Abstract
In Lao People's Democratic Republic (PDR), acute respiratory infections overburden the health care system, but viral etiology, genetic diversity, and seasonality, especially in light of the introduction of influenza vaccination in the country, are poorly understood. From August 2010 to April 2011, 309 outpatients were recruited at the Luang Prabang Provincial Hospital covering highland Lao communities. Nasopharyngeal swabs were screened for the presence of 13 respiratory viruses. At least one virus was detected in 69.6% and dual/triple viral infections in 12.9%/1.9% of the patients. Influenza A and B viruses combined were the most frequently detected pathogens, followed by human adenovirus and respiratory syncytial virus (RSV). The other viruses were detected in less than 10% of the patients. Phylogenetic analyses on a representative set of RSV strains revealed that, while otherwise very rare, the RSV‐B CB1/THB genotype cocirculated with other common genotypes. A single wave of influenza virus and RSV activity was observed during the rainy season, providing further support to influenza vaccination before the onset of the rains. This study provides recommendations for influenza vaccination that still needs optimization and highlights the need for revised guidelines for treatment and prevention of respiratory infections in Lao PDR, as well as for increased surveillance efforts.
Keywords: influenza virus, respiratory syncytial virus, respiratory tract, seasonal incidence
1. INTRODUCTION
In Lao People's Democratic Republic (PDR), a lower‐middle income country in Southeast Asia, acute respiratory infections are a leading cause of morbidity and mortality in the general population1 and pneumonia is a leading cause of death in children under 5 years old. As in other countries, respiratory infections represent a considerable burden to the health care system, for instance, up to 10% of outpatients or emergency admissions in Vientiane hospitals meet the influenza‐like illness (ILI) case definition.2
The etiology of respiratory diseases is complex. Influenza A and B viruses (family Orthomyxoviridae, genus Influenzavirus A and B) are frequently detected at all ages, but a variety of pathogens, including viruses and bacteria, cause similar symptoms that cannot be differentiated without laboratory confirmation. In Lao PDR, the influenza sentinel network was initiated only in 20073 and other respiratory viruses are normally not diagnosed.3, 4 In particular, respiratory syncytial virus (RSV; order Mononegavirales, family Pneumoviridae, genus Orthopneumovirus, and species human orthopneumovirus) is a leading cause of lower respiratory tract infections in infants and young children worldwide.5 A proper understanding of the etiology, seasonal patterns, and genetic diversity of respiratory viruses in Lao PDR is warranted. Notably, the success of the influenza vaccination recently introduced in the country relies on recommendations for optimal timing with respect to the season with highest influenza transmission. This is still poorly understood in subtropical countries. Nasopharyngeal swabs from patients with acute respiratory infections at the Luang Prabang Provincial Hospital in Northern Laos, catering to highland communities in Northern Laos, were screened for 13 viruses.
2. MATERIALS AND METHODS
2.1. Patients and specimen collection
Samples were collected at the Provincial Hospital in Luang Prabang, a town with a population of 56 000 located at an altitude of 700 m. The hospital provides primary care without particular specialization to the local population of the town and is a major referral center for the highland communities living throughout this mountainous province. Nasopharyngeal swabs were collected from 309 outpatients (51.1% male) presenting with acute respiratory illness with common cold symptoms with (287 of 309) or without fever. On average, 34 samples per month (range, 16 to 68) were conveniently collected between August 2010 and April 2011. The age of the participants ranged from 1 month to 63 years with 68.0% of the patients younger than 5 years old (median age, 2; mean age, 8). The study was approved by the Laos National Ethics Committee (336‐NECHR), and informed consent was obtained from all patients, their parents, or legal guardian prior to sample collection.
