Abstract
Enterovirus D68 (EV‐D68) is an emergent viral pathogen associated with mild to severe respiratory infections. In this study, we describe respiratory infections associated with EV‐D68 in Beijing over a 4 year period. Total nucleic acid was extracted from 7,945 clinical specimens collected between January 5, 2011 and July 30, 2015 in Beijing and used for detecting EV‐D68 and other enteroviruses by real‐time PCR. Overall, 555/7,945 (6.99%) specimens were enterovirus positive: 12/7,945 (0.2%) specimens were EV‐D68 positive. Of these patients, 11 were pediatric patients and 1 was a 76‐year‐old man. The main symptoms for the 12 EV‐D68 positive patients were fever (10/12, 83.3%) and cough (6/12, 50%). Ten EV‐D68 infection cases were identified in autumn or winter season. The phylogenetic relationships of the 12 EV‐D68 viral strains with other strains were analyzed based on the sequences of viral protein 1(VP1). The EV‐D68 strains from 2011 to 2013 belonged to groups 1 or 3, while all strains in 2014 were clustered into group 1 together with the strains circulating in the USA. In conclusion, EV‐D68 played a role in respiratory infections in Beijing during this period. In addition, the most common EV‐D68 strain detected was similar to that circulating in the USA in 2014. J. Med. Virol. 88:1529–1534, 2016 . © 2016 Wiley Periodicals, Inc.
Keywords: enterovirus D68, respiratory infection, VP1 sequence
INTRODUCTION
Enterovirus D68 (EV‐D68) belongs to the genus Enterovirus, species D of the picornaviridae family and was first identified in California in 1962 [Schieble et al., 1967]. It can cause mild to severe respiratory infections. More recently, circulation of three different EV‐D68 clades based on the phylogenetic analysis of VP1 sequences has been reported worldwide [Kaida et al., 2011; Rahamat‐Langendoen et al., 2011; Ikeda et al., 2012; Lauinger et al., 2012; Linsuwanon et al., 2012; Meijer et al., 2012; Piralla et al., 2012; Tokarz et al., 2012]. A nationwide outbreak of EV‐D68 associated with severe respiratory infection was reported in the USA from mid‐August 2014 to January 15, 2015 and a total of 1,153 people were affected during this epidemic (http://www.cdc.gov/non-polio-enterovirus/about/ev-d68.html). The EV‐D68 outbreaks in the USA have resulted in a worldwide public health concern with emergences of EV‐D68 infections in Denmark, Australia, France, and Italy [Bal et al., 2015; Esposito et al., 2015; Levy et al., 2015; Midgley et al., 2015]. In China, sporadic cases of EV‐D68 were reported between August 2006 and June 2012 [Xiang et al., 2012; Lu et al., 2014]. In a previous study, we identified one EVD68 strain in a respiratory specimen from a severe pneumonia case in Beijing in 2014 [Zhang et al., 2015]. There have been no further reports about the EV‐D68 infection in Beijing since then, especially after the US EV‐D68 pandemic in 2014.
In this study, we performed a retrospective surveillance study of respiratory infection cases and analyzed the epidemiological and clinical features of EV‐D68‐related diseases in Beijing, China during the 2011–2015 periods. All clinical specimens were tested in parallel for EV‐D68 and other enteroviruses with real‐time PCR. EV‐D68 positive specimens were also tested for other respiratory pathogens. Moreover, the phylogenetic relationships of EV‐D68 strains from Beijing, China and other available countries are also described in this study.
MATERIALS AND METHODS
In Beijing, the Respiratory Virus Surveillance System (RVSS) was established by the Beijing Center for Disease Prevention and Control in 2011. The RVSS now covers 30 sentinel hospitals distributed in all 16 districts of Beijing. This surveillance has obtained ethics approval from the Ethics Committee at Beijing Center for Disease Prevention and Control. Clinical specimens, including nasopharyngeal swabs and sputum were collected from respiratory infection cases and sent for laboratory diagnostic testing. A total of 7,945 clinical specimens from RVSS were collected and chosen for the EV‐D68 detection between January 5, 2011 and July 30, 2015.
