Abstract
Respiratory viruses (RVs) are ubiquitous pathogens that represent a major cause of community‐acquired pneumonia and chronic pulmonary diseases exacerbations. However, their contribution to acute respiratory failure events requiring intensive care unit admission in the era of rapid multiplex molecular assay deserves further evaluation. This study investigated the burden of viral infections in non immunocompromised patients admitted to the intensive care unit for acute respiratory failure using a multiplex molecular assay. Patients were investigated for RVs using immunofluoresence testing and a commercial multiplex molecular assay, and for bacteria using conventional culture. Half the patients (34/70, 49%) had a documented RVs infection. No other pathogen was found in 24 (71%) patients. Viral infection was detected more frequently in patients with obstructive respiratory diseases (64% vs. 29%; P = 0.0075). Multiplex molecular assay should be considered as an usefull diagnostic tool in patients admitted to the intensive care unit with acute respiratory failure, especially those with acute exacerbations of chronic obstructive pulmonary disease and asthma. J. Med. Virol. 86:1198–1202, 2014. © 2013 Wiley Periodicals, Inc.
Keywords: acute respiratory failure, pneumonia, polymerase chain reaction assay, respiratory viruses
Abbreviations
- CI
confidence interval
- hMPV
human metapneumovirus
- IF
immunofluorescence
- IQR
interquartile range
- NA
not available
- OR
odds ratio
- PIV
parainfluenza viruses
- RSV
respiratory syncytial virus
- RVs
respiratory virus(es).
INTRODUCTION
Respiratory viruses (RVs) represent one of the leading causes of community‐acquired pneumonia [File, 2003; de Roux et al., 2004] and a frequent trigger of exacerbations of asthma or chronic obstructive pulmonary disease. However, their contribution to acute respiratory failure requiring intensive care unit admission has not been extensively studied. The lack of rapid and sensitive diagnostic methods and the absence of effective antiviral treatment have limited the interest for RVs detection in routine. Recently, new multiplex molecular assays that allow the detection of up to 22 pathogens in few hours have been introduced [Liolios et al., 2001; van Elden et al., 2002; Reijans et al., 2008] and could impact patients care, in particular for the prescription of anti‐infective treatments [Woo et al., 1997; Barenfanger et al., 2000].
The aim of the study was to assess the burden of viral infections in non immunocompromised patients admitted to intensive care unit for acute respiratory failure using a multiplex molecular assay. The study also aimed to describe the clinical characteristics of viral infections and the bacterial co‐infections.
Patients and Methods
Seventy out of 207 non immunocompromised patients who were admitted with acute respiratory failure to our closed intensive care unit in a teaching hospital from January 2007 to July 2009 had a multiplex molecular assay performed on respiratory specimens and were included in the present study. The decision to perform virological screening was at the discretion of the attending physician. The institutional review board of the Clermont Ferrand teaching hospital approved this retrospective observational study and waived the need for informed consent. Acute respiratory failure was defined as a respiratory rate greater than 30 breaths per minute or respiratory distress symptoms or PaO2 on room air lower than 60 mmHg or a need for ventilatory support. Patients underwent noninvasive tests to look for infections, including non‐induced sputum examination for bacteria, mycobacteria, and fungi; blood cultures; and urine tests for bacterial antigens. Whenever possible, patients did not received antibiotics before sampling for bacterial culture. Bronchoscopy and bronchoalveolar lavage were performed when deemed appropriate by the attending physician. Bronchoalveolar lavage fluid was collected as described previously [Azoulay et al., 2008] and was used for bacterial, mycobacterial, and fungal cultures; detection of RVs antigen by immunofluorescence (IF); and cytological examination. Echocardiography, chest computed tomography, and thoracocentesis were also performed when deemed appropriate by the attending physician. IF (Argene, Verniolle, France) was performed routinely to test nasopharyngeal aspirates and bronchoalveolar lavage fluid for influenza A and B viruses; respiratory syncytial virus (RSV); parainfluenza viruses (PIV) 1, 2, and 3; and adenoviruses. Human metapneumovirus (hMPV) was sought starting in October 2007. All specimens were then stored frozen at minus 80°C until processing for multiplex molecular assay at the end of inclusion period. Total nucleic acids were purified from 200 µl of respiratory specimens by using the EasyMag System (Biomérieux, Marcy l'Etoile, France) and eluted in a final volume of 100 µl. Molecular investigation for RVs was carried out using the multiplex molecular assay, RespiFinder19 (Pathofinder, Maastricht, The Netherlands), that detects and differentiates 14 RVs including influenza viruses A and B; PIV‐1 to PIV‐4; RSV A and B; rhinovirus; human coronaviruses 229E, OC43, and NL63; hMPV; and adenovirus. Additionally, this multiplex molecular assay detected influenza A H5N1 and four bacteria (Chlamydophila pneumoniae, Mycoplasma pneumoniae, Legionella pneumophila, and Bordetella pertussis).
