Abstract
The spike glycoprotein of the Middle East respiratory coronavirus (MERS‐CoV) facilitates receptor binding and cell entry. During investigation of a multi‐facility outbreak of MERS‐CoV in Taif, Saudi Arabia, we identified a mixed population of wild‐type and variant sequences with a large 530 nucleotide deletion in the spike gene from the serum of one patient. The out of frame deletion predicted loss of most of the S2 subunit of the spike protein leaving the S1 subunit with an intact receptor binding domain. This finding documents human infection with a novel genetic variant of MERS‐CoV present as a quasispecies. J. Med. Virol. 89:542–545, 2017 . © 2016 Wiley Periodicals, Inc.
Keywords: coronavirus, MERS‐CoV, spike gene, genome
INTRODUCTION
Since first being recognized in 2012, Middle East respiratory syndrome coronavirus (MERS‐CoV) cases have continued to be reported from Saudi Arabia and neighboring countries. Although considerable progress has been made in our understanding of the epidemiology and clinical features of MERS‐CoV infection, less is known about this virus's capacity for genetic variation. Beginning in September 2014, an outbreak of MERS‐CoV was reported from multiple healthcare facilities located in Taif, in the Makkah Region of Saudi Arabia. An investigation of this outbreak by the Saudi Ministry of Health and U.S. Centers for Disease Control and Prevention (CDC) was recently reported [Assiri et al., 2016]. Among 38 laboratory‐confirmed MERS‐CoV cases, serum samples from 17 were sent to CDC for serologic and molecular evaluation. Spike gene sequences were obtained from 10 patients, including two patients with identical sequences with overlapping stays at a private hospital (hospital D). The first patient (#27) was a 75‐year‐old woman who was evaluated for severe acute respiratory symptoms at another hospital and transferred to the intensive care unit of hospital D on November 1 where she was confirmed positive for MERS‐CoV. On November 11, an 81‐year‐old inpatient (#30) staying on the same floor as patient #27, developed respiratory symptoms and also tested positive for MERS‐CoV. Both patients subsequently died. Further analysis of the serum sample from Patient #27 identified a second sequence with a large deletion in the spike gene. In this report, we describe this variant and speculate on its possible origins and implications for predicted spike protein function.
MATERIALS AND METHODS
Serum specimens from Taif patients were screened for MERS‐CoV by real‐time RT‐PCR and positive samples were further subjected to RT‐PCR and Sanger sequencing of the spike gene as previously reported [Assiri et al., 2016]. To confirm our finding of a deleted spike gene sequence amplified by primer set SF6/SR6 [Assiri et al., 2016] from patient #27, sample extracts were reamplified using a different primer set SF6/SSR6 [Assiri et al., 2016] using SuperScript III One‐Step RT‐PCR System with Platinum Taq DNA Polymerase (Thermo Fisher Scientific, Carlsbad, CA). The amplicons were sequenced on both Sanger (3130xl Genetic Analyzer, Fischer Scientific) and next generation (PacBio RS II, Pacific Biosciences, San Francisco, CA) platforms. Sequencher 4.8 software (Gene Codes, Ann Arbor, MI) was used for Sanger sequence assembly and editing. PacBio data analysis was performed using CLC Genomics Workbench v6 (Waltham, MA). Wild‐type and deleted genome sequences of the quasispecies were prepared from the serum of patient #27 using 20 overlapping primer sets [Assiri et al., 2016] and deposited in GenBank (KU710264; KU710265).
