Abstract
To determine the prevalence of human rhinovirus (HRV) infection in children with acute asthma exacerbations, investigation of HRV viral load and severity of asthma exacerbations is also required. Nasopharyngeal aspirates and swabs were collected and assessed for respiratory viruses. HRV‐positive samples were sequenced to identify types and determine viral load. Outpatients with asthma exacerbations underwent follow‐up evaluations, their swabs were collected and clinical outcomes were recorded at their next clinic visit 4 weeks later. One hundred forty‐three inpatients and 131 outpatients, including 88 patients with asthma exacerbations and 43 controls with stable asthma were recruited. HRV‐A was mainly detected in September and February (45.5% and 33.3%, respectively), while HRV‐C was mainly detected in November and April (70.0% and 55.6%, respectively). HRV‐C was the primary type and was primarily found in inpatients with severe asthma exacerbations. HRV‐A viral load in the group of inpatients with severe exacerbations was higher than in the mild and moderate groups (P < 0.001 and P = 0.022). The HRV‐A viral load of both inpatients and outpatients was higher than that of HRV‐C (P < 0.001 and P = 0.036). The main genotypes were HRV‐C53 and HRV‐A20 among inpatients, and this genotype caused more severe clinical manifestations. HRV persisted for no more than 4 weeks, and their symptoms or signs of disease were well‐controlled well. HRV‐C was most frequently detected in asthma exacerbations. HRV‐A with high viral load led to severe asthma exacerbations.
Keywords: asthma exacerbations, children, human rhinoviruses, type, viral load
1. INTRODUCTION
Asthma is a major childhood health risk. Acute asthma exacerbations remain a significant cause of morbidity in children and can lead to an accelerated decline in lung function,1, 2, 3 which emphasises the importance of finding an appropriate preventive treatment. Epidemiologic studies have detected viral infections in more than 80% of school children with asthma exacerbations.4 Of the respiratory viruses known to cause asthma exacerbations, between 60% and 70% are types of human rhinoviruses (HRV).5, 6 Moreover, one recent study showed that HRV was associated with a significantly increased risk of day‐to‐day asthma symptoms in children.7 Understanding the role of HRV in acute asthma exacerbations may lead to interventions that will improve asthma management.
HRV is a single‐stranded RNA virus belonging to the genus Enterovirus and the family Picornaviridae. There are more than 100 classical HRV serotypes, which are divided into groups A (HRV‐A) and B (HRV‐B). Because of developments in molecular methods, recent studies have discovered over 50 new HRV strains referred to as HRV‐C.8, 9, 10, 11, 12, 13, 14 Molecular epidemiological studies suggest that the dominant types are HRV‐A and HRV‐C, while HRV‐B is relatively rarely detected.12, 13, 14 HRV‐C accounts for the majority of asthma attacks in children and causes more severe attacks than other HRV types and other viruses.6, 15, 16, 17, 18, 19 In our previous study, we found that HRV infection was associated with asthma in our region.20 Some authors have demonstrated a close correlation between the HRV viral load and the disease severity in children with lower respiratory tract infections.21, 22, 23 However, there have been no reports on the correlation of the viral load of different HRV species with the severity of asthma exacerbations.
The objective of the present study is to determine the prevalence of HRV infection in children with acute asthma exacerbations and the virus detection after acute asthma acute attacks as well as to investigate the relationship between different HRV types, viral load and severity of acute asthma exacerbations.
2. MATERIALS AND METHODS
2.1. Study participants and sample collection
During the study from March 2012 through November 2015, we enrolled inpatients with acute asthma exacerbations from the Department of Respiratory Medicine and outpatients from November 2014 to December 2015 at the Children's Hospital of Chongqing Medical University in China. Inclusion criteria were: children with asthma exacerbations and symptom onset ≤3 days and children with stable asthma to make up the control group. Exclusion criteria were as follows: children with chronic pulmonary disease, cardiopathy, metabolic and genetic diseases, or immunosuppression. The diagnosis of asthma and the severity of asthma exacerbations were assessed according to the guidelines of the Global Initiative for Asthma.
Nasopharyngeal aspirates (NPAs) and swabs (NPSs) were collected when the patients were admitted to our department, and clinical outcomes were recorded. We conducted a follow‐up study among these outpatients with asthma exacerbations and their NPSs were collected, and clinical outcomes were recorded when they visited the clinic again after 4 weeks. This study was approved by the Ethics Committee of the Children's Hospital of Chongqing Medical University. The informed consent for participation in the study was obtained from patients or legal guardians.
2.2. Definition of the severity of asthma exacerbations
The severity of asthma exacerbations at presentation was determined using a previously validated reference standard for each subject by a paediatrician and divided into mild, moderate and severe groups.24 Asthma symptom control was assessed with the Childhood Asthma Control Test25 and divided into well‐controlled, not well‐controlled and poorly controlled groups according to the clinical symptom score.
2.3. HRV nucleotide sequence analysis and viral load detection
The 5′‐NCR (non‐coding region) and a partial sequence of the VP4/VP2 region was amplified by PCR as previously described.26, 27, 28 The PCR products were sequenced by Shanghai Majorbio Bio‐Pharm Technology. The nucleotide sequences were compared with reference HRV strains obtained from GenBank for typing and to distinguish human enterovirus (HEV).
Amplification of HRV RNA by real‐time fluorescence quantitative PCR (RT‐PCR) was performed with the 5′‐NCR primers and probe.29 RT‐PCR was performed using a TaKaRa OneStep PCR kit (Takara Biotechnology, Dalian, China).
2.4. Phylogenetic analysis
Phylogenetic analysis of HRV‐positive strains was performed as described previously using the neighbour‐joining (NJ) method,27, 30, 31 and the reliability of the tree was estimated with 1000 bootstrap replications. We performed a phylogenetic tree of the VP4/VP2 coding region and 5′‐NCR using the CLUSTAL W program and MEGA 5.05 software. Sequences of reference strains were obtained from the GenBank database.
2.5. Detection of other respiratory viruses
Respiratory syncytial virus types A and B (RSVA, RSVB), influenza virus A, B and C (IFVA, IFVB, IFVC), human coronaviruses (HCoV‐229E, HCoV‐OC43), human metapneumovirus (HMPV) and parainfluenza virus (PIV types 1‐4) were screened using nested PCR assays.32, 33 Adenovirus (ADV) was detected by means of multiplex PCR, and human bocavirus 1 (HBoV1) was screened using RT‐PCR.34, 35 All the specimens were analyzed using a commercial detection kit (TaKaRa Biotechnology, Dalian, China), according to the manufacturer's instructions.