Quantitative parameters were reported as median and interquartile range (IQR, 25th–75th percentiles) and were compared using the Mann–Whitney U test or the Wilcoxon test, as appropriate. Qualitative parameters were reported as number and percentage and were compared using the χ2 test or Fisher's exact test, as appropriate. Odds ratios (OR) and their 95% confidence intervals (95%CIs) were computed. P values less than 0.05 were considered significant. Statistical analyses were performed using Statview 5.0 (SAS Institute, Cary, NC).
RESULTS
Seventy patients (41 males and 29 females) with a median age of 62 (IQR, 52–76) years were included. Fourty‐seven (67%) were mechanically ventilated, 17 (24%) had circulatory insufficiency, and 12 (17%) needed renal replacement therapy. Median simplified acute physiology score II was 29 (20–35). Median intensive care unit length of stay was 5 (3–11) days and 14 (20%) patients died in the hospital. The main diagnosis at admission was acute exacerbation of chronic obstructive pulmonary disease or asthma for 39 patients, community acquired pneumonia for 14 patients, and non‐infectious lung disesases for 17 patients (Fig. 1).
Figure 1.
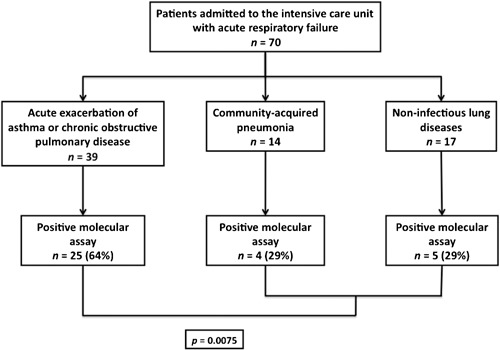
Etiologies of acute respiratory failure in study patients. Data are number (percent). Results are from the univariate analysis. Respiratory viruses isolated in patients with chronic obstructive respiratory diseases were: influenza A in 9 and B in 2; RSV in 3; hMPV in 4; adenovirus in 1; rhinovirus in 4; coronavirus NL63 in 1; coronavirus OC43 in 1; and coronavirus 229E in 1. One patient had a viral dual infection with influenza B and rhinovirus. Respiratory viruses isolated in patients with community‐acquired pneumonia were: influenza A in 1; RSV in 1; PIV 3 in 1; rhinovirus in 1; and coronavirus NL63 in 1. One patient had a dual viral infection with PIV3 and rhinovirus. Respiratory viruses isolated in patients with non‐infectious lung diseases were: influenza A in 1; adenovirus in 2; rhinovirus in 1; coronavirus NL63 in 1.
All study patients were investigated for RVs and results are depicted in Table I. Nine (13%) patients had a RVs detected by IF staining compared with 34 out of the 70 patients (49%) using multiplex molecular assay (P < 0.05). More than one third of molecular assay positive patients (13/34) were infected by an influenza virus. Two patients had two viruses detected that were rhinovirus and PIV3 in one and rhinovirus and influenza B in the other. Of note, no atypical bacteria was detected with multiplex molecular assay. Bacterial examination of a respiratory specimen was performed for 65 (93%) patients and was positive in 12 (17%). Streptococcus pneumoniae was the first isolated bacteria (n = 6). The positive molecular assay was the unique positive result among microbial investigations in 24 (34%) patients, including 19 patients with acute exacerbation of asthma or chronic obstructive pulmonary disease. IF staining was positive in only 5 of these 24 patients. Five (7%) patients had an isolated bacterial infection, and 7 (10%) had a bacterial and viral co‐infection (Table II).