RESULTS
As previously reported [Assiri et al., 2016], real‐time RT‐PCR testing of serum specimens from a MERS‐CoV outbreak in Taif identified two epidemiologically linked case‐patients (#27 and #30) with identical spike gene sequences. On further analysis, amplicons generated from patient #27 revealed a second smaller amplicon that was not present in the samples from patient #30 or other case‐patients (Fig. 1). Repeated RT‐PCR confirmed presence of the smaller product and Sanger sequencing identified a 530 nucleotide deletion which mapped to the region encoding subunit two of the spike protein gene (referred to from here forward as S530Δ). To confirm that this finding was not an artifact of our sequencing method, new amplicons generated from the serum sample were subjected to deep sequencing. Approximately, 22,000 reads were obtained providing a minimum coverage of a few hundred bases throughout, reaching a maximum of over 11,000× coverage at non‐deletion loci. A large population of reads with a relative coverage gap of 530 bases was obtained confirming the size and position of the predicted deletion. S530Δ was abundant in the cell‐free serum sample, with an approximate ratio of 4‐to‐1 deleted to intact sequence reads. No other sequence variants were detected. MERS‐CoV genome sequences were subsequently obtained from patient #27; genome sequencing was not attempted on the sample from patient #30 due to limited sample volume and low virus load.
Figure 1.
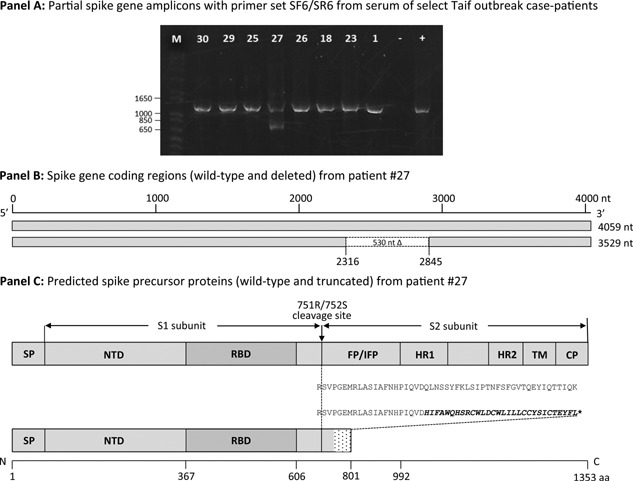
Schematic representation of MERS‐CoV spike gene coding region and predicted protein(s): SP, signal peptide; NTD, N‐terminal domain; RBD, receptor‐binding domain; FP, fusion peptide; IFP, internal fusion peptide; HR1, heptad repeat 1; HR2, heptad repeat 2; TM, transmembrane domain; CP, cytoplasmic domain (modified from Gao et al. [2013]). Panel A. MERS‐CoV spike gene amplicons generated by primer pair SF6/SR6 from serum of select Taif outbreak case‐patients (1). Lane designations: M, molecular weight marker; #, case‐patient identification numbers; −, negative template control; +, positive template control. Panel B. Wild‐type and variant spike gene with 530 nucleotide frame‐shift deletion mutation obtained from patient #27. Panel C. Wild‐type and truncated precursor proteins predicted for these genes. Stop codon (*).
The wild‐type MERS‐CoV spike precursor protein is comprised of 1353 residues that are organized into two subunits: an amino‐terminal subunit (S1, aa 1‐751) that contains the receptor binding domain (RBD), and a carboxy‐terminal subunit (S2, aa 752‐1353) that contains the putative fusion peptide, two heptad repeat domains and the transmembrane and intracellular domains (Fig. 1). Determinants of cellular tropism and interaction with the target cell receptor reside within the S1 domain, while mediators of membrane fusion are located within the S2 domain [Gao et al., 2013]. In the endoplasmic reticulum (ER)‐Golgi compartments of the infected cell, the precursor spike protein is first cleaved by a host protease such as furin at position 751R/752S into the S1 and S2 subunits that remain non‐covalently linked [Gierer et al., 2013; Millet and Whittaker, 2014]. Trimers of these proteins form with the S2 subunit embedded in the ER membrane and the S1 projecting outward. After release from the cell, the mature virus particle binds by the S1 RBD to the dipeptidyl peptidase‐4 (DPP4) receptor on the surface of a new cell [Raj et al., 2013]. A proteolytic cleavage at a second site in S2 located at position R887/S888 upstream from the putative fusion peptide then occurs that facilitates membrane fusion and virus entry into the host cell [Gao et al., 2013; Millet and Whittaker, 2014].
