Abstract
A number of human coronaviruses (HCoVs) were reported in the last and present centuries. Some outbreaks of which (eg, SARS and MERS CoVs) caused the mortality of hundreds of people worldwide. The problem of finding a potent drug against HCoV strains lies in the inability of finding a drug that stops the viral replication through inhibiting its important proteins. In spite of its limited efficacy and potential side effects, Ribavirin is extensively used as a first choice against HCoVs. Therefore, scientists reverted towards the investigation of different drugs that can more specifically target proteins. In this study, four anti‐HCV drugs (one approved by FDA and others under clinical trials) are tested against HCoV polymerases. Quantitative Structure‐Activity Relationship (QSAR) and molecular docking are both used to compare the performance of the selected nucleotide inhibitors to their parent nucleotides and Ribavirin. Both QSAR and molecular docking showed that IDX‐184 is superior compared to Ribavirin against MERS CoV, a result that was also reported for HCV. MK‐0608 showed a performance that is comparable to Ribavirin. We strongly suggest an in vitro study on the potency of these two drugs against MERS CoV.
Keywords: docking, HCV, human coronavirus, nucleotide inhibitor, polymerase, QSAR
1. INTRODUCTION
Middle East Respiratory Syndrome Coronavirus (MERS CoV) was first identified 4 years ago in the Kingdom of Saudi Arabia.1 This was the first emergence of a new epidemic outbreak since the Sever Acute Respiratory Syndrome Coronavirus (SARS CoV) that out broke in East Asia in late 2002.2, 3, 4, 5 While SARS fatality rate was 8%, MERS fatality rate was 36% (by 9th of November 2016, the number of laboratory confirmed infections was 1813 and the number of reported deaths was 645).2, 6 Six coronavirus strains were reported to be zoonotic (ie, transmission of infections from animals to humans is possible).7, 8, 9, 10, 11 These human coronaviruses were 229E and NL63 (belonging to Alphacoronavirus) and OC43, HKU1, SARS, and MERS (belonging to Betacoronavirus).5, 12 While the first four strains caused mild upper respiratory tract infections, like common cold, SARS and MERS both caused lower respiratory infections such as bronchitis, bronchiolitis, and pneumonia.3, 13 It was reported that SARS CoV and MERS CoV were hosted in bat through palm civet cat and dromedary camel, respectively.5, 14, 15, 16
Generally, coronaviruses are enveloped, positive‐sense, single‐stranded RNA (∼30 kb). Coronavirus genome is translated inside host cell into two groups of proteins; structural proteins, such as Spike (S), Nucleocapsid (N), Matrix (M) and Envelope (E), and non‐structural proteins such as RNA dependent RNA polymerase (nsp12) and Helicase (nsp13).17, 18 Coronaviruses enter host cells either through endosomal or non‐endosomal pathways.19 Endosomal entry of human coronaviruses takes place via four different host cell receptors; angiotensin‐converting enzyme 2 (receptor for each of SARS CoV and HCoV NL63), dipeptidyl peptidase 4 (receptor for MERS CoV), aminopeptidase N (receptor for HCoV 229E), and O‐acetylated sialic acid (receptor for HCoV OC43 and HCoV HKU1).5, 20, 21, 22, 23, 24 After cell entry, corona viral RNA is released into the cytoplasm in which the translation and replication occur. The translation of the Open Reading Frame 1a/b (ORF1a/b) yields two polypeptide chains (pp1a and pp1ab) that are further cleaved to form Non‐Structural Proteins (NSPs).25
Polymerases have conserved active site aspartates that take part in nucleotidyl transfer in different organisms from viruses to humans.26 The use of modified nucleotides to block the polymerization process was reported in the last two decades. The first FDA approved nucleotide inhibitor was Sofosbuvir (December 2013). Sofosbuvir was approved with Ribavirin and interferon or as interferon‐free regimen. In addition, it was also approved in combination with other target protein inhibitors.27, 28
Computer Aided Drug Design (CADD) is the utilization of computer software to mimic, visualize, and characterize the behavior of biological molecules. It often uses molecular modeling in conjunction with Quantitative Structure‐Activity Relationship (QSAR) in order to test the reactivity of a ligand and its binding pattern into protein active site.29, 30 Molecular docking is usually used in order to mimic the binding of a ligand into protein active site using a scoring function. More negative docking scores mean better binding of the drug to the active site of the target protein and hence, more inhibitory performance.31
In this study, combined QSAR and molecular docking are utilized to evaluate the binding of some nucleotide polymerase inhibitors (Sofosbuvir, IDX‐184, R7128, and MK‐0608) to HCoV polymerase active site.27, 32 These drugs are either FDA approved or under clinical trials. The study also compares the binding performance of these nucleotide inhibitors to that of native nucleotides and Ribavirin.