Table I.
Results of Viral Investigations
| Variables | Immunofluorescence | Multiplex molecular assay | P |
|---|---|---|---|
| Influenza | |||
| All | 5 | 13 | |
| A | 4 | 11 | |
| B | 1 | 2 | |
| RSV | 2 | 4 | |
| PIV | |||
| All | 0 | 1 | |
| 1 | NA | 0 | |
| 2 | NA | 0 | |
| 3 | NA | 1 | |
| 4 | NA | 0 | |
| hMPV | 2 | 4 | |
| Adenovirus | 0 | 3 | |
| Rhinovirus | NA | 6 | |
| Coronavirus | |||
| All | NA | 5 | |
| NL63 | NA | 3 | |
| OC43 | NA | 1 | |
| 229E | NA | 1 | |
| All virus | 9 | 36 | |
| All patients | 9 (13%) | 34 (49%) | <0.05 |
Data are number (percent). Results are from the univariate analysis.
hMPV, human metapneumovirus; NA, not available; PIV, parainfluenza virus; RSV, respiratory syncytial virus.
Table II.
Cross‐Results of Viral and Bacterial Investigations
| Study cohort (n = 70) | Flu (n = 13) | RSV (n = 4) | PIV‐3 (n = 1) | hMPV (n = 4) | Adenovirus (n = 3) | Rhinovirus (n = 6) | Corona–virus (n = 5) | Any virus (n = 34) | No virus (n = 36) | |
|---|---|---|---|---|---|---|---|---|---|---|
| Patients with culture | 65 (93%) | 13 (100%) | 3 (75%) | 1 (100%) | 4 (100%) | 2 (66%) | 5 (83%) | 5 (100%) | 31 (91%) | 34 (94%) |
| Bacteria in sputum | 12 (17%) | 2 (15%) | 1 (25%) | 1 (100%) | 3 (75%) | 0 | 1 (17%) | 0 | 7 (21%) | 5 (15%) |
| Streptococcus pneumoniae | 6 (9%) | 2 (15%) | 1 (25%) | 1 (100%) | 0 | 0 | 1 (17%) | 0 | 4 (12%) | 2 (6%) |
| Haemophilus Influenzae | 2 (3%) | 0 | 0 | 0 | 1 (25%) | 0 | 0 | 0 | 1 (3%) | 1 (3%) |
| Pseudomonas aeruginosa | 1 (1%) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 (3%) |
| Staphylococcus Aureus | 1 (1%) | 0 | 0 | 0 | 1 (25%) | 0 | 0 | 0 | 1 (3%) | 0 |
| Enterobacter Aerogenes | 1 (1%) | 0 | 0 | 0 | 1 (25%) | 0 | 0 | 0 | 1 (3%) | 0 |
| Acromobacte sp | 1 (1%) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 (3%) |
Data are number (percent).
hMPV, human metapneumovirus; NA, not available; PIV, parainfluenza virus; RSV, respiratory syncytial virus.
Compared to the patients with negative molecular assay, positive molecular assay patients (with or without bacterial co‐infection) were more often treated with inhaled corticosteroids (53% vs. 25%; P = 0.03; OR 3.38, 95% CI [1.23–9.28]), had more often bronchospasm on physical examination (62% vs. 31%; P = 0.017; OR 3.67, 95% CI [1.36–9.89]), had less interstitial infiltrates on chest radiography (15% vs. 42%; P = 0.026; OR 0.24, 95% CI [0.08–0.77]), and had a lower hospital mortality (9% vs. 33%; P = 0.027; OR 0.19, 95% CI [0.05–0.76]). Patients with acute exacerbation of asthma or chronic obstructive pulmonary disease were more prone to have a positive multiplex molecular assay compared to patients with community acquired pneumonia or non‐infectious lung diseases (64% vs. 29%; P = 0.0075; OR 4.36, 95% CI [1.58–12]). In this subgroup, no difference in baseline characteristics, symptoms, and outcome was found between positive and negative molecular assay patients.