2.2. Viral nucleic acid detection
All samples (n = 309) were tested by two independent laboratories (Lab1 and Lab2). Nucleic acids were purified using Qiagen Viral RNA Mini Kits (Qiagen, Venlo, The Netherlands; Lab1) or NucliSENS easyMAG system (bioMérieux, Bruxelles, Belgium; Lab2). In Lab1, after reverse transcription with SuperScript III Reverse Transcriptase (Life Technologies, Merelbeke, Belgium), sample screening was performed using in‐house polymerase chain reactions (PCRs) for influenza A virus,6, 7 RSV,8 parainfluenzaviruses 1‐4 (PIV1‐4),9, 10, 11 human metapneumovirus (hMPV),12 human coronaviruses 229E (hCoV‐229E) and OC43 (hCoV‐OC43).13 The same pathogens as well as influenza B virus, human adenovirus (hAdV), human bocavirus (hBoV), human rhinovirus (hRV), and human coronavirus HKU1 (hCoV‐HKU1) were detected by Lab2 using the FTD respiratory pathogens 21 kit (Fast‐track Diagnostics, Esch‐sur‐Alzette, Luxembourg). All samples were tested by the two laboratories to compare detection rates of the in‐house PCRs against commercial tests and to broaden the range of pathogens detected. Results of conventional PCRs were visualized on an agarose gel, and positivity was confirmed by sequencing on an ABI 3130 Avant Capillary Sequencer (Applied Biosystems, Nieuwerkerk, The Netherlands) using the PCR primers also as sequencing primers. The ectodomain of the G‐gene of RSV was also amplified as described before14 and sequenced (GenBank accession numbers MG773863‐MG773881).
2.3. Data analyses
Genetic distances and phylogenetic trees were calculated using maximum composite likelihood model with 500 bootstrap values in MEGA6.15 Onset and offset of the monsoon season in Luang Prabang were retrieved from Annual Mekong Flood Reports.16, 17 Descriptive statistical analyses were performed using SigmaPlot 12.5, using χ 2 test and z test for low proportions, Mann‐Whitney rank‐sum test for virological status according to age, Fisher exact test for seasonality of virus‐positive detections according to age groups, and McNemar test for assessing differences between laboratories. Comparison of detection rates between this study and a previous study carried out in Vientiane Capital3 was performed using z test.
3. RESULTS
3.1. Prevalence of respiratory pathogens
At least one virus was detected in 69.6% (215 of 309) of the patients by at least one of the two laboratories. Influenza A and B viruses (combined) were the most frequently detected pathogens (79 of 309, 25.6%; Table 1), with influenza B viruses (37 of 79, 46.8%) dominating, followed by A(H1N1)pdm09 (21 of 79, 26.6%) and A(H3N2) viruses (12 of 79, 15.2%; nine of 42 influenza A strains not characterized; Figure 1A). hAdV (40 of 309, 12.9%) and RSV (31 of 309, 10.0%) were also frequently detected, while the other viruses were detected in less than 10% of the patients, and PIV4 was not detected at all (Table 1).
Table 1.
Virus detections by laboratory, patient age group, and number of viral mixed infections
| Virus | No. of positive samples per diagnostic laboratory | No. of positive patients per age group, n (%) | Total no. of positive detections (%) | No. of viral mixed infections | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Lab1 | Lab2 | P a | 0‐4 | 5‐15 | 15‐50 | >50 | P b | |||
| N | 309 | 309 | – | 210 | 35 | 58 | 6 | – | 309 | |
| Influenza A | 39 | 42 | 0.450 | 18 (8.6) | 6 (17.1) | 16 (27.6) | 2 (33.3) | <0.001 | 42 (13.6) | 6 |
| Influenza B | nd c | 37 | – | 12 (5.7) | 10 (28.6) | 15 (25.9) | 0 | <0.001 | 37 (12.0) | 4 |
| RSV | 29 | 26 | 0.450 | 26 (12.4) | 4 (11.4) | 1 (1.7) | 0 | 0.152 | 31 (10.0) | 7 |
| PIV1 | 9 | 7 | 0.617 | 8 (3.8) | 0 (0.0) | 2 (3.4) | 0 | 0.323 | 10 (3.2) | 6 |
| PIV2 | 3 | 1 | 0.617 | 3 (1.4) | 1 (2.9) | 0 | 0 | 0.428 | 4 (1.3) | 2 |
| PIV3 | 11 | 10 | 1.000 | 11 (5.2) | 1 (2.9) | 0 | 0 | 0.273 | 12 (3.9) | 7 |
| PIV4 | 0 | 0 | – | 0 | 0 | 0 | 0 | – | 0 | 0 |
| hMPV | 16 | 19 | 0.248 | 14 (6.7) | 5 (14.3) | 0 | 0 | 0.173 | 19 (6.1) | 6 |
| hCOV‐OC43 | 5 | 11 | 0.041 | 10 (4.8) | 1 (2.9) | 0 | 0 | 0.558 | 11 (3.6) | 6 |
| hCoV‐229E | 0 | 2 | – | 1 (0.5) | 1 (2.9) | 0 | 0 | 0.546 | 2 (0.6) | 2 |
| hCoV‐HKU1 | nd | 15 | – | 11 (5.2) | 2 (5.7) | 2 (3.4) | 0 | 0.929 | 15 (4.9) | 6 |
| hRV | nd | 22 | – | 19 (9.0) | 1 (2.9) | 2 (3.4) | 0 | 0.050 | 22 (7.1) | 9 |
| hAdV | nd | 40 | – | 37 (17.6) | 3 (8.6) | 0 (0.0) | 0 | 0.008 | 40 (12.9) | 22 |
| hBoV | nd | 22 | – | 22 (10.5) | 0 | 0 (0.0) | 0 | 0.002 | 22 (7.1) | 15 |
| Total d | – | – | – | 150 (71.4) | 27 (77.1) | 36 (62.1) | 2 (33.3) | 0.878 | 215 (69.6) | 46 |
McNemar test.