2.6. Statistical analysis
Data were analyzed using the SPSS 17.0 software package. Categorical variables were compared using the chi‐squared or Fisher's exact test, and continuous variables were compared using Student's t‐test or the nonparametric Mann‐Whitney U‐test. Two‐sided P‐values of <0.05 were considered statistically significant.
3. RESULTS
3.1. Population demographics
A total of 143 inpatients with acute asthma exacerbations and another 131 outpatients, including 88 patients with asthma exacerbations and 43 controls with stable asthma were enrolled in this study. The age of inpatients ranged from 17 to 170 months (median, 45 months), and the male to female ratio was 84:59. The age of outpatients ranged from 22 to 144 months (median, 64 months), and the male to female ratio was 81:50. The percentage of mild asthma exacerbations was 21.7% (31/143), moderate was 53.1% (76/143) and severe was 25.2% (36/143) of inpatients. In outpatients, 20.5% (18/88) of exacerbations were mild, 58.0% (51/88) were moderate and 21.6% (19/88) were severe.
3.2. Virus detection
Among inpatients, at least one virus was detected in 121 (84.6%) cases. HRV was identified in 72 samples (50.3%), and 65.3% (47/72) of the cases were identified as single HRV infections. Other common viruses included 36 RSV cases (25.2%), 13 PIV cases (9.1%), 12 HBoV1 cases (8.4%), 9 IFVA cases (6.3%), 6 ADV cases (4.2%), 2 HMPV cases (1.4%), and 8 HEV cases (5.6%). Among outpatients, respiratory viruses were detected in 45 (34.4%) subjects, and 40 were single infections. The rate of viral infections was 43.2% (38/88) in the group with asthma exacerbations. The HRV‐positive rate in the group with asthma exacerbations was 15.9% (14/88). The detection rate of both total virus and HRV of inpatients was significantly higher than that of outpatients (84.6% vs 43.2%; P < 0.001 and 50.3% vs 15.9%; P < 0.001). The detection rate of both total virus and HRV of inpatients was significantly higher than that of the stable asthma group (84.6% vs 16.3%; P < 0.001 and 50.3% vs 2.3%; P < 0.001). Furthermore, both the rate of total virus and HRV‐positive status in outpatients with asthma exacerbation was higher than that of the stable asthma group (43.2% vs 16.3%; P = 0.002 and 15.9% vs 2.3%; P = 0.045) (Table 1).
Table 1.
Specific detected virus among all patients
| Inpatients | Outpatients | ||
|---|---|---|---|
| Virus | Asthma exacerbation (n = 143) N (%) | Asthma exacerbation (n = 88) N (%) | Stable asthma (n = 43) N (%) |
| HRV | 72 (50.3) | 14 (15.9) | 1 (2.3) |
| HRV‐A | 25 (17.5) | 3 (3.4) | 1 (2.3) |
| HRV‐B | 5 (3.5) | 1 (1.1) | 0 (0) |
| HRV‐C | 42 (29.4) | 9 (10.2) | 0 (0) |
| Untyped | 0 (0) | 1 (1.1) | 0 (0) |
| RSV | 36 (25.2) | 4 (4.5) | 0 (0) |
| RSVA | 23 (16.1) | 3 (3.4) | 0 (0) |
| RSVB | 13 (9.1) | 1 (1.1) | 0 (0) |
| PIV | 13 (9.1) | 9 (10.2) | 3 (7.0) |
| PIV3 | 8 (5.6) | 5 (5.7) | 3 (7.0) |
| PIV4 | 5 (3.5) | 4 (4.5) | 0 (0) |
| IFV | 9 (6.3) | 3 (3.4) | 1 (2.3) |
| IFVA | 9 (6.3) | 1 (1.1) | 0 (0) |
| IFVB | 0 (0) | 2 (2.3) | 1 (2.3) |
| HBoV1 | 12 (8.4) | 3 (3.4) | 2 (4.7) |
| ADV | 6 (4.2) | 1 (1.1) | 0 (0) |
| HMPV | 2 (1.4) | 1 (1.1) | 0 (0) |
| HCoV | 0 (0) | 1 (1.1) | 0 (0) |
| HEV | 8 (5.6) | 2 (2.3) | 0 (0) |
| Total | 121 (84.6) | 38 (43.2) | 7 (16.3) |
HRV, human rhinovirus; RSV, respiratory syncytial virus; PIV, parainfluenza virus; IFV, influenza virus; HBoV1, human bocavirus type 1; ADV, adenovirus; HMPV, human metapneumovirus; HCoV, human coronaviruses; HEV, human enterovirus.
3.3. HRV types and asthma exacerbations
In hospitalized children, the VP4/VP2 region was successfully amplified in 56 samples: HRV‐A was identified in 35.7% of samples (20/56), HRV‐B in 8.9% (5/56), and HRV‐C in 55.4% (31/56). The number of HRV‐C‐positive cases was more than those of HRV‐A and HRV‐B (P = 0.037 and P < 0.001). The HRV‐C detection rate in severe groups was significantly higher than that of the mild group (36.1% vs 6.5%; P = 0.004). The rates of HRV‐A and HRV‐B‐positivity rated showed no different in the three severity groups (P = 0.596 and P = 0.731). The VP4/VP2 sequence was amplified in one outpatient, while others could not be obtained because of low amplicon yield, and 5′‐NCR sequences from the remaining HRV‐positive specimens were used for typing. The majority of those in the group with acute asthma exacerbations (64.3%; 9/14) was infected with an HRV‐C, 21.4% (3/14) was infected with HRV‐A, only one patient was infected with HRV‐B and one was untyped (Table 2). Because of the small number of outpatients with HRV in the different severity groups, comparisons were not performed.
Table 2.
HRV types and severity of acute asthma exacerbations in children
| Inpatients (n = 143) N (%) | Outpatients (n = 88) N (%) | ||||||
|---|---|---|---|---|---|---|---|
| Severity | HRV‐A | HRV‐B | HRV‐C | Severity | HRV‐A | HRV‐B | HRV‐C |
| Mild (n = 31) | 6 (19.4) | 1 (3.2) | 2 (6.5) | Mild (n = 18) | 0 (0) | 0 (0) | 4 (22.2) |
| Moderate (n = 76) | 9 (11.8) | 2 (2.6) | 16 (21.1) | Moderate (n = 51) | 1 (2.0) | 1 (2.0) | 3 (5.9) |
| Severe (n = 36) | 5 (13.9) | 2 (5.6) | 13 (36.1) * | Severe (n = 19) | 2 (10.5) | 0 (0) | 2 (10.5) |
HRV indicates human rhinovirus.