DISCUSSION
In this study, RVs were detected by multiplex molecular assay in about half the patients admitted to the intensive care unit with acute respiratory failure and were the only pathogen detected in 71% of the RVs infected patients. The multiplex molecular assay had a higher sensitivity than IF staining, accordingly to previous studies [Liolios et al., 2001; van Elden et al., 2002; Legoff et al., 2005; Reijans et al., 2008]. The seemingly higher prevalence of RVs compared to previous reports [Carrat et al., 2006; Daubin et al., 2006] may be due in part to the high sensitivity of multiplex molecular assay, but also to specific characteristics of our study population. In this cohort, patients with acute exacerbation of asthma or chronic obstructive pulmonary disease were more prone to viral infection compared to the other patients. Respiratory viruses were the only microbial documentation in most cases. We believe this clearly emphasizes the major role of RVs as a trigger of exacerbations in patients with chronic underlying respiratory conditions [Couch et al., 1997; Lieberman et al., 2002; Sethi, 2011]. However, the present results do not allow any firm conclusion regarding the exact significance of RVs detection by molecular screening during acute respiratory failure. The detection of RVs by molecular assay alone has been associated with lower virus titers and with fewer reported respiratory symptoms compared to concomitant detection by both molecular assay and conventional methods [Jansen et al., 2011]. It suggests that molecular methods may allow the detection of mildly symptomatic stages of RVs infections or even asymptomatic viral shedding. Thus, we cannot exclude that some of our patients exhibited asymptomatic viral shedding concomitant with respiratory failure of any other etiology. Finally, more than one third of the study patients were infected by influenza viruses most of which were missed by IF assay. First, this underlines the role of influenza in severe exacerbations of chronic obstructive pulmonary disease and asthma and thus the critical importance to promote vaccination in these population. Second, these results support the need for a rapid influenza diagnosis to ensure proper infection control measures.
This study reports lower mortality rates in patients with positive multiplex molecular assay compared to those with negative assays. This apparent better prognosis of virus‐associated respiratory disorders, as described in the study by Daubin et al. [2006], likely reflects the better prognosis of acute exacerbations of chronic respiratory conditions compared to the other etiologies of acute respiratory failure.
The rapid detection of isolated viral infection could be considered to shorten or even withhold antibiotic therapy in patients with exacerbation of chronic obstructive pulmonary disease or asthma. In these patients, a positive molecular assay may give a plausible etiology for the acute respiratory failure event thus allowing to stop antibiotics in patients with negative bacterial investigation or even to withhold antibiotics in those with no clinical and radiological signs of pneumonia. The short delay about a few hours to render results of multiplex molecular assay could help clinicians to move toward this objective. However, intensivists may be reluctant to stop antibiotics in the settings of acute respiratory failure and the impact of molecular testing for RVs on antibiotics consumption deserves further investigation.
Finally, no atypical bacteria was detected and despite the size‐population limitation this has to also be taken into consideration for probabilist antibiotic treatment.
The monocenter retrospective design, the low sample size and the decision to perform virological screening let at the discretion of the attending physician are important limitations that hamper the interpretation of the clinical impact of RVs detection in our study. In particular, the selection process of our study patients may have introduced a major bias. On one hand, attending physician may have been more prone to perform virological assays in patients at higher probability of RVs infection (i.e., contact with infected people or flu‐like syndrome). On the other hand, they may have excluded patients with well‐defined etiology for acute respiratory failure thus missing potential concomitant RVs infections. The net effect of this on the observed RVs infection frequency will remain difficult to determine and hampers the interpretation of the present results.
This study demonstrates the high diagnostic yield for virus detection using a single multiplex molecular assay in patients admitted to the intensive care unit with acute respiratory failure, and especially those with acute exacerbation of obstructive respiratory diseases. This diagnostic tool providing results in a few hours allows to reconsider RVs detection as part of the etiologic investigations in patients with acute respiratory failure, especially for those with acute exacerbations of asthma, or chronic obstructive pulmonary disease. Whether a rapid diagnosis translates into clinical benefit with reduced antibiotics and more appropriate and cost‐effectiveness care deserves further investigation.