Mann‐Whitney rank‐sum test.
Not done.
Total number of patients per age group category positive for ≥1 virus.
hAdV, human adenovirus; hBoV, human bocavirus; hCoV, human coronaviruses; hMPV, human metapneumovirus; hRV, human rhinovirus; PIV, parainfluenzaviruses; RSV, respiratory syncytial virus.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
Figure 1.
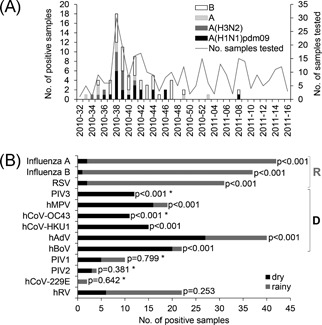
Seasonal occurrence of influenza A and B viruses in Lao People's Democratic Republic (color‐coded bars) and numbers of samples tested (line) by calendar weeks (A); virus occurence by dry and rainy season (B). *P values by χ 2 tests or z tests. D, significantly more frequent during dry season; hAdV, human adenovirus; hBoV, human bocavirus; hCoV, human coronaviruses; hMPV, human metapneumovirus; hRV, human rhinovirus; PIV, parainfluenzaviruses; R, significantly more frequent during rainy season; RSV, respiratory syncytial virus
When comparing the results obtained by the two laboratories, more samples were positive for RSV and PIV1‐3 in Lab1 compared to Lab2, while more samples were positive for influenza A, hMPV, hCoV‐OC43, and hCoV‐229E in Lab2 (Table 1). Discrepant results were observed only in weak positive samples. A statistical difference between the results obtained by the two laboratories was only found for hCoV‐OC43 (P = 0.041; Table 1), suggesting that most in‐house PCRs were performing well and can be more sustainably used in resource‐limited countries compared to commercial PCRs.
3.2. Mixed infections
Dual and triple infections were detected in 12.9% (40 of 309) and 1.9% (6 of 309) of the patients (Table 1). All viruses were involved in at least one mixed infection, but hAdV and hBoV were implicated in 47.8% (22 of 46) and 32.6% (15 of 46) of the mixed infections. The combinations of viruses varied greatly between patients, with 60.9% (28 of 46) of mixed infections being due to a unique virus combination. Mixed hAdV‐hBoV infections were observed in nine patients.
3.3. Age‐specific distribution
All viruses combined, the virological status (ie, positivity for ≥1 virus) of the patients was not significantly associated with age. However, patients with viral mixed infections were significantly younger than those with a single infection (median age, 19.5 vs 28.0 months; P = 0.015). PIV2, PIV3, hMPV, hCoV‐OC43, hCoV‐229E, hAdV, and hBoV were also exclusively found in children under 15 years old (Table 1), whereas influenza viruses were detected in all age groups and RSV (n = 1), PIV1 (n = 2), hCoV‐HKU1 (n = 2), and hRV (n = 2) were only sporadically found in the 15‐ to 50‐year age group (Table 1). Influenza A (median age, 8.5 vs 2.0 years; P < 0.001) or influenza B (median age, 7.0 vs 2.0 years; P < 0.001) positive patients were significantly older than those who tested negative, whereas hAdV (median age, 15.5 vs 24.0 months; P = 0.008) and hBoV (median age, 12.5 vs 24.0 months; P = 0.002) positive patients were significantly younger than negative patients.