There were more HRV‐C positive inpatients in the severe asthma exacerbation than in the mild exacerbation group (P = 0.004).
3.4. Clinical features of HRV positive and negative patients
The percentage of severe acute asthma exacerbations in the HRV‐positive group was higher than that of the HRV‐negative group (P = 0.032), while the number of patients with fever was less than that in the HRV‐negative group (P = 0.007). The white blood cell counts and neutrophil percentages were both significantly higher in the HRV‐positive group (P = 0.041 and P = 0.043, respectively). The other features between the HRV‐positive and HRV‐negative groups had no statistically significant difference. All the clinical features between single HRV‐A‐positive and HRV‐C‐positive groups had no statistically significant difference (Table 3). The percentage of familial eczema and neutrophils were significantly higher in the group of outpatients with HRV infection than in the group without HRV infection (P = 0.049 and P = 0.030, respectively). The clinical characteristics of HRV‐A and HRV‐C‐positive outpatients showed that the percentage of allergic rhinitis in the HRV‐C‐positive group was significantly higher than that of the HRV‐A group (P = 0.045) (Table 4).
Table 3.
Comparison of clinical features between HRV‐positive and HRV‐negative inpatients with asthma exacerbations
| HRV N (%) | HRV single infection N (%) | |||||
|---|---|---|---|---|---|---|
| Variable | Single infection (n = 47) | Negative (n = 71) | P | HRV‐A (n = 16) | HRV‐C (n = 29) | P |
| Male:female | 23:24 | 47:24 | 0.062 | 5:11 | 16:13 | 0.124 |
| Age (median, month) | 40 | 43 | 0.658 | 48 | 37 | 0.109 |
| History of eczema | 13 (27.7) | 26 (36.6) | 0.311 | 4 (25.0) | 8 (27.6) | 1.000 |
| History of allergic conditions | 8 (17.0) | 18 (25.4) | 0.285 | 3 (18.8) | 5 (17.2) | 1.000 |
| Familial asthma | 28 (59.6) | 43 (60.6) | 0.858 | 10 (62.5) | 16 (55.2) | 0.634 |
| Cough | 47 (100) | 71 (100) | ‐ | 16 (100) | 29 (100) | ‐ |
| Rhinorrhea | 8 (17.0) | 12 (16.9) | 0.986 | 5 (31.3) | 3 (10.3) | 0.177 |
| Expectoration | 34 (72.3) | 47 (66.2) | 0.481 | 11 (68.8) | 21 (72.4) | 1.000 |
| Wheezing | 41 (87.2) | 58 (81.7) | 0.422 | 14 (87.5) | 26 (89.7) | 1.000 |
| Fever | 14 (29.8) | 39 (54.9) | 0.007 | 5 (31.3) | 9 (31.0) | 1.000 |
| Severe exacerbation | 16 (34.0) | 12 (16.9) | 0.032 | 5 (31.3) | 10 (34.5) | 0.826 |
| Length of hospitalization (median, day) | 5 (3‐8) | 5 (2‐16) | 0.388 | 5 (3‐6) | 5 (4‐8) | 0.250 |
| White blood cell (×109/L) | 12.38 ± 5.13 | 10.77 ± 4.87 | 0.041 | 12.16 ± 5.71 | 12.58 ± 5.05 | 0.868 |
| Neutrophils (%) | 64.70 ± 20.27 | 56.44 ± 21.95 | 0.043 | 64.50 ± 21.18 | 65.31 ± 19.85 | 0.915 |
| Lymphocyte (%) | 30.60 ± 18.50 | 39.03 ± 21.19 | 0.031 | 29.63 ± 18.42 | 30.62 ± 18.47 | 0.859 |
| CRP > 8 mg/L (%) | 6 (12.8) | 8 (11.3) | 0.805 | 3 (18.8) | 3 (10.3) | 0.737 |
| Platelet (×109/L) | 333 ± 102 | 321 ± 94 | 0.308 | 319 ± 106 | 337 ± 102 | 0.887 |
HRV indicates human rhinovirus. Categorical variables were compared using the chi‐squared test, and the continuous variables were compared using the Student's t‐test or the nonparametric Mann‐Whitney U‐test. Two‐sided P‐values of < 0.05were considered statistically significant.
Table 4.
Comparison of clinical features between HRV‐positive and HRV‐negative outpatients with asthma exacerbations
| HRV N (%) | HRV‐positive N (%) | |||||
|---|---|---|---|---|---|---|
| Variable | Positive (n = 14) | Negative (n = 74) | P | HRV‐A (n = 3) | HRV‐C (n = 9) | P |
| Male:female | 8:6 | 49:25 | 0.729 | 2:1 | 5:4 | 1.000 |
| Age (median, month) | 60 | 64 | 0.941 | 54 | 56 | 0.926 |
| History of allergic rhinitis | 12 (85.7) | 49 (66.2) | 0.257 | 1 (33.3) | 9 (100) | 0.045 |
| History of eczema | 5 (35.7) | 24 (32.4) | 1.000 | 1 (33.3) | 4 (44.4) | 1.000 |
| Familial allergic rhinitis | 3 (21.4) | 10 (13.5) | 0.723 | 1 (33.3) | 2 (22.2) | 1.000 |
| Familial eczema | 3 (21.4) | 3 (4.1) | 0.049 | 1 (33.3) | 2 (22.2) | 1.000 |
| Familial asthma | 1 (7.1) | 12 (16.2) | 0.641 | 0 (0) | 1 (11.1) | 1.000 |
| Passive smoking | 6 (42.9) | 18 (24.3) | 0.271 | 0 (0) | 5 (55.6) | 0.310 |
| Cough | 13 (92.9) | 71 (95.9) | 0.507 | 3 (100) | 8 (88.9) | 1.000 |
| Expectoration | 6 (42.9) | 22 (29.7) | 0.513 | 2 (66.7) | 3 (33.3) | 0.735 |
| Wheezing | 13 (92.9) | 57 (77.0) | 0.324 | 3 (100) | 8 (88.9) | 1.000 |
| Fever | 1 (7.1) | 1 (1.4) | 0.294 | 0 (0) | 1 (11.1) | 1.000 |
| Tachypnea | 6 (42.9) | 14 (18.9) | 0.107 | 0 (0) | 5 (55.6) | 0.310 |
| White blood cell (×109/L) | 10.22 ± 3.66 | 9.85 ± 3.06 | 0.698 | 9.49 ± 2.76 | 11.26 ± 3.47 | 0.307 |
| Neutrophils (%) | 56.87 ± 13.93 | 46.95 ± 12.63 | 0.030 | 60.67 ± 17.21 | 58.43 ± 11.39 | 0.540 |
| Eosinophils (%) | 6.26 ± 3.68 | 6.48 ± 3.94 | 0.791 | 8.33 ± 6.81 | 5.39 ± 2.30 | 0.757 |
| FEV1% predicted | 91.68 ± 12.78 | 92.69 ± 14.12 | 0.913 | 92.70 ± 0 | 95.30 ± 12.89 | 0.655 |
| PEF% predicted | 93.35 ± 11.39 | 80.25 ± 19.47 | 0.147 | 75.30 ± 0 | 96.20 ± 12.08 | 0.180 |
| Positive skin tests | 7 (50.0) | 42 (58.1) | 0.431 | 1 (33.3) | 6 (66.7) | 0.735 |
FEV1, forced expiratory volume in one second; PEF, peak expiratory flow. Two‐sided P‐values of < 0.05were considered statistically significant.