Total nucleic acid (RNA and DNA) was extracted from the clinical specimens by Thermo Scientific™ KingFisher™ Flex Magnetic Particle Processors (Thermo Fisher, Waltham, MA). EV‐D68 and other enteroviruses were simultaneously detected in one tube with a commercial real‐time RT‐PCR kit (Duplex Real‐time PCR Detection Kit for Enterovirus 68, Jiangsu uninovo biological technology company, China). A panel of respiratory pathogens including influenza virus A (pandemic H1N1 influenza virus, seasonal influenza A virus H3N2) and B, respiratory syncytial virus, parainfluenza virus 1–4, adenovirus, rhinovirus, human metapneumovirus, human coronavirus (NL63, OC43, 229E, and HKU1), human bocavirus, mycoplasma, and chlamydia were also investigated in the EV‐D68 positive specimens with commercial real‐time RT‐PCR kits (Multiplex Combined Real‐time PCR Detection Kit for Respiratory Viruses, Jiangsu uninovo biological technology company, China).
The VP1 fragment of EV‐D68 positive specimens was amplified and sequenced as previously described [Tokarz et al., 2012]. The consensus VP1 sequences were assembled using DNASTAR software, Version 5.0 (DNASTAR, Inc., Madison, WI) and aligned with MEGA software, version 6.0 [Tamura et al., 2013]. The phylogenetic relationships of representative VP1 sequences of EV‐D68 from Beijing, China and other available countries (The USA, Republic of South Africa, Japan, France, Netherlands, Thailand, Senegal, and Gambia) were estimated by using the maximum‐likelihood method with 1,000 bootstrap replicates with MEGA software, version 6.0. The evolutionary distances were estimated for EV‐D68 VP1 sequences from 13 Beijing EV‐D68 strains, the prototype EV‐D68 (Fermon strain, AY426531) and one reference strain circulating in the USA in 2014 (US/MO/14‐18948 strain, KM851226) using Maximum Composite Likelihood model (500 bootstrap replicates) with MEGA, version 6.0.
RESULTS
In total, 7,945 clinical specimens from RVSS were collected and chosen for the EV‐D68 retrospective study from January 5, 2011 to July 30, 2015 (shown in Table I). Totally, 555/7,945 (6.99%) specimens were enterovirus positive. Of the 555 enterovirus positive specimens, 444 (28 in 2011–2012, 353 in 2013 and 94 in 2014–2015) were from pediatric cases and 78 (2 in 2011–2012 and 76 in 2014–2015) from adult cases. 12/7,945 (0.20%) specimens were EV‐D68 positive: 11 were from pediatric cases (median age 8 years; range from 3 years to 15 years) and 1 from an adult case (aged 76). The rates for EV‐D68 were 0.3% in the pediatric group (11/3,457) and 0.02% in the adult group (1/4,488), respectively. The rates for each age group were also shown in Table I. In the 12 EV‐D68 positive cases, no other respiratory pathogens were detected in the clinical specimens.
Table I.
Clinical Samples Screened for Enterovirus and EV‐D68 From January 5, 2011 to July 20, 2015 in Beijing, China
| Year | Age group a | No. of clinical specimens | No. of enterovirus positives (%) | No. of EV‐D68 positives(%) |
|---|---|---|---|---|
| January 5, 2011 to January 18, 2012 | Pediatric case | 451 | 28 (6.2) | 3 (0.7) |
| Adult case | 188 | 2 (1.1%) | 1 (0.5) | |
| April 28, 2013 to October 22, 2013 b | Pediatric case | 2,112 | 353 (16.7%) | 3 (0.1%) |
| August 1, 2014 to July 30, 2015 | Pediatric case | 894 | 96 (10.7%) | 5 (0.6%) |
| Adult case | 4,300 | 76 (1.8%) | 0 | |
| Total | 7,945 | 555 (7.0%) | 12 (0.2%) |
Pediatric cases ≦14 years old and adult cases > 14 years old.