3.4. Seasonal distribution
Influenza A and B viruses were almost exclusively detected during a single wave of activity between August and December 2010 (weeks 33 to 49; peak in week 38). A(H1N1)pdm09, A(H3N2), and B viruses cocirculated during the peak of the epidemic, but A(H3N2) was no longer found after week 41 (Figure 1A). Influenza A and B viruses were significantly more often detected during the rainy season (1 May 2010 to 15 December 2010 in Luang Prabang),17 similarly to RSV (Figure 1B). In contrast, PIV3, hMPV, hCoV‐OC43, hCoV‐HKU1, hAdV, and hBoV prevalences were significantly higher during the dry season (16 December 2010 to 27 April 2011;16 Figure 1B). No difference in seasonality depending on the age group was observed.
3.5. Phylogenetic analysis of RSV
Partial G protein sequences were obtained for 24 of 31 RSV‐positive samples and characterized as RSV‐A (1 of 24, 4.2%; genotype NA1; Figure S1) and RSV‐B (23 of 24, 95.8%; Figure 2). The majority (20 of 23, 87.0%) of RSV‐B strains grouped within genotype BA9 and were highly similar to each other (genetic distance of 0% to 1.6%). The remaining RSV‐B strains (3 of 23, 13.0%; genetic distance 0%) clustered together with sequences from China, Thailand, and India, previously attributed to a new CB118 or THB19 genotype, here named CB1/THB. Sequences from this genotype were detected as early as 2000‐2004 in China (exact date unknown20) but mainly after 2009. The genetic distances between sequences of the CB1/THB genotype ranged from 0% to 4.3%. Three strains from China (BJ/20265, BJ/23183, and BJ/23484) had a three‐amino‐acid insertion that was not found in any other sequence of the CB1/THB genotype.
Figure 2.
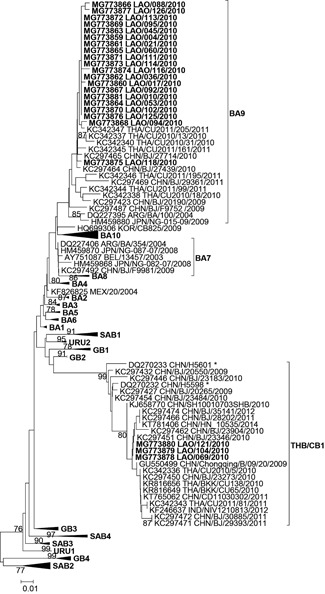
Phylogenetic analyses of RSV‐B strains. Strain name is as follows: GenBank accession number followed by three‐letter country isocode/strain/collection year. Sequences from this study in boldface. Only bootstrap values greater than 70% are shown. *Strains isolated between 2000 and 2004 in China (no exact date available)
4. DISCUSSION
In this provincial hospital catering to the needs of the town of Luang Prabang and to the highland communities in northern Lao PDR, essentially 70% of all outpatients presenting with respiratory symptoms recruited were below the age of 5 and 70% of the participating patients suffered from a viral infection. Despite the low prevalence of hRV, the incidence of viral infections was significantly higher than in ILI patients in Vientiane capital3 (2007‐2008; 321 of 543, 59.1%; P = 0.003). Detection rates are influenced by various factors, such as inclusion criteria, case definitions, age of participants as well as cultural and ecological differences. When only children under 5 years were compared in both locations, the incidence of viral infections was similar (150 of 210, 71.4% in the current study; 118 of 169, 69.8%;3 P = 0.820), suggesting higher variabilities among older age groups.
Seasonality of virus circulation may also contribute to differences in disease incidence between studies. In Luang Prabang in 2010, the incidence of influenza A and B viruses was significantly associated with the rainy season; in 2010, the influenza season presented as a single wave with a peak in September and declined in October‐November, similar to other regions in Lao PDR during the same year.2, 21, 22 Also RSV was mostly found during the rainy season in Luang Prabang in 2010, similarly to Thailand,23 Vietnam,24 Cambodia25 and to Vientiane in 2009‐20104 and 2014.26 In contrast, other respiratory viruses (hAdV, hBoV, hMPV, hCoV‐HKU1, hCoV‐OC43, and PIV3) occurred more often during the dry season, causing respiratory disease mainly when influenza and RSV were less present, while PIV1‐2, hRV, and hCoV‐229E were detected throughout the year, contributing to the background incidence of respiratory infections.2 The time covered by the current study may have somewhat biased seasonality patterns and will require confirmation over longer time periods.