The deleted gene would predict an 801 amino acid truncated protein prematurely terminating at an out‐of‐frame stop codon (Fig. 1). This protein would contain the entire N‐terminal S1 subunit, including the virus RBD, 20 in‐frame residues immediately C‐terminal to the R751/S752 protease cleavage site and 30 out‐of‐frame non‐spike residues. All key components of the membrane fusion architecture of the S2 subunit located anterior to the premature stop codon, including the proposed fusion peptide (aa 949‐970) [Ou et al., 2016], would be predicted to be lost. The 30 non‐spike residues (HIFAWQHSRCWLDCWLILLCCYSICTEYFL) at the C‐terminus of the truncated protein include 14 hydrophobic residues (Ala, Phe, Gly, Ile, Leu, Met, Val, or Trp) and five cysteines.
DISCUSSION
RNA viruses are prone to rapid expansion of genomic variants or quasispecies that may aid virus escape from immunesurveillance and expand tissue tropism and host range [Duarte et al., 1994]. Well documented among coronaviruses generally, MERS‐CoV quasispecies have been identified in naturally [Briese et al., 2014] and experimentally [Borucki et al., 2016] infected dromedary camels and SARS‐CoV in humans [Tang et al., 2006], but no similar instances have been reported for human MERS‐CoV infections. Moreover, although naturally occurring deletion mutations of varying size and location have been previously identified among human coronaviruses, including SARS‐CoV [Chiu et al., 2005], HCoV‐OC43 [Vijgen et al., 2005], and MERS‐CoV [Lamers et al., 2016], most have been restricted to the nucleocapsid or non‐structural accessory protein genes located near the 3′‐end of the viral genome. With the exception of a single codon deletion (residue 1293) in the spike transmembrane domain that was reported for a MERS‐CoV derived from a dromedary camel [Chu et al., 2014], no naturally occurring deletions in the spike gene have been previously reported from human derived virus.
Modification of the coronavirus spike protein through natural and experimentally induced mutations has been shown to change cell and organ tropism leading in some cases to changes in virus pathogenicity and host range [Rasschaert et al., 1990; Wesley et al., 1991; Vijgen et al., 2005; Brandão et al., 2006; Terada et al., 2012]. Although most spike gene deletion mutations have been found in the S1 region, some studies have also documented mutations in the S2 region with similar effects. For example, specific mutations introduced into the S2 region of feline coronavirus have been shown to change virus tropism from the gut epithelium to macrophages with associated changes in pathogenicity from a mild enteric infection to fatal immune‐mediated disease, respectively [Rottier et al., 2005].
The spike gene deletion described here would most likely to render the virus defective, either non‐infectious or with substantially reduced infectivity. Loss of the S2 subunit would likely disrupt membrane anchoring of the spike protein and prevent fusion of the virus and host cell. Propagation of defective viruses requires a helper virus to compensate for lost function. In the case of S530Δ, it is interesting to speculate how this mutation might conversely help sustain wild‐type MERS‐CoV infection. One hypothetical outcome of loss of the S2 transmembrane domain would be to yield a free S1 subunit with a “sticky” hydrophobic tail that might lead to aggregated/misfolded protein due to the additional disulfide bounds that might form. Assuming that S530Δ still forms stable trimer complexes that retain biding affinity for DPP4, this form of the spike protein might act as a “decoy,” blocking spike‐specific MERS‐CoV neutralizing antibodies. A similar concept has been hypothesized as an immune escape strategy used by Ebola virus, termed “antigenic subversion” [Mohan et al., 2012]. It has been shown that infection of susceptible cells with MERS‐CoV can be inhibited with a soluble form of the DPP4 receptor [Raj et al., 2013]. Conversely, free spike protein fragments could theoretically bind and block anti‐RBD neutralizing antibodies. Arguing against this hypothesis is that anti‐MERS‐CoV antibodies were not detected (titer <400) in this patient by a sensitive enzyme immunoassay [Assiri et al., 2016] and the paucity of other reports of MERS‐CoV spike gene deletions suggest that this event is rare and not of deliberate design.