3.5. Seasonality of HRV infections and asthma exacerbations
The frequency of asthma exacerbations among HRV‐positive inpatients was calculated as 48.0% (12/25) in spring, 44.0% (11/25) in summer, 60.6% (43/71) in autumn, and 27.3% (6/22) in winter during the study period by merging the same month. HRV‐A was mainly detected in September and February (45.5% and 33.3%, respectively), while HRV‐C was mainly detected in November and April (70.0% and 55.6%, respectively) (Fig. 1).
Figure 1.
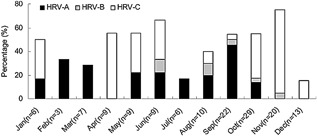
Seasonal incidence (rate of positive samples) by month of HRV‐A, HRV‐B, and HRV‐C. Data was combined from 3 years and bar graph represents the detection rate of the three HRV types
3.6. HRV viral load and severity of asthma exacerbations
The genome viral load of 62 inpatients infected with HRV has been detected, and 10 cases have not been detected using the 5′‐NCR primers and probe. The median age of these inpatients was 45 months, and the male to female ratio was 32:30. The genome viral load ranged from 2.2 to 2.9 × 107 copies/mL. However, there was no significant difference of HRV total viral load among the different severities of asthma exacerbations (Fig. 2A). The viral load of HRV‐A in the severe group was higher than that in the mild group (mean, 2.5 × 105 copies/mL vs 1.7 × 103 copies/mL; P < 0.001) and the moderate group (mean, 2.5 × 105 copies/mL vs 2.1 × 103 copies/mL; P = 0.022) (Fig. 2B). However, the HRV‐C viral load among the three groups showed no significant difference (Fig. 2C). Moreover, the viral loads of HRV‐A and HRV‐B were significantly higher than the HRV‐C viral load regardless of the severity (mean, 8.5 × 103 copies/mL vs 2.5 × 102 copies/mL; P < 0.001; mean, 1.6 × 104 copies/mL vs 2.5 × 102 copies/mL; P = 0.014) (Fig. 2D). The viral load of HRV‐A was higher than HRV‐C in the group with severe asthma exacerbations (mean, 2.5 × 105 copies/mL vs 1.6 × 102 copies/mL; P < 0.0001) (Fig. 2E). Because of the small number of outpatients with HRV in the different severity groups, the comparisons between different HRV types viral load and severity of disease were excluded. Even so, the total viral load of HRV was compared. And the HRV‐A total viral load was significantly higher than that of HRV‐C (mean, 9.4 × 103 copies/mL vs 7.1 × 102 copies/mL; P = 0.036).
Figure 2.
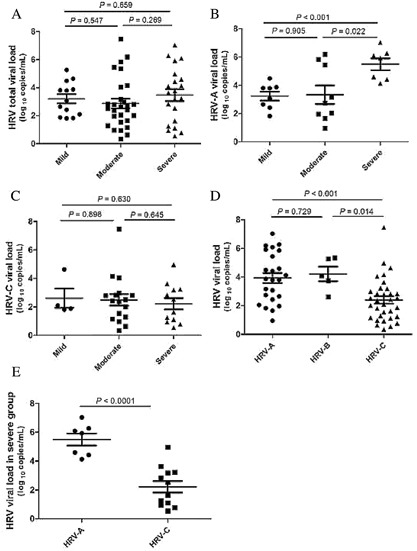
Relationship between HRV viral load and severity of asthma exacerbations among inpatients. A: HRV total viral load among the three groups shows no significant difference. B: HRV‐A viral load in severe group was significantly higher than in the mild and moderate groups (P < 0.001 and P = 0.022). C: HRV‐C viral load the among three groups shows no significant difference. D: HRV‐A and HRV‐B viral load were significantly higher than HRV‐C regardless of the severity (P < 0.001 and P = 0.014). E: In the severe group, HRV‐A viral load was significantly higher than HRV‐C (P < 0.0001). The comparison of viral load was conducted by nonparametric Mann‐Whitney U‐test
3.7. Phylogenetic analysis
Figure 3 shows the phylogenetic tree of inpatients based on the nucleotide sequences of the VP4/VP2 coding region. The 56 HRV‐positive samples from inpatients were grouped into 37 HRV genotypes, 31 HRV‐C samples were grouped into 19 HRV‐C genotypes, 20 HRV‐A samples were grouped into 13 HRV‐A genotypes, and 5 HRV‐B samples were grouped into 5 HRV‐B genotypes. There were six (10.7%) strains which could be designated into novel genotypes and temporarily named HRV‐C53 and four (7.1%) HRV‐A20 strains, the two predominant genotypes, followed by HRV‐C21 three (5.4%), HRV‐A49 three (5.4%) (Fig. 3). The clinical characteristics of HRV‐C53 and HRV‐A20‐positive inpatients are summarized in Table 5, the severity of their asthma exacerbations was moderate or severe and there were five cases of dyspnea. Figure 4 shows the phylogenetic tree of outpatients based on the nucleotide sequences of the 5′‐NCR, and all the HRV‐positive samples were grouped into 15 genotypes. One sample was amplified by the VP4/VP2 region and divided into HRV‐C22 (Accession number: JN621242). The distribution of outpatients’ genotypes had no obvious regularity and neither HRV‐C53 nor HRV‐A20 was found.