In 2012, Lao PDR started to offer seasonal influenza vaccination in April‐May to pregnant women, the elderly, patients with comorbidities, and health care personnel.27 Based on retrospective surveillance data from 2008‐201128 and 2010‐201429 with increased transmission in August‐December or September‐November, respectively, the optimal timing for vaccination was recommended in April‐June.28, 29 In our 2010‐2011 study, increased transmission of influenza viruses was observed between August and December, indicating that, if already implemented, vaccination during the recommended period would likely have had a beneficial effect on the 2010 influenza season. As surveillance efforts increase worldwide, it becomes apparent that influenza seasonality in the (sub)tropics are country or even region‐specific30 and may also differ between years. For instance, two waves of increased influenza transmission were observed in Lao PDR in 2014 and 2015 (http://www.who.int/flunet), complicating the current vaccination timing. Offering vaccination twice a year (eg, April‐May and October‐November) to the general or even a selected population would be challenging in practice, as already attempted in 2012 in Peru,29 a country that has influenza seasons varying by climate zones.31 Thus, continuing surveillance efforts in Lao PDR are warranted to characterize potential regional and annual variations of influenza seasons, and to estimate the influenza disease burden due to waning of vaccine‐induced antibodies at the time of a secondary influenza transmission period to optimize vaccination recommendations. In the meantime, the target population such as pregnant women enrolled in antenatal care programs could be readily vaccinated at any time of the year, provided that the most recent vaccine formulation is available.
In this study, RSV strains circulating in Lao PDR were characterized for the first time, with the detection of the common genotypes of RSV‐A NA1, RSV‐B BA9, and the rare CB1/THB genotype. Cocirculation of different genotypes in a country during RSV season is not uncommon.32, 33 Most RSV genotypes are often shared between various regions of the world. For instance, after its first characterization in Canada in 2010,34 the RSV‐A ON1 genotype rapidly spread throughout the world, with at least 21 countries reporting it by 2014.35 Luang Prabang attracts a lot of foreign tourists; therefore, the presence of the global genotypes RSV‐A NA1 and RSV‐B BA918, 25 is not surprising. In contrast, genotype RSV‐B CB1/THB has only very rarely been detected in Asia (China, Thailand, India, and Lao PDR; see Figure 2), as early as 2000‐2004 but mainly between 2009 and 2012.18, 19, 20 This may be indicative of differences in virulence or cross‐protective immunogenicity with other genotypes. Further genotyping of RSV strains, especially from the Greater Mekong subregion, is required to better understand the particular epidemiology of the CB1/THB genotype.
5. CONCLUSIONS
In conclusion, 70% of acute respiratory infections in outpatients in Provincial Hospital in Northern Laos had a viral etiology. While some viruses contributed to the background incidence throughout the year, others were mainly detected during the dry season (PIV3, hMPV, hCoV‐OC43, hCoV‐HKU1, hAdV, and hBoV). Influenza and RSV infections presented as a single wave during the wet season, providing further support to a single vaccination against influenza before the onset of the rains. While otherwise very rare, the RSV‐B CB1/THB genotype cocirculated with other common genotypes. Antibiotics should be prescribed with care in the case of uncomplicated respiratory infections36 and the timing of vaccination against influenza may be further optimized. Revised guidelines for the treatment and prevention of respiratory infections in Lao PDR, as well as increased surveillance efforts, are therefore needed.
CONFLICTS OF INTEREST
The authors declare that there are no competing conflicts of interest.
Supporting information
Supporting information
Suppl. Figure S1. Phylogenetic analyses of RSV‐A strains. Strain name is as follows: GenBank accession number followed by 3‐letter country isocode/strain/collection year. Sequences from this study in boldface. Only bootstrap values > 70% are shown
ACKNOWLEDGMENTS
The authors thank the study volunteers for their participation and the staff from Luang Prabang Provincial Hospital for sample collection. They also thank S. Wolter and R. Sinner for their technical help. This study was supported by the Ministère des Affaires étrangères et européennes, Luxembourg and the Luxembourg Institute of Health through “PaReCIDS” project.
Snoeck CJ, Ponghsavath V, Luetteke N, et al. Etiology of viral respiratory infections in Northern Lao People's Democratic Republic. J Med Virol. 2018;90:1553–1558. 10.1002/jmv.25237
References
REFERENCES
- 1. World Health Organization. WHO Country Cooperation Strategy for the Lao People's Democratic Republic, 2012‐2015. 2011: 46. http://www.wpro.who.int/laos/publications/country_cooperation_lao_2012-2015/en/. Accessed 10 July 2017.