This study had several limitations. Different specimen types from different time‐points were not available from this patient or other patients in the predicted transmission chain preventing determination S530Δ persistence in the patient or capacity for transmission. Limited available serum also prevented culture attempts, which would have allowed assessment of virus viability and direct assessment of protein form and function. Nevertheless, our finding provides new insights into the capacity of MERS‐CoV for genetic variation that may have unforeseen public health implications.
CDC Disclaimer: The findings and conclusions in this report are those of the author(s) and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
REFERENCES
- Assiri A, Abedi GR, Bin Saeed AA, Abdalla MA, al‐Masry M, Choudhry AJ, Lu X, Erdman DD, Tatti K, Binder AM, Rudd J, Tokars J, Miao C, Alarbash H, Nooh R, Pallansch M, Gerber SI, Watson JT. 2016. Multifacility outbreak of middle east respiratory syndrome in taif, Saudi Arabia. Emerg Infect Dis 22:32–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assiri AM, Midgley CM, Abedi GR, Bin Saeed A, Almasri MM, Lu X, Al‐Abdely HM, Abdalla O, Mohammed M, Algarni HS, Alhakeem RF, Sakthivel SK, Nooh R, Alshayab Z, Alessa M, Srinivasamoorthy G, AlQahtani SY, Kheyami A, HajOmar WH, Banaser TM, Esmaeel A, Hall AJ, Curns AT, Tamin A, Alsharef AA, Erdman D, Watson JT, Gerber SI. 2016. Epidemiology of a novel recombinant MERS‐CoV in humans in Saudi Arabia. J Infect Dis 214:712–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borucki MK, Lao V, Hwang M, Gardner S, Adney D, Munster V, Bowen R, Allen JE. 2016. Middle East respiratory syndrome coronavirus intra‐host populations are characterized by numerous high frequency variants. PLoS ONE 11:e0146251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandão PE, Gregori F, Richtzenhain LJ, Rosales CA, Villarreal LY, Jerez JA. 2006. Molecular analysis of Brazilian strains of bovine coronavirus (BCoV) reveals a deletion within the hypervariable region of the S1 subunit of the spike glycoprotein also found in human coronavirus OC43. Arch Virol 151:1735–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briese T, Mishra N, Jain K, Zalmout IS, Jabado OJ, Karesh WB, Daszak P, Mohammed OB, Alagaili AN, Lipkin WI. 2014. Middle East respiratory syndrome coronavirus quasispecies that include homologues of human isolates revealed through whole‐genome analysis and virus cultured from dromedary camels in Saudi Arabia. MBio 5:e01146–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu RW, Chim SS, Tong YK, Fung KS, Chan PK, Zhao GP, Lo YM. 2005. Tracing SARS‐coronavirus variant with large genomic deletion. Emerg Infect Dis 11:168–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu DK, Poon LL, Gomaa MM, Shehata MM, Perera RA, Abu Zeid D, El Rifay AS, Siu LY, Guan Y, Webby RJ, Ali MA, Peiris M, Kayali G. 2014. MERS coronaviruses in dromedary camels, Egypt. Emerg Infect Dis 20:1049–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte EA, Novella IS, Weaver SC, Domingo E, Wain‐Hobson S, Clarke DK, Moya A, Elena SF, de la Torre JC, Holland JJ. 1994. RNA virus quasispecies: Significance for viral disease and epidemiology. Infect Agents Dis 3:201–214. [PubMed] [Google Scholar]