All from pediatric cases (≦14 years old).
The demographic and clinical characteristics of the 12 EV‐D68 positive patients are shown in Table II. Seven were male patients and five were female. Eleven of twelve were pediatric patients and the other one was a 76‐year‐old man. Nine patients were admitted to the hospital and remaining three were treated as outpatients. The main symptoms for the 12 EV‐D68 patients were fever (10/12, 83.3%) and cough (6/12, 50%). No patients were admitted to the intensive care unit. Among the 12 EV‐D68 patients, 10 were detected in the autumn or winter (September to January). The other two were detected in the summer (July and August).
Table II.
Demographic and Clinical Characteristics of 12 EV‐D68 Positive Patients
| Patient | Laboratory code | Age (years) | Sex | Data of symptom onset | Clinical symptom | Admission to hospital |
|---|---|---|---|---|---|---|
| 1 a | 2011‐21186 | 15 | Male | November 4, 2011 | Fever, sputum production, chest ache | Yes |
| 2 | 2011‐21282 | 10 | Female | November 18, 2011 | Fever | Yes |
| 3 | 2011‐21286 | 4 | Male | November 26, 2011 | Fever | Yes |
| 4 | 2012‐12225 | 76 | Male | January 6, 2012 | Fever | Yes |
| 5 | 2013‐0720‐6 | 5 | Male | July 20, 2013 | Cough | No |
| 6 | 2013‐0825‐6 | 6 | Male | August 25, 2013 | Fever | No |
| 7 | 2013‐1017‐26 | 10 | Female | October 17, 2013 | Cough, fever, sputum production, sneezing | No |
| 8 | 2014‐R0672 | 10 | Female | September 12, 2014 | Cough, fever | Yes |
| 9 | 2014‐R970 | 11 | Male | September 14, 2014 | Fever | Yes |
| 10 | 2014‐R1011 | 3 | Female | October 7, 2014 | Cough, fever, sore throat, sputum production, chest ache | Yes |
| 11 | 2014‐R1153 | 8 | Female | September 21, 2014 | Cough, sputum production | Yes |
| 12 | 2014‐R1357 | 8 | Male | October 10, 2014 | Cough, fever, sore throat | Yes |
Collect the clinical specimen from pediatric clinic.
The phylogenetic relationships of the 12 EV‐D68 strains from Beijing, China and other local strains were analyzed based on the sequences of VP1 and shown in Figure 1. The 12 EV‐D68 strains belonged into groups 1 (n = 8) and 3 (n = 4). Of note, the EV‐D68 strains sampled in 2011–2013 were distributed into either group 1 or group 3 whereas all EV‐D68 strains sampled in 2014 were clustered into group 1 together with the strains circulating in the USA. The other EV‐D68 strains from China sampled between 2008 and 2012 also belonged to either group 1 or 3.
Figure 1.
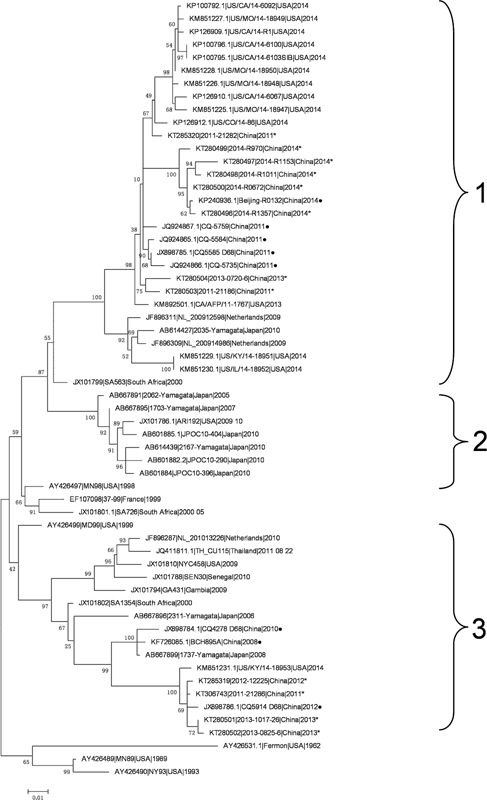
Phylogenetic analysis of 12 EV‐D68 strains from Beijing with other strains from different geographic locations. The phylogenetic relationships of representative viral protein 1 (VP1) sequences from different countries were estimated by using the maximum‐likelihood method with 1,000 bootstrap replicates using MEGA 6.0 (http://www.megasoftware.net). Bootstrap values are indicated on each tree. The scale bar represents nucleotide substitutions per site. The 12 EV‐D68 strains generated by this study are indicated with an asterisk. The VP1 sequences of other EV‐D68 strains in China are indicated with a solid circle. GenBank accession numbers and the country of origin are shown for each EV‐D68 strain.