2. MATERIALS AND METHODS
This study utilizes the medicinal chemistry platform, SCIGRESS 3.0 (Fujitsu, Poland)33 installed on Dell Precision T3500 workstation to examine the molecular docking of nucleotides (ATP, UTP, CTP, and GTP), their corresponding nucleotide inhibitors (MK‐0608, Sofosbuvir, R7128, and IDX‐184, respectively) and Ribavirin to different polymerases of HCoV strains.
2.1. Geometry optimization
Structures of the nucleotides and nucleotide inhibitors are first sketched using SCIGRESS tools,33 followed by geometry optimization using Molecular Mechanics force field (MM3 method). The structures are further optimized using semi‐empirical Parameterization Method 6 (PM6 in water). Optimized structures are then examined by calculating the Infra‐Red (IR) vibrational spectra using PM6 method in order to ensure that they are real (no negative vibrations). Finally, structures are quantum mechanically optimized according to the Density Functional Theory (DFT) using B88‐LYP functional (DZVP basis set). This is followed by the calculation of IR vibrational spectra at the same level. At this stage, structures that are finally optimized become ready for QSAR and docking calculations.
2.2. Quantitative Structure Activity Relationships (QSAR) parameters
SCIGRESS 3.0 software is used to calculate the QSAR parameters in order to compare the activity and stability of the investigated nucleotides (ATP, UTP, CTP, and GTP) to their activated modifications (MK‐0608, Sofosbuvir, R7128, and IDX‐184, respectively) in addition to the Ribavirin tri‐phosphate. QSAR parameters are calculated for the geometry‐optimized structures. The QSAR parameters include total energy (Kcal/mol), heat of formation (Kcal/mol), dielectric energy (Kcal/mol), steric energy (Kcal/mol), dipole moment (Debye), electron affinity (eV), ionization potential (eV), Log P, molar refractivity, polarizability (Å3), solvent accessible surface area (Å2), and the frontier energy gap (LUMO‐HOMO) (eV).
2.3. Modeling of HCoV polymerases
Human Coronavirus (HCoV) polymerase sequences for the six human strains of coronaviruses (HKU1, MERS, SARS, OC43, NL63, and 229E) are downloaded from National Center for Biotechnology Information (NCBI) protein database (https://www.ncbi.nlm.nih.gov/protein).28 Nineteen polymerase sequences (with unique 307 amino acids) having conserved active site amino acids (D255 and D256) are selected from the given strains as follows: four from HKU1, two from MERS, one from SARS, six from OC43, four from NL63, and two from 229E. Multiple sequence alignment is performed using CLUSTAL OMEGA web server (https://www.ebi.ac.uk/Tools/msa/clustalo/) of the European Bioinformatics Institute (EMBL‐EBI).34, 35 The alignment is presented with the help of ESPript 3.0 web server (http://espript.ibcp.fr/ESPript/ESPript/).36 I‐TASSER web server (http://zhanglab.ccmb.med.umich.edu/I-TASSER/) is used to generate the 3D structures of all of the downloaded polymerase sequences.37 Five models are generated for each sequence. Model validation is performed using version four of Structure Analysis and Verification Server (SAVES) (https://services.mbi.ucla.edu/SAVES/). Four programs are used for the validation of the present models; PROCHECK, Verify 3D, PROVE, and ERRAT along with the c‐score of I‐TASSER modeling web server.38, 39 The 1NB6 (chain B) PDB file model shows that the interaction of nucleotides or drugs with the active site of polymerases involves binding to two divalent metal ions (Mg+2 or Mn+2) of the two aspartates in the conserved active site. This model is adopted for the docking calculations in this work.40 In order to be ready for the docking step, protein models are minimized using MM3 implemented on SCIGRESS 3.0.30, 32, 33
2.4. Molecular docking
The optimized structures of both, the drugs and nucleotides are used in the docking calculations. Nineteen protein models are docked with ATP, GTP, CTP, UTP, Sofosbuvir, IDX‐184, MK‐0608, R7128, and Ribavirin. Docking calculations are performed using SCIGRESS 3.0 software assuming that the ligand is flexible and that the active site is rigid.41 Docking experiments are performed three times and the average values for docking scores are recorded. Duncan multiple range tests are performed on the obtained data in order to point out significantly different means of docking scores using SPSS software version 17.