- Gao J, Lu G, Qi J, Li Y, Wu Y, Deng Y, Geng H, Li H, Wang Q, Xiao H, Tan W, Yan J, Gao GF. 2013. Structure of the fusion core and inhibition of fusion by a heptad repeat peptide derived from the S protein of Middle East respiratory syndrome coronavirus. J Virol 87:13134–13140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierer S, Bertram S, Kaup F, Wrensch F, Heurich A, Krämer‐Kühl A, Welsch K, Winkler M, Meyer B, Drosten C, Dittmer U, von Hahn T, Simmons G, Hofmann H, Pöhlmann S. 2013. The spike protein of the emerging betacoronavirus EMC uses a novel coronavirus receptor for entry, can be activated by TMPRSS2, and is targeted by neutralizing antibodies. J Virol 87:5502–5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers MM, Raj VS, Shafei M, Ali SS, Abdallh SM, Gazo M, Nofal S, Lu X, Erdman DD, Koopmans MP, Abdallat M, Haddadin A, Haagmans BL. 2016. Circulation of MERS‐CoV deletions variants in humans, Jordan, 2015. Emerg Infect Dis 22:716–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet JK, Whittaker GR. 2014. Host cell entry of Middle East respiratory syndrome coronavirus after two‐step, furin‐mediated activation of the spike protein. Proc Natl Acad Sci USA 111:15214–15219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan GS, Li W, Ye L, Compans RW, Yang C. 2012. Antigenic subversion: A novel mechanism of host immune evasion by Ebola virus. PLoS Pathog 8:e1003065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou X, Zheng W, Shan Y, Mu Z, Dominguez SR, Holmes KV, Qian Z. 2016. Identification of the fusion peptide‐containing region in betacoronavirus spike glycoproteins. J Virol 90:5586–5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj VS, Mou H, Smits SL, Dekkers DH, Muller MA, Dijkman R, Muth D, Demmers JA, Zaki A, Fouchier RA, Thiel V, Drosten C, Rottier PJ, Osterhaus AD, Bosch BJ, Haagmans BL. 2013. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus‐EMC. Nature 495:251–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasschaert D, Duarte M, Laude H. 1990. Porcine respiratory coronavirus differs from transmissible gastroenteritis virus by a few genomic deletions. J Gen Virol 71:2599–2607. [DOI] [PubMed] [Google Scholar]
- Rottier PJ, Nakamura K, Schellen P, Volders H, Haijema BJ. 2005. Acquisition of macrophage tropism during the pathogenesis of feline infectious peritonitis is determined by mutations in the feline coronavirus spike protein. J Virol 79:14122–14130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang JW, Cheung JLK, Chu IMT, Sung JJY, Peiris M, Chan PKS. 2006. The large 386‐nt deletion in SARS‐associated coronavirus: Evidence for quasispecies? J Infect Dis 194:808–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terada Y, Shiozaki Y, Shimoda H, Mahmoud HY, Noguchi K, Nagao Y, Shimojima M, Iwata H, Mizuno T, Okuda M, Morimoto M, Hayashi T, Tanaka Y, Mochizuki M, Maeda K. 2012. Feline infectious peritonitis virus with a large deletion in the 5'‐terminal region of the spike gene retains its virulence for cats. J Gen Virol 93:1930–1934. [DOI] [PubMed] [Google Scholar]
- Vijgen L, Keyaerts E, Moës E, Thoelen I, Wollants E, Lemey P, Vandamme AM, Van Ranst M. 2005. Complete genomic sequence of human coronavirus OC43: Molecular clock analysis suggests a relatively recent zoonotic coronavirus transmission event. J Virol 79:1595–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesley RD, Woods RD, Cheung AK. 1991. Genetic analysis of porcine respiratory coronavirus, an attenuated variant of transmissible gastroenteritis virus. J Virol 65:3369–3373. [DOI] [PMC free article] [PubMed] [Google Scholar]