- 2. Khamphaphongphane B, Ketmayoon P, Lewis HC, et al. Epidemiological and virological characteristics of seasonal and pandemic influenza in Lao PDR, 2008‐2010. Influenza Other Respir Viruses. 2013;7(3):304‐311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Vongphrachanh P, Simmerman JM, Phonekeo D, et al. An early report from newly established laboratory‐based influenza surveillance in Lao PDR. Influenza Other Respir Viruses. 2010;4(2):47‐52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sentilhes AC, Choumlivong K, Celhay O, et al. Respiratory virus infections in hospitalized children and adults in Lao PDR. Influenza Other Respir Viruses. 2013;7(6):1070‐1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhang Y, Yuan L, Zhang Y, Zhang X, Zheng M, Kyaw MH. Burden of respiratory syncytial virus infections in China: Systematic review and meta‐analysis. J Glob Health. 2015;5(2):020417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ducatez MF, Olinger CM, Owoade AA, et al. Avian flu: multiple introductions of H5N1 in Nigeria. Nature. 2006;442(7098):37‐37. [DOI] [PubMed] [Google Scholar]
- 7. Ward CL, Dempsey MH, Ring CJ, et al. Design and performance testing of quantitative real time PCR assays for influenza A and B viral load measurement. J Clin Virol. 2004;29(3):179‐188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cane PA, Pringle CR. Respiratory syncytial virus heterogeneity during an epidemic: analysis by limited nucleotide sequencing (SH gene) and restriction mapping (N gene). J Gen Virol. 1991;72(Pt 2):349‐357. [DOI] [PubMed] [Google Scholar]
- 9. Aguilar JC, Pérez‐Breña MP, García ML, Cruz N, Erdman DD, Echevarría JE. Detection and identification of human parainfluenza viruses 1, 2, 3, and 4 in clinical samples of pediatric patients by multiplex reverse transcription‐PCR. J Clin Microbiol. 2000;38(3):1191‐1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Echevarría JE, Erdman DD, Swierkosz EM, Holloway BP, Anderson LJ. Simultaneous detection and identification of human parainfluenza viruses 1, 2, and 3 from clinical samples by multiplex PCR. J Clin Microbiol. 1998;36(5):1388‐1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Karron RA, Froehlich JL, Bobo L, Belshe RB, Yolken RH. Rapid detection of parainfluenza virus type 3 RNA in respiratory specimens: use of reverse transcription‐PCR‐enzyme immunoassay. J Clin Microbiol. 1994;32(2):484‐488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Maertzdorf J, Wang CK, Brown JB, et al. Real‐time reverse transcriptase PCR assay for detection of human metapneumoviruses from all known genetic lineages. J Clin Microbiol. 2004;42(3):981‐986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vabret A, Mouthon F, Mourez T, Gouarin S, Petitjean J, Freymuth F. Direct diagnosis of human respiratory coronaviruses 229E and OC43 by the polymerase chain reaction. J Virol Methods. 2001;97(1‐2):59‐66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sato M, Saito R, Sakai T, et al. Molecular epidemiology of respiratory syncytial virus infections among children with acute respiratory symptoms in a community over three seasons. J Clin Microbiol. 2005;43(1):36‐40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30(12):2725‐2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mekong River Commission. Annual Meking Flood Report 2011, 2014:72. http://www.mrcmekong.org/assets/Publications/basin-reports/Annual-Mekong-Flood-Report-2011.pdf. Accessed 10 August 2017.
- 17. Mekong River Commission . Annual Mekong Flood Report 2010, 2011:76. http://www.mrcmekong.org/assets/Publications/basin-reports/Annual-Mekong-Flood-Report-2010.pdf. Accessed 10 August 2017.