After the removal of water and ligands (except Mg+2), addition of missing H atoms and geometry optimization using MM3 force field, Hepatitis C Virus NS5b RdRp (PDB ID 2XI3) is docked using the same ligands for comparison.
3. RESULTS AND DISCUSSION
3.1. Sequence comparison and analysis
Figure 1 shows the alignment of 19 HCoV polymerase sequences from different HCoV strains and the solved structure of Hepatitis C Virus polymerase (PDB ID 2XI3). The secondary structure of HCV polymerase is presented in the top of sequences in order to highlight the conservation of the active site. Two consecutive aspartates protrude from the beta turn structure between ß9 and ß10. The conservation of amino acids in the ß10 structure with that of HCV is obvious. This is an important observation that the present work is based on. The conservation of the active site amino acids and the backbone that holds these amino acids suggests the same manner of interactions as in HCV and hence inhibition of HCoV polymerases is possible using nucleotide inhibitors.
Figure 1.
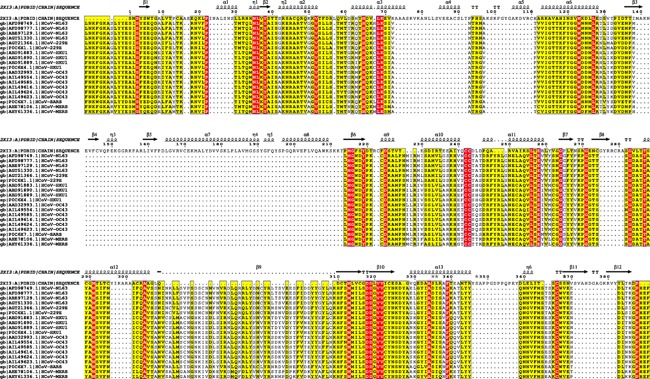
Sequence alignment for the 19 HCoV polymerases along with HCV NS5b RdRp (from the PDB file 2XI3). The alignment is performed using CLUSTAL omega web server and visualized using ESPript software 3.0. The conserved amino acids are highlighted in red. The active site environment amino acids (4Å around the two active site aspartates D255 and D256) are all conserved except for amino acid number 291 which is Glutamate (E) in all HCoV types other than NL63 and 229E where it is Threonine (T)
A 4Å region around the conserved active site aspartates (D255 and D256) is selected to perform the docking.41, 42 The active site of a randomly selected model contains the following amino acids; R50, W112, Y114, P115, C117, N190, I252, L253, S254, M289, S290, and E291 or T291. These amino acids are conserved except for amino acid number 291 that is Glutamate (E) in all Betacoronavirus and Threonine (T) in NL63, and 229E.
3.2. Quantitative Structure‐Activity Relationships (QSAR)
Tables 1 and 2 show the calculated values of some Quantitative Structure‐Activity Relationship (QSAR) parameters. These values are calculated for the DFT (B88LYP functional) optimized structures of the nucleotides (ATP, UTP, CTP, and GTP), their analogues (MK‐0608, Sofosbuvir, R7128, and IDX‐184, respectively) and Ribavirin.
Table 1.
QSAR parameters representing the stability of the structures calculated for the nucleotides, their modification analogues and Ribavirin
| Nucleotides and drugs | Steric energy (kcal/mol) | Dielectric energy (kcal/mol) | Heat of formation (kcal/mol) | Energy gap (LUMO‐HOMO) |
|---|---|---|---|---|
| ATP | 27.8 | −3.283 | −697.35 | 3.452 |
| UTP | 7.163 | −3.381 | −849.40 | 3.744 |
| CTP | −20.896 | −4.004 | −788.90 | 3.729 |
| GTP | −27.431 | −3.706 | −753.24 | 3.122 |
| MK‐0608 triphosphate | 37.503 | −3.283 | −706.43 | 3.705 |
| Sofosbuvir triphosphate | −0.464 | −3.381 | −848.97 | 3.785 |
| R7128 triphosphate | −5.574 | −4.004 | −788.10 | 3.621 |
| IDX‐184 triphosphate | −17.548 | −3.706 | −756.93 | 3.201 |
| Ribavirin triphosphate | 68.154 | −3.427 | −753.76 | 3.773 |
HOMO, highest occupied molecular orbital; LUMO, lowest unoccupied molecular orbital.