All authors contributed to the study design, data collection, data analysis, and to the preparation of the manuscript.
Conflict of interest: All authors report no conflict relevant to this study.
REFERENCES
- Azoulay E, de Miranda S, Bele N, Schlemmer B. 2008. Diagnostic strategy for acute respiratory failure in patients with haematological malignancy. Rev Mal Respir 25:433–449. [DOI] [PubMed] [Google Scholar]
- Barenfanger J, Drake C, Leon N, Mueller T, Troutt T. 2000. Clinical and financial benefits of rapid detection of respiratory viruses: An outcomes study. J Clin Microbiol 38:2824–2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrat F, Leruez‐Ville M, Tonnellier M, Baudel JL, Deshayes J, Meyer P, Maury E, Galimand J, Rouzioux C, Offenstadt G. 2006. A virologic survey of patients admitted to a critical care unit for acute cardiorespiratory failure. Intensive Care Med 32:156–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couch RB, Englund JA, Whimbey E. 1997. Respiratory viral infections in immunocompetent and immunocompromised persons. Am J Med 102:2–9; discussion 25‐26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daubin C, Parienti JJ, Vincent S, Vabret A, du Cheyron D, Ramakers M, Freymuth F, Charbonneau P. 2006. Epidemiology and clinical outcome of virus‐positive respiratory samples in ventilated patients: A prospective cohort study. Crit Care 10:R142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Roux A, Marcos MA, Garcia E, Mensa J, Ewig S, Lode H, Torres A. 2004. Viral community‐acquired pneumonia in nonimmunocompromised adults. Chest 125:1343–1351. [DOI] [PubMed] [Google Scholar]
- File TM. 2003. Community‐acquired pneumonia. Lancet 362:1991–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen RR, Wieringa J, Koekkoek SM, Visser CE, Pajkrt D, Molenkamp R, de Jong MD, Schinkel J. 2011. Frequent detection of respiratory viruses without symptoms: Toward defining clinically relevant cutoff values. J Clin Microbiol 49:2631–2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legoff J, Guerot E, Ndjoyi‐Mbiguino A, Matta M, Si‐Mohamed A, Gutmann L, Fagon JY, Belec L. 2005. High prevalence of respiratory viral infections in patients hospitalized in an intensive care unit for acute respiratory infections as detected by nucleic acid‐based assays. J Clin Microbiol 43:455–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman D, Gelfer Y, Varshavsky R, Dvoskin B, Leinonen M, Friedman MG. 2002. Pneumonic vs nonpneumonic acute exacerbations of COPD. Chest 122:1264–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liolios L, Jenney A, Spelman D, Kotsimbos T, Catton M, Wesselingh S. 2001. Comparison of a multiplex reverse transcription‐PCR‐enzyme hybridization assay with conventional viral culture and immunofluorescence techniques for the detection of seven viral respiratory pathogens. J Clin Microbiol 39:2779–2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijans M, Dingemans G, Klaassen CH, Meis JF, Keijdener J, Mulders B, Eadie K, van Leeuwen W, van Belkum A, Horrevorts AM, Simons G. 2008. RespiFinder: A new multiparameter test to differentially identify fifteen respiratory viruses. J Clin Microbiol 46:1232–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethi S. 2011. Molecular diagnosis of respiratory tract infection in acute exacerbations of chronic obstructive pulmonary disease. Clin Infect Dis 52:S290–S295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Elden LJ, van Kraaij MG, Nijhuis M, Hendriksen KA, Dekker AW, Rozenberg‐Arska M, van Loon AM. 2002. Polymerase chain reaction is more sensitive than viral culture and antigen testing for the detection of respiratory viruses in adults with hematological cancer and pneumonia. Clin Infect Dis 34:177–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo PC, Chiu SS, Seto WH, Peiris M. 1997. Cost‐effectiveness of rapid diagnosis of viral respiratory tract infections in pediatric patients. J Clin Microbiol 35:1579–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]