Figure 3.
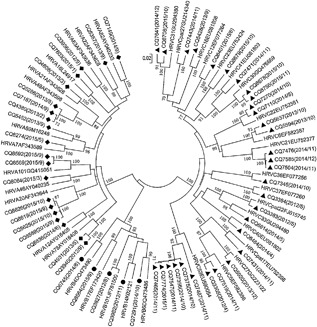
Phylogenetic analysis of the HRV VP4/VP2 coding region from inpatients using the neighbour‐joining method with 1000 bootstrap replicates with MEGA5.05; branches showing >70% bootstrap support are indicated. The strains in this study are marked with CQ (Chongqing) and HRV‐A labeled ♦, HRV‐B labeled ●, and HRV‐C labeled ▴
Table 5.
Clinical features of inpatients with HRV‐C53 and HRV‐A20 infections
| Sample | Genotype | Age/sex (year‐month) | Sample collection time | Symptoms | Severity |
|---|---|---|---|---|---|
| CQ7356 | C53 | 2‐7/Male | 2014/10 | Fever/cough/wheezing/dyspnea | Severe |
| CQ7375 | C53 | 3‐4/Female | 2014/10 | Fever/cough/expectoration/wheezing | Moderate |
| CQ7465 | C53 | 2‐9/Male | 2014/11 | Fever/cough/expectoration/wheezing/dyspnea | Severe |
| CQ7467 | C53 | 2‐9/Female | 2014/11 | Cough/expectoration/wheezing/dyspnea | Severe |
| CQ7547 | C53 | 2‐1/Female | 2014/11 | Fever/cough/expectoration/wheezing/dyspnea/diarrhoea | Severe |
| CQ7774 | C53 | 2‐3/Male | 2015/01 | Cough/expectoration/wheezing | Moderate |
| CQ8589 | A20 | 7‐0/Female | 2015/09 | Cough/expectoration/wheezing | Moderate |
| CQ8619 | A20 | 4‐0/Female | 2015/09 | Cough/wheezing/dyspnea | Severe |
| CQ8625 | A20 | 2‐7/Female | 2015/10 | Cough/expectoration/wheezing | Moderate |
| CQ8626 | A20 | 3‐0/Male | 2015/10 | Fever/cough/expectoration/wheezing/diarrhoea | Moderate |
CQ indicates Chongqing.
Figure 4.
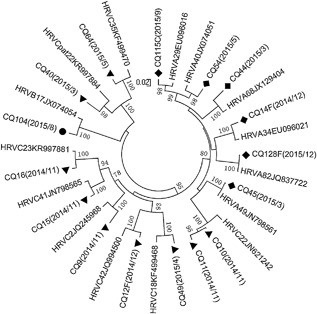
Phylogenetic analysis of the HRV 5′‐NCR region in outpatients using the neighbor‐joining method with 1000 bootstrap replicates with MEGA5.05; branches showing >70% bootstrap support are indicated. The strains in this study are marked with CQ (Chongqing), F (follow‐up samples) and C (control group samples) and HRV‐A labeled ♦, HRV‐B labeled, ● and HRV‐C labeled ▴
3.8. Follow‐up of outpatients
All the HRV‐positive outpatients with acute asthma exacerbations participated in the follow‐up, and only one case was infected with RSVA within 4 weeks after first the visit. The remaining patients tested negative, and the severity of the disease was well‐controlled. Another 23 patients were without HRV during the first visit, while after 4 weeks, five cases of acute asthma exacerbations appeared again and among them, four patients were infected with HRV (Table 6).
Table 6.
Details of follow‐up after 4 weeks in outpatients with asthma exacerbations
| Sample | Age/sex (year‐month) | First visit pathogen | Follow‐up of condition | Follow‐up of pathogen |
|---|---|---|---|---|
| CQ2 | 1‐10/Female | HRV‐C | Well controlled | Negative |
| CQ9 | 4‐0/Male | HRV‐C | Well controlled | Negative |
| CQ10 | 4‐4/Female | HRV‐C | Well controlled | Negative |
| CQ11 | 5‐4/Male | HRV‐C | Re‐exacerbation | RSVA |
| CQ15 | 4‐8/Female | HRV‐C | Well controlled | Negative |
| CQ16 | 8‐0/Male | HRV‐C | Well controlled | Negative |
| CQ40 | 3‐8/Female | HRV‐C | Well controlled | Negative |
| CQ49 | 7‐8/Female | HRV‐C | Well controlled | Negative |
| CQ64 | 7‐2/Male | HRV‐C/IFVB | Well controlled | Negative |
| CQ44 | 3‐7/Male | HRV‐A | Well controlled | Negative |
| CQ45 | 12‐11/Female | HRV‐A | Well controlled | Negative |
| CQ54 | 4‐6/Female | HRV‐A/HBoV1 | Well controlled | Negative |
| CQ104 | 5‐11/Female | HRV‐B | Well controlled | Negative |
| CQ29 | 8‐7/Male | HRV‐untyped | Well controlled | Negative |
| CQ14 | 8‐0/Female | RSVA | Re‐exacerbation | HRV‐A |
| CQ21 | 12‐0/Female | IFVA | Well controlled | Negative |
| CQ96 | 7‐2/Female | PIV3 | Well controlled | Negative |
| CQ98 | 11‐3/Male | PIV3 | Well controlled | Negative |
| CQ24 | 2‐10/Female | PIV4 | Well controlled | Negative |
| CQ23 | 5‐2/Female | RSVA/PIV4 | Well controlled | Negative |
| CQ12 | 5‐11/Male | Negative | Re‐exacerbation | HRV‐C/IFVA |
| CQ120 | 5‐0/Female | Negative | Re‐exacerbation | HRV‐untyped |
| CQ128 | 8‐9/Female | Negative | Re‐exacerbation | HRV‐A/RSVA |
| CQ31 | 2‐9/Female | Negative | Re‐exacerbation | Negative |
| CQ34 | 1‐5/Male | Negative | Well controlled | Negative |
| CQ37 | 4‐1/Male | Negative | Well controlled | Negative |
| CQ42 | 8‐7/Female | Negative | Well controlled | Negative |
| CQ51 | 8‐7/Female | Negative | Well controlled | Negative |
| CQ60 | 3‐3/Male | Negative | Well controlled | Negative |
| CQ72 | 5‐9/Female | Negative | Well controlled | Negative |
| CQ73 | 3‐9/Female | Negative | Well controlled | Negative |
| CQ112 | 5‐4/Male | Negative | Well controlled | Negative |
| CQ116 | 5‐2/Female | Negative | Well controlled | Negative |
| CQ117 | 2‐5/Male | Negative | Well controlled | Negative |
| CQ119 | 4‐0/Female | Negative | Well controlled | Negative |
| CQ125 | 7‐11/Female | Negative | Well controlled | Negative |
| CQ126 | 4‐7/Female | Negative | Well controlled | Negative |
4. DISCUSSION