The evolutionary distances for the 12 EV‐D68 VP1 sequences are shown in Table III. One of EV‐D68 strains from our previous study was used as a reference for the estimation of evolutionary distance together with the prototype EV‐D68 (Fermon strain, AY426531) and one reference strain circulating in the USA in 2014 (US/MO/14‐18948 strain, KM851226). The average evolutionary distance of the 12 EV‐D68 strains and three reference strains was 0.089. Compared with the prototype EV‐D68, the evolutionary distances of 13 Beijing EV‐D68 strains ranged from 0.145 to 0.173. In contrast, compared to the USA reference strain from 2014, the evolutionary distances of 13 Beijing EV‐D68 strains ranged from 0.015 (in 2011) to 0.148 (in 2013).
Table III.
Evolutionary Distances for EV‐D68 VP1 Sequences
| Stains | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AY426531.1|Fermon|USA|1962 | |||||||||||||||
| KM851226|US/MO/14‐18948|USA|2014 | 0.151 | ||||||||||||||
| KP240936.1|Beijing‐R0132|China|2014 | 0.157 | 0.042 | |||||||||||||
| KT280496|2014‐R1357|China|2014 | 0.159 | 0.039 | 0.004 | ||||||||||||
| KT280497|2014‐R1153|China|2014 | 0.166 | 0.051 | 0.014 | 0.017 | |||||||||||
| KT280498|2014‐R1011|China|2014 | 0.158 | 0.046 | 0.009 | 0.011 | 0.012 | ||||||||||
| KT280499|2014‐R970|China|2014 | 0.164 | 0.043 | 0.014 | 0.017 | 0.025 | 0.019 | |||||||||
| KT280500|2014‐R0672|China|2014 | 0.156 | 0.039 | 0.002 | 0.004 | 0.012 | 0.007 | 0.012 | ||||||||
| KT280501|2013‐1017‐26|China|2013 | 0.168 | 0.148 | 0.146 | 0.149 | 0.158 | 0.152 | 0.158 | 0.149 | |||||||
| KT280502|2013‐0825‐6|China|2013 | 0.164 | 0.143 | 0.142 | 0.145 | 0.154 | 0.147 | 0.153 | 0.145 | 0.004 | ||||||
| KT280504|2013‐0720‐6|China|2013 | 0.145 | 0.035 | 0.038 | 0.036 | 0.048 | 0.041 | 0.039 | 0.036 | 0.145 | 0.141 | |||||
| KT285319|2012‐12225|China|2012 | 0.173 | 0.146 | 0.139 | 0.142 | 0.152 | 0.145 | 0.151 | 0.142 | 0.01 | 0.012 | 0.144 | ||||
| KT306743|2011‐21286|China|2011 | 0.17 | 0.145 | 0.138 | 0.141 | 0.148 | 0.144 | 0.149 | 0.141 | 0.008 | 0.01 | 0.142 | 0.004 | |||
| KT285320|2011‐21282|China|2011 | 0.146 | 0.015 | 0.035 | 0.033 | 0.043 | 0.038 | 0.038 | 0.033 | 0.14 | 0.135 | 0.026 | 0.136 | 0.134 | ||
| KT280503|2011‐21186|China|2011 | 0.148 | 0.029 | 0.032 | 0.03 | 0.042 | 0.037 | 0.038 | 0.03 | 0.143 | 0.139 | 0.017 | 0.139 | 0.138 | 0.02 | □ |
DISCUSSION
In recent years, outbreaks caused by EV‐D68 respiratory infection have been reported in many parts of the world [Kaida et al., 2011; Ikeda et al., 2012; Tokarz et al., 2012; Xiang et al., 2012; Renois et al., 2013; Lu et al., 2014; Levy et al., 2015; Midgley et al., 2015]. To understand the EV‐D68 circulation in Beijing, we investigated the clinical specimens from patients with respiratory infections for EV‐D68 during the period 2011–2015. In this study, the rate of EV‐D68 (0.20%) in Beijing is lower than the rates reported in Italy (0.24%) [Piralla et al., 2014], the Netherlands (0.95%) [Poelman et al., 2015], Philippines (1.10%) [Furuse et al., 2015], Germany (7.70%) [Reiche et al., 2015], Denmark (10.80%) [Midgley et al., 2015], and Norway (11.00%) [Bragstad et al., 2015]. The low rate of EV‐D68 detected in this study may be a consequence of the sampling method as the clinical specimens were collected from all age groups. Adults accounted for 56.5% of the respiratory infection cases while EV‐D68 mainly affects children. In addition, this could be a result of sampling time frame with no EV‐D68 infections detected in Beijing during the period of January to July of 2015, which is not the EV‐D68 circulating season.
