benkirane, M. , Transy, C. , Landau, N.R. , Kim, B. , Margottin‐Goguet, F. (2012) SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nat. Immunol. 13, 223–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wu, L. (2013) Cellular and biochemical mechanisms of the retroviral restriction factor SAMHD1. ISRN Biochem. 2013, pii: 728392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Beloglazova, N. , Flick, R. , Tchigvintsev, A. , Brown, G. , Popovic, A. , Nocek, B. , Yakunin, A.F. (2013) Nuclease activity of the human SAMHD1 protein implicated in the Aicardi‐Goutieres syndrome and HIV‐1 restriction. J. Biol. Chem. 288, 8101–8110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ryoo, J. , Choi, J. , Oh, C. , Kim, S. , Seo, M. , Kim, S.Y. , Seo, D. , Kim, J. , White, T.E. , Brandariz‐Nuñez, A. , Diaz‐Griffero, F. , Yun, C.H. , Hollenbaugh, J.A. , Kim, B. , Baek, D. , Ahn, K. (2014) The ribonuclease activity of SAMHD1 is required for HIV‐1 restriction. Nat. Med. 20, 936–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Baldauf, H.‐M. , Pan, X. , Erikson, E. , Schmidt, S. , Daddacha, W. , Burggraf, M. , Schenkova, K. , Ambiel, I. , Wabnitz, G. , Gramberg, T. , Panitz, S. , Flory, E. , Landau, N.R. , Sertel, S. , Rutsch, F. , Lasitschka, F. , Kim, B. , Konig, R. , Fackler, O.T. , Keppler, O.T. (2012) SAMHD1 restricts HIV‐1 infection in resting CD4(+) T cells. Nat. Med. 18, 1682–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pauls, E. , Ruiz, A. , Riveira‐Muñoz, E. , Permanyer, M. , Badia, R. , Clotet, B. , Keppler, O.T. , Ballana, E. , Este, J.A. (2014) p21 regulates the HIV‐1 restriction factor SAMHD1. Proc. Natl. Acad. Sci. USA 111, E1322–E1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. White, T.E. , Brandariz‐Nuñez, A. , Valle‐Casuso, J.C. , Amie, S. , Nguyen, L.A. , Kim, B. , Tuzova, M. , Diaz‐Griffero, F. (2013) The retroviral restriction ability of SAMHD1, but not its deoxynucleotide triphosphohydrolase activity, is regulated by phosphorylation. Cell Host Microbe 13, 441–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. St Gelais, C. , de Silva, S. , Amie, S.M. , Coleman, C.M. , Hoy, H. , Hollenbaugh, J.A. , Kim, B. , Wu, L. (2012) SAMHD1 restricts HIV‐1 infection in dendritic cells (DCs) by dNTP depletion, but its expression in DCs and primary CD4+ T‐lymphocytes cannot be upregulated by interferons. Retrovirology 9, 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Erikson, E. , Adam, T. , Schmidt, S. , Lehmann‐Koch, J. , Over, B. , Goffinet, C. , Harter, C. , Bekeredjian‐Ding, I. , Sertel, S. , Lasitschka, F. , Keppler, O.T. (2011) In vivo expression profile of the antiviral restriction factor and tumor‐targeting antigen CD317/BST‐2/HM1.24/tetherin in humans. Proc. Natl. Acad. Sci. USA 108, 13688–13693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Allred, D.C. , Clark, G.M. , Elledge, R. , Fuqua, S.A. , Brown, R.W. , Chamness, G.C. , Osborne, C.K. , McGuire, W.L. (1993) Association of p53 protein expression with tumor cell proliferation rate and clinical outcome in node‐negative breast cancer. J. Natl. Cancer Inst. 85, 200–206. [DOI] [PubMed] [Google Scholar]
- 27. Berger, A. , Sommer, A.F. , Zwarg, J. , Hamdorf, M. , Welzel, K. , Esly, N. , Panitz, S. , Reuter, A. , Ramos, I. , Jatiani, A. , Mulder, L.C. , Fernandez‐Sesma, A. , Rutsch, F. , Simon, V. , König, R. , Flory, E. (2011) SAMHD1‐deficient CD14+ cells from individuals with Aicardi‐Goutières syndrome are highly susceptible to HIV‐1 infection. PLoS Pathog. 7, e1002425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schmidt, S. , Fritz, J.V. , Bitzegeio, J. , Fackler, O.T. , Keppler, O.T. (2011) HIV‐1 Vpu blocks recycling and biosynthetic transport of the intrinsic immunity factor CD317/tetherin to overcome the virion release restriction. MBio 2, e00036–e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Keppler, O.T. , Yonemoto, W. , Welte, F.J. , Patton, K.S. , Iacovides, D. , Atchison, R.E. , Ngo, T. , Hirschberg, D.L. , Speck, R.F. , Goldsmith, M.A. (2001) Susceptibility of rat‐derived cells to replication by human immunodeficiency virus type 1. J. Virol. 75, 8063–8073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Laguette, N. , Benkirane, M. (2012) How SAMHD1 changes our view of viral restriction. Trends Immunol. 33, 26–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Grody, W.W. , Fligiel, S. , Naeim, F. (1985) Thymus involution in the acquired immunodeficiency syndrome. Am. J. Clin. Pathol. 84, 85–95. [DOI] [PubMed] [Google Scholar]