The structures are optimized quantum mechanically using DFT (B88LYP functional). Each nucleotide analogue was compared to its parent nucleotide and the best drug values presented in Bold. In addition, each nucleotide analogue was compared to Ribavirin and the best drug values are underlined.
Table 2.
QSAR parameters representing the reactivity of the structures calculated for the nucleotides, their modifications analogues and Ribavirin
| Nucleotides and drugs | Electron affinity (eV) | Dipole moment (Debye) | Log P | Solvent accessible surface area (Å2) | Polarizability (Å3) | Molar refractivity |
|---|---|---|---|---|---|---|
| ATP | 2.035 | 1.866 | 0.337 | 393.59 | 30.373 | 94.699 |
| UTP | 2.197 | 7.795 | −0.061 | 384.163 | 27.802 | 88.675 |
| CTP | 2.158 | 7.380 | 0.141 | 367.959 | 27.444 | 88.785 |
| GTP | 1.939 | 5.373 | −0.068 | 399.688 | 31.663 | 95.838 |
| MK‐0608 triphosphate | 2.050 | 9.109 | 0.415 | 397.394 | 31.402 | 99.337 |
| Sofosbuvir triphosphate | 2.221 | 8.057 | 0.542 | 388.528 | 27.82 | 86.983 |
| R7128 triphosphate | 2.228 | 8.949 | 0.823 | 395.047 | 28.048 | 91.73 |
| IDX‐184 triphosphate | 1.901 | 8.516 | 0.01 | 418.933 | 32.636 | 100.476 |
| Ribavirin triphosphate | 2.126 | 2.953 | 0.251 | 370.762 | 25.956 | 85.228 |
Log P is the logarithm of the partition coefficient, which measure the distribution of the compound in lipid/water system.
The structures are optimized quantum mechanically using DFT (B88LYP functional). Each nucleotide analogue was compared to its parent nucleotide and the best drug values represented in Bold. In addition, each nucleotide analogue was compared to Ribavirin and the best drug values are underlined.
Table 1 shows some parameters that reflect the stability of the structures (dielectric energy, steric energy, heat of formation, and the frontier energy gap [LUMO‐HOMO energy]) while Table 2 shows other parameters that reflect the reactivity of the structures (dipole moment, electron affinity, molar refractivity, polarizability, Log P, and solvent accessible surface area).29
Nucleotide analogues are probably all similar to the parent nucleotides in their stability. The parameters showing favorable values are in bold numbers (Table 1). On the other hand, the nucleotide analogues are more stable compared to Ribavirin in the majority of the stability parameters presented in Table 1 (underlined values). This is apparent from steric energy, heat of formation and energy gap parameters that have lower (better) values for nucleotide analogues compared to Ribavirin.
Similar to Tables 1 and 2 presents a comparison of each nucleotide analogue to its parent nucleotide and each drug to Ribavirin, but this time in terms of parameters describing structural reactivity instead of structural stability. Table 2 reveals that the nucleotide analogues are more reactive than their parent nucleotides (bold values in Table 2). This can be brought from the higher (better) values of dipole moment, solvent accessible surface area, and polarizability. Almost all nucleotide analogues values are better (more reactive) than Ribavirin (underlined values in Table 2). This can be recognized from the higher (better) values of dipole moment, surface accessible surface area, polarizability, and molar refractivity. Moreover, Log P shows that the studied compounds are more hydrophobic compared to parent nucleotides and ribavirin. This may facilitate passage through membranes.
3.3. Molecular docking
ATP, UTP, CTP, GTP, MK‐0608, Sofosbuvir, R7128, IDX‐184, and ribavirin all in its active form are docked into the active site of the 19 protein models for HCoV strains in addition to the solved structure of HCV NS5b RdRp (PDB ID 2XI3).
The average values of the docking scores are tested for significant differences using Duncan's multiple range test in SPSS 17.0 software package. For the 19 HCoV models, the docking score values show statistically insignificant differences between parent nucleotides and their analogue drugs. This implies a possible equal competition in binding to the HCoV active sites. Nevertheless, few models show significantly better docking for some nucleotides (eg, CTP has six significantly better docking score values compared to R7128, while ATP, UTP and GTP, each has three significantly better values compared to Mk‐0608, Sofosbuvir and IDX‐184, respectively. Most of these nucleotides are from the strain NL63).