In total, 11/12 EV‐D68 patients were pediatric cases presenting with mild clinical symptoms and none were admitted to the ICU. Similar phenomena were observed in the Germany and the Netherlands [Meijer et al., 2014; Reiche et al., 2015]. All of 12 EV‐D68 patients were without any underlying disease. Many EV‐D68 pediatric patients in the USA in 2014, had asthma or a history of wheezing, and were admitted to the ICU requiring bilevel positive airway pressure ventilation. This indicates that children with underlying disease are more likely to be seriously affected by EV‐D68 infection.
Ten out of twelve EV‐D68 strains were identified in autumn or winter and the others, in summer. The EV‐D68 strains circulating in autumn were described in Italy, Japan, the Netherlands, and the USA [Khetsuriani et al., 2006; Kaida et al., 2011; Rahamat‐Langendoen et al., 2011; Tokarz et al., 2012; Piralla et al., 2014]. Our study indicates that EV‐D68 could also cause the respiratory infection in the summer and winter. The phylogenetic relationship between EV‐D68 strains circulating in Beijing, China, and other countries showed that EV‐D68 strains from Beijing circulating in 2011 to 2013 belonged to group 1 or 3, while all of the strains in 2014 belonged to group 1 together with the strains circulating in the USA in 2014. In 2011, some other EV‐D68 strains from China were clustered into group 1 together with the strains circulating in the USA. The Beijing EV‐D68 strains in group 1 were also shown to be closely related to the USA reference strain in 2014 by evolutionary distance estimation. These results demonstrated that EV‐D68 strains similar to those circulating in the USA in 2014 already existed in China from 2011 and became predominant strains circulating in Beijing in 2014.
The EV‐D68 strains circulating in Beijing in 2014 were also shown to be highly homologous to the EV‐D68 strains isolated from an AFP case in the USA (Accession number: KM892501). AFP cases associated with EV‐D68 infections were observed in Norway, Australia, France, and the USA [Ayscue et al., 2014; Lang et al., 2014; Bragstad et al., 2015; Levy et al., 2015]. Therefore, greater attention should be given to EV‐D68 infections as the cause of AFP. The ongoing screens for EV‐D68 in respiratory infections and AFP cases will provide novel information to track the EV‐D68 spreading in the world and help evaluate the sequence diversity in circulating EV‐D68 strains.
ACKNOWLEDGMENTS
The authors would like to thank sentinel hospitals of RVSS in Beijing for collecting samples and investigating cases. The authors wish to acknowledge Dr. Jean Lutamyo Mbisa and Kevin E. Brown of Public Health England for their reading throughout the original manuscript.