HRV is an important cause of both upper and lower respiratory illnesses as well as acute exacerbations of childhood asthma. In our study, HRV was the most common viral pathogen that induced asthma exacerbations in children. This finding is consistent with previous studies,6, 11, 15 and the detection rate of HRV in asthma exacerbation patients was significantly higher than that in the stable asthma group. The detection rate of HRV was 50.3% among inpatients, while some authors reported the rate to be between 60% and 70%. The reason for the difference was that their studies were performed among school‐aged children, and the region was different.5, 6 Among the inpatients, 34.7% (25/72) were diagnosed with HRV co‐infection, 52% (13/25) of cases were diagnosed as co‐infection with RSV, 24% (6/25) of cases were diagnosed as co‐infection with HBoV1, 16% (4/25) of cases were diagnosed as PIV, 8% (2/25) of cases were diagnosed as ADV, 8% (2/25) of cases were diagnosed as IFVA and 4% (1/25) of cases were diagnosed as HEV. The median age was 43 months, and 64% (16/25) were in the moderate to severe category groups. The incidence of HRV in outpatients was lower than that in hospitalized children, but the methods of virus detection are consistent, which may be because the severity of outpatients was relatively mild or onset of other causes, such as medication, is irregular. The detection rate of RSV was 25.2% (36/143) among inpatients, and 13 cases were diagnosed as co‐infection with HRV, 7 cases were diagnosed as co‐infection with other viruses (PIV/ADV/HEV/IFVA) and others were diagnosed as single infections. The median age was 33 months, and 69.5% (25/36) were in the moderate to severe category groups.
Previous studies have demonstrated that neutrophils are the predominant inflammatory cell in the airways of patients with acute asthma exacerbation, and experimental HRV infection has been shown to increase airway neutrophilic inflammation in asthmatic subjects.36, 37, 38 Our clinical and laboratory results, combined with haemograms, indicated that acute viral infections, especially HRV, need to be considered during popular seasons in children with acute asthma attacks, even though the percentage of neutrophils does not exceed 70%.
Although this study only included data from 3 years, we showed both annual and seasonal variation in HRV types. The seasonal prevalence of asthma exacerbations was consistent with HRV infection and occurred mainly in autumn. Our results were similar to those presented in earlier reports, which have associated the September peak in asthma hospitalizations with rhinovirus circulation.39, 40 Our subtropical climate was the reason for the difference.
HRV‐C attracts special clinical interest because they can cause more severe illnesses, requiring hospitalization in infants and children, compared with the HRV‐A or HRV‐B, and are closely associated with acute asthma exacerbations.15, 41, 42 Our study has demonstrated that the HRV‐C detection rate was greater than 50% among inpatients and outpatients, which supports the hypothesis that HRV‐C was to the most common cause of acute asthma attacks in children. Among inpatients, HRV‐C was responsible for not only the majority of acute asthma attacks compared to other HRV species or other known viruses, but was also more frequently involved in severe asthma attacks.
The phylogenetic analysis showed that HRV‐C53 and HRV‐A20 were the predominant strains among inpatients. The HRV‐C53 genotype mainly occurred in October and November 2014, HRV‐A20 occurred in September and October 2015 in Chongqing, China and caused more severe clinical manifestations. HRV‐C53 was once known as HRV‐Cpat10, which was provisionally assigned types (designated “pat”), and we did not find other literature reporting the relationships between asthma attacks and HRV‐C53 or HRV‐Cpat10.27, 28 However, the two genotypes were not found in outpatients, and these results supported that HRV‐C53 and HRV‐A20 were more common in severe asthma attacks.
Some observations suggested that people with asthma might be at risk for higher viral loads and symptoms affecting their respiratory tract during HRV infection.43 Our study confirmed that HRV‐A viral load in the severe asthma exacerbations group was significantly higher than that of the mild and moderate groups, while HRV‐C was not. This result indicates that HRV‐A viral load is associated with the severity of asthma exacerbations but HRV‐C is not. Additionally, in severe asthma exacerbations, the HRV‐A viral load was higher than that of HRV‐C, which suggested that the mechanisms of the exacerbations were not similar.
We further compared clinical features between single HRV‐A positive and HRV‐C positive inpatients and no difference was found between the two groups. However, the number of outpatients with allergic rhinitis increased in the HRV‐C infection group and whether indicated those asthmatic children who complicated with allergic rhinitis were tend to infect HRV‐C even appear exacerbations. The findings highlight the fact that physicians and parents should provide preventive management to those children during the peak season of HRV‐C infections.
HRV‐C species do not grow in standard cell cultures used for virus isolation, which limits the research on their biological characteristics. According to our understanding of the mechanism, it is possible that the HRV‐C receptor is distinct from the intercellular adhesion molecule 1 (ICAM‐1) and low‐density lipoprotein receptor (LDLR) family members used by HRV‐A.44, 45 The latest research has reported that human cadherin‐related family member 3 (CDHR3), a member of the cadherin family of transmembrane proteins, mediates HRV‐C entry into host cells, and an asthma‐related mutation (rs6967330) in this gene is associated with enhanced viral binding and replication. CDHR3 mutations result in susceptibility to childhood asthma with severe exacerbations.46, 47 These findings suggest that CDHR3 is a functional receptor for HRV‐C and explain asthma exacerbations are commonly induced by HRV‐C. Additionally, different HRV types may have different replications and induce diversified cytokines and chemokines, which contribute to varying illness severity.48 Further study is needed to investigate the mechanism of HRV‐A and HRV‐C resulting in different illness severities.