The docking scores of most of the investigated drugs show no statistically significant superiority over Ribavirin for almost all of the HCoV models. Fortunately, IDX‐184 is an exception. IDX‐184 shows seven significantly better results compared to Ribavirin; for HKU1 (ABD91889.1, ABD91883.1, and P0C6 × 4.1), OC43 (AIL49623.1 and AIL49554.1), and NL63 (ABE97129.1 and AFD98777.1). On the other hand, MK‐0608 shows three significantly better docking scores compared to ribavirin; for HKU1 (ABD91889.1), OC43 (AIL49554.1), and NL63 (ABE97129.1). MK‐0608 shows three significantly better results compared to Ribavirin; for HKU1 (ABD91889.1), OC43 (AIL49554.1), and NL63 (ABE97129.1). R7128 shows only one significantly better value for NL63 (ABE97129.1) compared to Ribavirin.
Figure 2 shows the average docking score values for all HCoV strains and HCV polymerases docked by nucleotide inhibitors and Ribavirin with the error bars presenting the standard deviations. HCoV strains are presented in colored circles while a diamond presents HCV. SARS and MERS are presented in larger gray and orange circles respectively for clarification. MERS CoV, OC43 CoV, HKU1 CoV, 229E CoV, and NL63 CoV have the same rhythm of average docking scores ranking. They show priority for IDX‐184, followed by MK‐0608 and Ribavirin. On the other hand, SARS CoV shows priority for both MK‐0608 and Ribavirin followed by IDX‐184. The average docking scores for SARS CoV and MERS CoV are better than that of HCV, suggesting potent inhibitory performance of IDX‐184 and MK‐0608 against MERS CoV and SARS CoV as reported for HCV.
Figure 2.
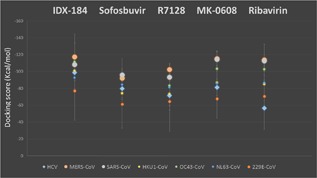
Average docking score values calculated for the HCoV (colored circles) and experimental solved HCV (diamond) polymerases. MERS and SARS CoVs are represented by large circles for clarification. The error bars represent the standard deviations of means. Docking is performed using SCIGRESS 3.0 software
3.4. HCV versus MERS CoV
From the two MERS CoV models, the model generated from the sequence ID AHY61336.1 is selected for further investigation after performing the docking study. The selection is based on the validation data calculated with SAVES server. The preferable, allowed and disallowed regions of the Ramachandran plot are 77.05%, 15.08,% and 7.87%, respectively, for the selected model versus 72.46%, 13.77%, and 13.77%, respectively, for the other model. Moreover, the overall quality factor is 89.726% for the selected model versus 85.324% for the other one.
Figure 3 shows the interactions that take place between the ligand GTP and both HCV (solved structure PDB ID 2XI3) and MERS CoV (docked structure with docking score of −118.98 ± 24.43 Kcal/mol) polymerases. The same interaction pattern occurred in both cases. In case of HCV, GTP interacts with Arg48, Lys141, Arg158, Asp220, Asp225, and Asp318. On the other hand, GTP forms polar contacts with Asp113, Leu253, Ser254, Asp255, Asp256, and Phe288 of MERS CoV polymerase. Both HCV and MERS CoV polymerases forms coordination bonds through Mg+2 with GTP and this facilitates the binding. These types of interactions suggest strong binding and hence potent inhibition performance of the nucleotide inhibitors, which are used as anti‐HCV drugs, against MERS CoV and other HCoV polymerases. Figure 3 is produced using PyMOL software (1.7.6) [PyMOL Molecular Graphics System, 2016].
Figure 3.