- 32. Haynes, B.F. , Hale, L.P. , Weinhold, K.J. , Patel, D.D. , Liao, H.X. , Bressler, P.B. , Jones, D.M. , Demarest, J.F. , Gebhard‐Mitchell, K. , Haase, A.T. , Bartlett, J.A. (1999) Analysis of the adult thymus in reconstitution of T lymphocytes in HIV‐1 infection. J. Clin. Invest. 103, 921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cribier, A. , Descours, B. , Valadão, A.L. , Laguette, N. , Benkirane, M. (2013) Phosphorylation of SAMHD1 by cyclin A2/CDK1 regulates its restriction activity toward HIV‐1. Cell Reports 3, 1036–1043. [DOI] [PubMed] [Google Scholar]
- 34. Guarner, J. , Falcón‐Escobedo, R. (2009) Comparison of the pathology caused by H1N1, H5N1, and H3N2 influenza viruses. Arch. Med. Res. 40, 655–661. [DOI] [PubMed] [Google Scholar]
- 35. Guo, Y. , Korteweg, C. , McNutt, M.A. , Gu, J. (2008) Pathogenetic mechanisms of severe acute respiratory syndrome. Virus Res. 133, 4–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Johnson, J.E. , Gonzales, R.A. , Olson, S.J. , Wright, P.F. , Graham, B.S. (2007) The histopathology of fatal untreated human respiratory syncytial virus infection. Mod. Pathol. 20, 108–119. [DOI] [PubMed] [Google Scholar]
- 37. Leyva‐Grado, V.H. , Hai, R. , Fernandes, F. , Belicha‐Villanueva, A. , Carter, C. , Yondola, M.A. (2014) Modulation of an ectodomain motif in the influenza A virus neuraminidase alters tetherin sensitivity and results in virus attenuation in vivo. J. Mol. Biol. 426, 1308–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yondola, M.A. , Fernandes, F. , Belicha‐Villanueva, A. , Uccelini, M. , Gao, Q. , Carter, C. , Palese, P. (2011) Budding capability of the influenza virus neuraminidase can be modulated by tetherin. J. Virol. 85, 2480–2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schaller, T. , Bauby, H. , Hue, S. , Malim, M.H. , Goujon, C. (2014) New insights into an X‐traordinary viral protein. Front. Microbiol. 5, 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Welbourn, S. , Dutta, S.M. , Semmes, O.J. , Strebel, K. (2013) Restriction of virus infection but not catalytic dNTPase activity is regulated by phosphorylation of SAMHD1. J. Virol. 87, 11516–11524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pauls, E. , Jimenez, E. , Ruiz, A. , Permanyer, M. , Ballana, E. , Costa, H. , Nascimiento, R. , Parkhouse, R.M. , Peña, R. , Riveiro‐Muñoz, E. , Martinez, M.A. , Clotet, B. , Esté, J.A. , Bofill, M. (2013) Restriction of HIV‐1 replication in primary macrophages by IL‐12 and IL‐18 through the upregulation of SAMHD1. J. Immunol. 190, 4736–4741. [DOI] [PubMed] [Google Scholar]
- 42. Allouch, A. , David, A. , Amie, S.M. , Lahouassa, H. , Chartier, L. , Margottin‐Goguet, F. , Barré‐Sinoussi, F. , Kim, B. , Sáez‐Cirión, A. , Pancino, G. (2014) Reply to Pauls et al.: p21 is a master regulator of HIV replication in macrophages through dNTP synthesis block. Proc. Natl. Acad. Sci. USA 111, E1325–E1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Thiele, H. , du Moulin, M. , Barczyk, K. , George, C. , Schwindt, W. , Nürnberg, G. , Frosch, M. , Kurlemann, G. , Roth, J. , Nürnberg, P. , Rutsch, F. (2010) Cerebral arterial stenoses and stroke: novel features of Aicardi‐Goutieres syndrome caused by the Arg164X mutation in SAMHD1 are associated with altered cytokine expression. Hum. Mutat. 31, E1836–E1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Abdel‐Salam, G.M. , El‐Kamah, G.Y. , Rice, G.I. , El‐Darouti, M. , Gornall, H. , Szynkiewicz, M. , Aymard, F. , Zaki, M.S. , Abdel‐Aleem, A.K. , Lebon, P. , Crow, Y.J. (2010) Chilblains as a diagnostic sign of aicardi‐goutières syndrome. Neuropediatrics 41, 18–23. [DOI] [PubMed] [Google Scholar]
- 45. Van Heteren, J.T. , van der Knaap, M.S. , Poll The, B.W. , Kuijpers, T.W. (2007) Plasmacytoid dendritic cells and interferon‐alpha in Aicardi‐Goutieres syndrome. Neuropediatrics 38, 269–275. [DOI] [PubMed] [Google Scholar]
- 46. Ramantani, G. , Häusler, M. , Niggemann, P. , Wessling, B. , Guttmann, H. , Mull, M. , Tenbrock, K. , Lee‐Kirsch, M.A. (2011) Aicardi‐Goutières syndrome and systemic lupus erythematosus (SLE) in a 12‐year‐old boy with SAMHD1 mutations. J. Child Neurol. 26, 1425–1428. [DOI] [PubMed] [Google Scholar]
- 47. Lee‐Kirsch, M.A. (2010) Nucleic acid metabolism and systemic autoimmunity revisited. Arthritis Rheum. 62, 1208–1212. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data