Through the follow‐up among outpatients with asthma exacerbations, HRV persisted for no more than 4 weeks in the nasopharynx, and their symptoms or signs of disease appeared well‐controlled. Some authors have reported that HRV infections in infants rarely result in persistence of RNA beyond 30 days.49 Further studies of large samples are required to confirm these findings. Some asthma exacerbation cases may be caused by other respiratory viruses or may present without viral infection at the first visit, while asthma attacks caused by HRV infections may appear again after 4 weeks.
In summary, the virus most frequently detected in asthma exacerbations appears to be HRV, primarily HRV‐C, which commonly occurs in severe asthma exacerbations. HRV‐A with high viral load leads to severe asthma exacerbations, and HRV‐C viral load and disease severity have no significant correlation. The novel genotypes HRV‐C53 and HRV‐A20 have been found to be predominant strains that cause more severe clinical manifestations. Additionally, we followed up on HRV changes in the airways of asthmatic children, and HRV RNA was found to persist for no longer than 4 weeks. Overall, our results provide some theoretical evidence for the prevention of childhood asthma exacerbations and improving asthma management.
CONFLICTS OF INTEREST
The authors have no conflicts of interest to declare.
ACKNOWLEDGMENTS
We acknowledge the assistance of the patients and their caregivers involved in the study, the staff of the Department of Respiratory Medicine of Children's Hospital of Chongqing, and the contributions of the State Key Laboratory on the Safety of Pathogenic Microorganisms of the Academy of Military Medical Science in Beijing, the Key Laboratory of Developmental Diseases in Childhood at Chongqing Medical University, and the Ministry of Education.
AUTHORS’ CONTRIBUTIONS
E.M.L. conceived and designed this study and revised the manuscripts. S.Y.Z., L.L.W., L.R., J.L., and W.L. collected the samples and performed the experiments. S.Y.Z. analyzed the data and wrote the paper.
Zheng S‐Y, Wang L‐L, Ren L, Luo J, Liao W, Liu E‐M. Epidemiological analysis and follow‐up of human rhinovirus infection in children with asthma exacerbation. J Med Virol. 2017;90: 219–228. 10.1002/jmv.24850
REFERENCES
- 1. Busse WW, Lemanske RF. Asthma. N Engl J Med. 2001; 344:350–362. [DOI] [PubMed] [Google Scholar]
- 2. Garbino J, Gerbase MW, Wunderli W, et al. Lower respiratory viral illnesses: improved diagnosis by molecular methods and clinical impact. Am J Respir Crit Care Med. 2004; 170:1197–1203. [DOI] [PubMed] [Google Scholar]
- 3. Glezen WP, Greenberg SB, Atmar RL, Piedra PA, Couch RB. Impact of respiratory virus infections on persons with chronic underlying conditions. JAMA. 2000; 283:499–505. [DOI] [PubMed] [Google Scholar]
- 4. Johnston SL, Pattemore PK, Sanderson G, et al. Community study of role of viral infections in exacerbations of asthma in 9‐11 year old children. BMJ. 1995; 310:1225–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Johnston SL. Innate immunity in the pathogenesis of virus induced asthma exacerbations. Proc Am Thorac Soc. 2007; 4:267–270. [DOI] [PubMed] [Google Scholar]
- 6. Kim WK, Gern JE. Updates in the relationship between human rhinovirus and asthma. Allergy Asthma Immunol Res. 2012; 4:116–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tovey ER, Stelzer‐Braid S, Toelle BG, et al. Rhinoviruses significantly affect day‐to‐day respiratory symptoms of children with asthma. J Allergy Clin Immunol. 2015; 135:663–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pelon W, Mogabgab WJ, Phillips IA, Pierce WE. A cytopathogenic agent isolated from naval recruits with mild respiratory illnesses. Proc Soc Exp Biol Med. 1957; 94:262–267. [DOI] [PubMed] [Google Scholar]
- 9. Lamson D, Renwick N, Kapoor V, et al. MassTag polymerase‐chain‐reaction detection of respiratory pathogens, including a new rhinovirus genotype, that caused influenza‐like illness in New York State during 2004–2005. J Infect Dis. 2006; 194:1398–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Palmenberg AC, Spiro D, Kuzmickas R, et al. Sequencing and analyses of all known human rhinovirus genomes reveal structure and evolution. Science. 2009; 324:55–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Olenec JP, Kim WK, Lee WM, et al. Weekly monitoring of children with asthma for infections and illness during common cold seasons. J Allergy Clin Immunol. 2010; 125:1001–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Arakawa M, Okamoto‐Nakagawa R, Toda S, et al. Molecular epidemiological study of human rhinovirus species A, B and C from patients with acute respiratory illnesses in Japan. J Med Microbiol. 2012; 61:410–419. [DOI] [PubMed] [Google Scholar]
- 13. Kiyota N, Kushibuchi I, Kobayashi M, et al. Genetic analysis of the VP4/VP2 coding region in human rhinovirus species C inpatients with acute respiratory infection in Japan. J Med Microbiol. 2013; 62:610–617. [DOI] [PubMed] [Google Scholar]
- 14. Kiyota N, Kobayashi M, Tsukagoshi H, et al. Genetic analysis of human rhinovirus species A to C detected inpatients with acute respiratory infection in Kumamoto prefecture, Japan 2011–2012. Infect Genet Evol. 2014; 21:90–102. [DOI] [PubMed] [Google Scholar]
- 15. Bizzintino J, Lee WM, Laing IA, et al. Association between human rhinovirus C and severity of acute asthma in children. Eur Respir J. 2011; 37:1037–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lau SK, Yip CC, Tsoi HW, et al. Clinical features and complete genome characterization of a distinct human rhinovirus (HRV) genetic cluster, probably representing a previously undetected HRV species, HRV‐C, associated with acute respiratory illness in children. J Clin Microbiol. 2007; 45:3655–3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Renwick N, Schweiger B, Kapoor V, et al. A recently identified rhinovirus genotype is associated with severe respiratory‐tract infection in children in Germany. J Infect Dis. 2007; 196:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lauinger IL, Bible JM, Halligan EP, et al. Patient characteristics and severity of human rhinovirus infections in children. J Clin Virol. 2013; 58:216–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Linder JE, Kraft DC, Mohamed Y, et al. Human rhinovirus C: age, season, and lower respiratory illness over the past 3 decades. J Allergy Clin Immunol. 2013; 131:69–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Xiao QY, Zheng SY, Zhou LL, et al. Impact of human rhinovirus types and viral load on the severity of illness in hospitalized children with lower respiratory tract infections. Pediatr Infect Dis J. 2015; 34:1187–1192. [DOI] [PubMed] [Google Scholar]