REFERENCES
- Ayscue P, Van Haren K, Sheriff H, Waubant E, Waldron P, Yagi S, Yen C, Clayton A, Padilla T, Pan C, Reichel J, Harriman K, Watt J, Sejvar J, Nix WA, Feikin D, Glaser C; Centers for Disease Control and Prevention (CDC). 2014. Acute flaccid paralysis with anterior myelitis—California, June 2012–June 2014. MMWR Morb Mortal Wkly Rep 63:903–906. [PMC free article] [PubMed] [Google Scholar]
- Bal A, Schuffenecker I, Casalegno JS, Josset L, Valette M, Armand N, Dhondt PB, Escuret V, Lina B. 2015. Enterovirus D68 nosocomial outbreak in elderly people, France, 2014. Clin Microbiol Infect 21:e61–e62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bragstad K, Jakobsen K, Rojahn AE, Skram MK, Vainio K, Holberg‐Petersen M, Hungnes O, Dudman SG, Kran AM. 2015. High frequency of enterovirus D68 in children hospitalised with respiratory illness in Norway, autumn 2014. Influenza Other Respir Viruses 9:59–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito S, Zampiero A, Ruggiero L, Madini B, Niesters H, Principi N. 2015. Enterovirus D68‐associated community‐acquired pneumonia in children living in Milan, Italy. J Clin Virol 68:94–96. [DOI] [PubMed] [Google Scholar]
- Furuse Y, Chaimongkol N, Okamoto M, Imamura T, Saito M, Tamaki R, Saito M; Tohoku‐RITM Collaborative Research Team, Lupisan SP, Oshitani H. 2015. Molecular epidemiology of enterovirus d68 from 2013 to 2014 in Philippines. J Clin Microbiol 53:1015–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda T, Mizuta K, Abiko C, Aoki Y, Itagaki T, Katsushima F, Katsushima Y, Matsuzaki Y, Fuji N, Imamura T, Oshitani H, Noda M, Kimura H, Ahiko T. 2012. Acute respiratory infections due to enterovirus 68 in Yamagata, Japan between 2005 and 2010. Microbiol Immunol 56:139–143. [DOI] [PubMed] [Google Scholar]
- Kaida A, Kubo H, Sekiguchi J, Kohdera U, Togawa M, Shiomi M, Nishigaki T, Iritani N. 2011. Enterovirus 68 in children with acute respiratory tract infections, Osaka, Japan. Emerg Infect Dis 17:1494–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khetsuriani N, Lamonte‐Fowlkes A, Oberst S, Pallansch MA. 2006. Centers for Disease Control and Prevention. Enterovirus surveillance—United States, 1970–2005. MMWR Surveill Summ 55:1–20. [PubMed] [Google Scholar]
- Lang M, Mirand A, Savy N, Henquell C, Maridet S, Perignon R, Labbe A, Peigue‐Lafeuille H. 2014. Acute flaccid paralysis following enterovirus D68 associated pneumonia, France, 2014. Euro Surveill 19:20952. [DOI] [PubMed] [Google Scholar]
- Lauinger IL, Bible JM, Halligan EP, Aarons EJ, MacMahon E, Tong CY. 2012. Lineages, sub‐lineages and variants of enterovirus 68 in recent outbreaks. PLoS ONE 7:e36005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy A, Roberts J, Lang J, Tempone S, Kesson A, Dofai A, Daley AJ, Thorley B, Speers DJ. 2015. Enterovirus D68 disease and molecular epidemiology in Australia. J Clin Virol 69:117–121. [DOI] [PubMed] [Google Scholar]
- Linsuwanon P, Puenpa J, Suwannakarn K, Auksornkitti V, Vichiwattana P, Korkong S, Theamboonlers A, Poovorawan Y. 2012. Molecular epidemiology and evolution of human enterovirus serotype 68 in Thailand, 2006–2011. PLoS ONE 7:e35190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu QB, Wo Y, Wang HY, Wei MT, Zhang L, Yang H, Liu EM, Li TY, Zhao ZT, Liu W, Cao WC. 2014. Detection of enterovirus 68 as one of the commonest types of enterovirus found in patients with acute respiratory tract infection in China. J Med Microbiol 63:408–414. [DOI] [PubMed] [Google Scholar]