- 18. Cui G, Zhu R, Qian Y, et al. Genetic variation in attachment glycoprotein genes of human respiratory syncytial virus subgroups a and B in children in recent five consecutive years. PLoS One. 2013;8(9):e75020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Auksornkitti V, Kamprasert N, Thongkomplew S, et al. Molecular characterization of human respiratory syncytial virus, 2010‐2011: identification of genotype ON1 and a new subgroup B genotype in Thailand. Arch Virol. 2014;159(3):499‐507. [DOI] [PubMed] [Google Scholar]
- 20. Deng JZR, Quian Y, Zhoa L, Wang F. Sequence analysis of G glycoprotein of human respiratory syncytial virus subtype B strains isolated from children with acute respiratory infections in Beijing, China in years 2000–2004. Chin J Microbiol Immunol. 2006;26(1):1‐5. [Google Scholar]
- 21. Phommasack B, Moen A, Vongphrachanh P, et al. Capacity building in response to pandemic influenza threats: Lao PDR case study. Am J Trop Med Hyg. 2012;87(6):965‐971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mayxay M, Castonguay‐Vanier J, Chansamouth V, et al. Causes of non‐malarial fever in Laos: a prospective study. Lancet Glob Health. 2013;1(1):e46‐e54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yoshihara K, Le MN, Okamoto M, et al. Association of RSV‐A ON1 genotype with increased pediatric acute lower respiratory tract infection in Vietnam. Sci Rep. 2016;6:27856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bryant JE, Binh BTL, Trinh HN, et al. Direct whole‐genome deep‐sequencing of human respiratory syncytial virus A and B from Vietnamese children identifies distinct patterns of inter‐ and intra‐host evolution. J Gen Virol. 2015;96(12):3470‐3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Arnott A, Vong S, Mardy S, et al. A study of the genetic variability of human respiratory syncytial virus (HRSV) in Cambodia reveals the existence of a new HRSV group B genotype. J Clin Microbiol. 2011;49(10):3504‐3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nguyen VH, Dubot‐Pérès A, Russell FM, et al. Acute respiratory infections in hospitalized children in Vientiane, Lao PDR ‐ the importance of respiratory syncytial virus. Sci Rep. 2017;7(1):9318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Centers for Disease Control and Prevention . Lao People's Democratic Republic Launches Seasonal Flu Vaccination Program. 2012; https://www.cdc.gov/flu/spotlights/vaccination-program-launch-lao.htm. Accessed 21 July 2017.
- 28. Saha S, Chadha M, Al Mamun A, et al. Influenza seasonality and vaccination timing in tropical and subtropical areas of southern and south‐eastern Asia. Bull World Health Organ. 2014;92(5):318‐330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hirve S, Newman LP, Paget J, et al. Influenza seasonality in the tropics and subtropics ‐ when to vaccinate? PLoS One. 2016;11(4):e0153003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hirve S Seasonal Influenza Vaccine Use in Low and Middle Income Countries in the Tropics and Subtropics. A systematic review. 2015. 96. http://apps.who.int/iris/bitstream/handle/10665/188785/9789241565097_eng.pdf;jsessionid=2315C0E35018ADBCFE0B2BA99A495162?sequence=1. Accessed 15 May 2018.
- 31. Laguna‐Torres VA, Gómez J, Ocaña V, et al. Influenza‐like illness sentinel surveillance in Peru. PLoS One. 2009;4(7):e6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Espinosa Y, San Martín C, Torres A, Farfán M, Torres J, Avadhanula V, Piedra P, Tapia L, et al. Genomic loads and genotypes of respiratory syncytial virus: viral factors during lower respiratory tract infection in Chilean hospitalized infants. Int J Mol Sci. 2017;18(3):654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tuan TA, Thanh TT, Hai NT, et al. Characterization of hospital and community‐acquired respiratory syncytial virus in children with severe lower respiratory tract infections in Ho Chi Minh City, Vietnam, 2010. Influenza Other Respir Viruses. 2015;9(3):110‐119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Eshaghi A, Duvvuri VR, Lai R, et al. Genetic variability of human respiratory syncytial virus A strains circulating in Ontario: a novel genotype with a 72 nucleotide G gene duplication. PLoS One. 2012;7(3):e32807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Duvvuri VR, Granados A, Rosenfeld P, Bahl J, Eshaghi A, Gubbay JB. Genetic diversity and evolutionary insights of respiratory syncytial virus A ON1 genotype: global and local transmission dynamics. Sci Rep. 2015;5:14268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Quet F, Vlieghe E, Leyer C, et al. Antibiotic prescription behaviours in Lao People's Democratic Republic: a knowledge, attitude and practice survey. Bull World Health Organ. 2015;93(4):219‐227. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information
Suppl. Figure S1. Phylogenetic analyses of RSV‐A strains. Strain name is as follows: GenBank accession number followed by 3‐letter country isocode/strain/collection year. Sequences from this study in boldface. Only bootstrap values > 70% are shown