- 21. Pretorius MA, Tempia S, Treurnicht FK, et al. Genetic diversity and molecular epidemiology of human rhinoviruses in South Africa. Influenza Other Respir Viruses. 2014; 8:567–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Takeyama A, Hashimoto K, Sato M, et al. Rhinovirus load and disease severity in children with lower respiratory tract infections. J Med Virol. 2012; 84:1135–1142. [DOI] [PubMed] [Google Scholar]
- 23. Esposito S, Daleno C, Scala A, et al. Impact of rhinovirus nasopharyngeal viral load and viremia on severity of respiratory infections in children. Eur J Clin Microbiol Infect Dis. 2014; 33:41–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Qureshi F, Pestian J, Davis P, Zaritsky A. Effect of nebulized ipratropium on the hospitalization rates of children with asthma. N Engl J Med. 1998; 339:1030–1035. [DOI] [PubMed] [Google Scholar]
- 25. Liu AH, Zeiger RS, Sorkness CA, et al. The Childhood Asthma Control Test: retrospective determination and clinical validation of a cut point to identify children with very poorly controlled asthma. J Allergy Clin Immunol. 2010; 126:267–273. [DOI] [PubMed] [Google Scholar]
- 26. Kiang D, Kalra I, Yagi S, et al. Assay for 5′ noncoding region analysis of all human rhinovirus prototype strains. J Clin Microbiol. 2008; 46:3736–3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McIntyre CL, Knowles NJ, Simmonds P. Proposals for the classification of human rhinovirus species A, B and C into genotypically assigned types. J Gen Virol. 2013; 94:1791–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Simmonds P, McIntyre CL, Savolainen‐Kopra C, Tapparel C, Mackay IM, Hovi T. Proposals for the classification of human rhinovirus species C into genotypically assigned types. J Gen Virol. 2010; 91:2409–2419. [DOI] [PubMed] [Google Scholar]
- 29. Granados A, Luinstra K, Chong S, et al. Use of an improved quantitative polymerase chain reaction assay to determine differences in human rhinovirus viral loads in different populations. Diagn Microbiol Infect Dis. 2012; 74:384–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980; 16:111–120. [DOI] [PubMed] [Google Scholar]
- 31. Saitou N, Nei M. The neighbor‐joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987; 4:406–425. [DOI] [PubMed] [Google Scholar]
- 32. Coiras MT, Aguilar JC, García ML, Casas I, Pérez‐Breña P. Simultaneous detection of fourteen respiratory viruses in clinical specimens by two multiplex reverse transcription nested‐PCR assays. J Med Virol. 2004; 72:484–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Coiras MT, Perez‐Brena P, Garcia ML, Casas I. Simultaneous detection of influenza A, B, and C viruses, respiratory syncytial virus, and adenoviruses in clinical samples by multiplex reverse transcription nested‐PCR assay. J Med Virol. 2003; 69:132–144. [DOI] [PubMed] [Google Scholar]
- 34. Xu W, McDonough MC, Erdman DD. Species‐specific identification of human adenoviruses by a multiplex PCR assay. J Clin Microbiol. 2000; 38:4114–4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Allander T, Jartti T, Gupta S, et al. Human bocavirus and acute wheezing in children. Clin Infect Dis. 2007; 44:904–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Norzila MZ, Fakes K, Henry RL, Simpson J, Gibson PG. Interleukin‐8 secretion and neutrophil recruitment accompanies induced sputum eosinophil activation in children with acute asthma. Am J Respir Crit Care Med. 2000; 161:769–774. [DOI] [PubMed] [Google Scholar]
- 37. Message SD, Laza‐Stanca V, Mallia P, et al. Rhinovirus‐induced lower respiratory illness is increased in asthma and related to virus load and Th1/2 cytokine and IL‐10 production. Proc Natl Acad Sci USA. 2008; 105:13562–13567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fleming H, Little F, Schnurr D, et al. Rhinovirus‐16 colds in healthy and in asthmatic subjects: similar chages in upper and lower airways. Am J Respir Crit Care Med. 1999; 160:100–108. [DOI] [PubMed] [Google Scholar]
- 39. Johnston NW, Johnston SL, Duncan JM, et al. The September epidemic of asthma exacerbations in children: a search for etiology. J Allergy Clin Immunol. 2005; 115:132–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sears MR, Johnston NW. Understanding the September asthma epidemic. J Allergy Clin Immunol. 2007; 120:526–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Miller EK, Khuri‐Bulos N, Williams JV, et al. Human rhinovirus C associated with wheezing in hospitalized children in the Middle East. J Clin Virol. 2009; 46:85–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Drysdale SB, Alcazar M, Wilson T, et al. Respiratory outcome of prematurely born infants following human rhinovirus A and C infections. Eur J Pediatr. 2014; 173:913–919. [DOI] [PubMed] [Google Scholar]
- 43. Kennedy JL, Shaker M, McMeen V, et al. Comparison of viral load in individuals with and without asthma during infections with rhinovirus. Am J Respir Crit Care Med. 2014; 189:532–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Greve JM, Davis G, Meyer AM, et al. The major human rhinovirus receptor is ICAM‐1. Cell. 1989; 56:839–847. [DOI] [PubMed] [Google Scholar]
- 45. Hofer F, Gruenberger M, Kowalski H, et al. Members of the low density lipoprotein receptor family mediate cell entry of a minor‐group common cold virus. Proc Natl Acad Sci USA. 1994; 91:1839–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bochkov YA, Watters K, Ashraf S, et al. Cadherin‐related family member 3, a childhood asthma susceptibility gene product, mediates rhinovirus C binding and replication. Proc Natl Acad Sci USA. 2015; 112:5485–5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bønnelykke K, Sleiman P, Nielsen K, et al. A genome‐wide association study identifies CDHR3 as a susceptibility locus for early childhood asthma with severe exacerbations. Nat Genet. 2014; 46:51–55. [DOI] [PubMed] [Google Scholar]
- 48. Nakagome K, Bochkov YA, Ashraf S, et al. Effects of rhinovirus species on viral replication and cytokine production. J Allergy Clin Immunol. 2014; 134:332–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Loeffelholz MJ, Trujillo R, Pyles RB, et al. Duration of rhinovirus shedding in the upper respiratory tract in the first year of life. Pediatrics. 2014; 134:1144–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]