- Meijer A, Benschop KS, Donker GA, van der Avoort HG. 2014. Continued seasonal circulation of enterovirus D68 in the Netherlands, 2011–2014. Euro Surveill 19:1–6. [DOI] [PubMed] [Google Scholar]
- Meijer A, van der Sanden S, Snijders BEP, Jaramillo‐Gutierrez G, Bont L, van der Ent CK, Overduin P, Jenny SL, Jusic E, van der Avoort HGAM, Smith GJD, Donker GA, Koopmans MPG. 2012. Emergence and epidemic occurrence of enterovirus 68 respiratory infections in The Netherlands in 2010. Virology 423:49–57. [DOI] [PubMed] [Google Scholar]
- Midgley SE, Christiansen CB, Poulsen MW, Hansen CH, Fischer TK. 2015. Emergence of enterovirus D68 in Denmark, June 2014 to February 2015. Euro Surveill 20:1–6. [DOI] [PubMed] [Google Scholar]
- Piralla A, Girello A, Grignani M, Gozalo‐Margüello M, Marchi A, Marseglia G, Baldanti F. 2014. Phylogenetic characterization of enterovirus 68 strains in patients with respiratory syndromes in Italy. J Med Virol 86:1590–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piralla A, Lilleri D, Sarasini A, Marchi A, Zecca M, Stronati M, Baldanti F, Gerna G. 2012. Human rhinovirus and human respiratory enterovirus (EV68 and EV104) infections in hospitalized patients in Italy, 2008–2009. Diagn Microb Infect Dis 73:162–167. [DOI] [PubMed] [Google Scholar]
- Poelman R, Schölvinck EH, Borger R, Niesters HG, van Leer‐Buter C. 2015. The emergence of enterovirus D68 in a Dutch University Medical Center and the necessity for routinely screening for respiratory viruses. J Clin Virol 62:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahamat‐Langendoen J, Riezebos‐Brilman A, Borger R, van der Heide R, Brandenburg A, Schölvinck E, Niesters HGM. 2011. Upsurge of human enterovirus 68 infections in patients with severe respiratory tract infections. J Clin Virol 52:103–106. [DOI] [PubMed] [Google Scholar]
- Reiche J, Böttcher S, Diedrich S, Buchholz U, Buda S, Haas W, Schweiger B, Wolff T. 2015. Low‐level circulation of enterovirus D68‐associated acute respiratory infections, Germany, 2014. Emerg Infect Dis 21:837–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renois F, Bouin A, Andreoletti L. 2013. Enterovirus 68 in pediatric patients hospitalized for acute airway diseases. J Clin Microbiol 51:640–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schieble JH, Fox VL, Lennette EH. 1967. A probable new human picornavirus associated with respiratory diseases. Am J Epidemiol 85:297–310. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokarz R, Firth C, Madhi SA, Howie SRC, Wu WY, Sall AA, Haq S, Briese T, Lipkin WI. 2012. Worldwide emergence of multiple clades of enterovirus 68. J Gen Virol 93:1952–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Z, Gonzalez R, Wang Z, Ren L, Xiao Y, Li J, Li Y, Vernet G, Paranhos‐Baccalà G, Jin Q, Wang J. 2012. Coxsackievirus A21, enterovirus 68, and acute respiratory tract infection, China. Emerg Infect Dis 18:821–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Ren L, Luo M, Li A, Gong C, Chen M, Yu X, Wu J, Deng Y, Huang F. 2015. Enterovirus D68‐associated severe pneumonia, China, 2014. Emerg Infect Dis 21:916–918. [DOI] [PMC free article] [PubMed] [Google Scholar]