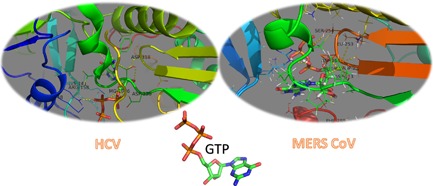
3D structures of solved HCV NS5b polymerase (PDB ID 2XI3) with GTP and the docked GTP to MERS CoV selected model. Polar interactions occur in both cases with the active site amino acids (Asp318 for HCV & Asp255 and Asp256 for MERS CoV) and the active site pocket amino acids. In addition, metal interactions mediate ligand binding to the polymerases. Structures are presented using PyMOL software where proteins are represented by colored cartoon and the drugs are represented by atom colored licorice (N, blue; O, red; C, green; H, white; and P, orange)
Figure 4 shows the 2D ligand interaction diagrams plotted using Maestro (Schrödinger Release 2016‐2: Maestro, Schrödinger, LLC, New York, NY, 2016) for the selected MERS CoV model. Ligands (IDX‐184, MK‐0608, Sofosbuvir, R7128, and Ribavirin) are docked to the polymerase active site pocket using SIGRESS 3.0 software. These drugs form H‐bonds with the active site pocket amino acids in addition to the metal interactions. The H‐bonds are formed with both backbone structures (solid arrows in Fig. 4) and side chains (dotted arrows). IDX‐184 and R7128 did not form any H‐bonds with MERS CoV polymerase, yet both have metal interactions that stabilize the complexes having docking scores of −140.4 and −116.8 Kcal/mol, respectively. On the other hand, MK‐0608 and Sofosbuvir both form one H‐bond with MERS CoV polymerase active site. The amino acids involved in H‐bond formation are Lys116 (backbone) and Arg50 (side chain), respectively. Metal interactions are also present with docking scores of −126.4 and −97.8 Kcal/mol for MK‐0608 and Sofosbuvir, respectively. Ribavirin forms two H‐bonds with MERS CoV polymerase through Arg50 (side chain) and Lys116 (backbone). This is accompanied by metal interactions and the docking score is −139.0 Kcal/mol.
Figure 4.
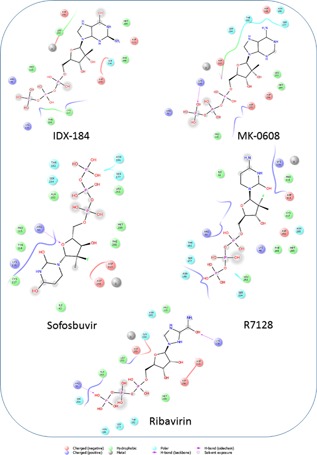
2D ligand interaction diagrams plotted by Maestro software for the docked nucleotide inhibitors and Ribavirin into the active site pocket of MERS CoV polymerase. Major interactions are through the formation of H‐bonds (violet arrows) with the active site pocket amino acids in addition to metal interactions that stabilize the complexes. Water exposed groups (OH and NH2 groups surrounded by gray smudge) suggest water‐ligand interactions in the active site vicinity
On the other hand, HCV docking results show nearly similar results (Fig. S1). MK‐0608, Sofosbuvir and Ribavirin each form one H‐bond with HCV polymerase through Asp90 (side chain), Asp89 (backbone), and Asp89 (backbone), respectively. The docking scores are −81.1, −79.8, and −56.5 Kcal/mol, respectively. Similar to the case of MERS CoV, no H‐bond is formed with IDX‐184 and R7128 and the docking scores are −98.9 and −71.5 Kcal/mol, respectively. Metal interactions are reported for R7128 and Ribavirin only.
The formation of H‐bonds (in both cases of HCV and MERS CoV) provides stability to the protein/ligand complex and hence inhibiting viral polymerase function, since the polymerase active site will not be free to bind to the nucleotides for the polymerization process to be completed. The gray smudges in Figs. 4 and S1 indicate water exposed atoms or groups. Some OH and NH2 groups are water exposed and hence formation of water‐ligand interactions in the active site vicinity may occur and consequently adds to the stabilization of the protein/ligand complex. This may be proved by studying the dynamics of binding.43
The same manner of interactions occurs in other HCoV strains and the amino acids that are involved in the formation of H‐bonds with ligands are Lys40, Lys46, Arg48, Arg50, Thr51, Asp113, Tyr114, Lys116, Ser177, Asn186, Tyr223, Tyr227, Ser245, Asp255, and Gly257. These amino acids comprise the active site pocket of the polymerase. The repeated amino acids that form H‐bonds in different strains of the virus are Arg48, Arg50, Lys116, Tyr227, and Asp255.
Based on the above data from both QSAR and molecular docking one can conclude that anti‐HCV polymerase drugs could be effective potent inhibitors against HCoV polymerases including the most recent outbreak of MERS CoV. This inhibition takes place through the formation of H‐bonds between the nucleotide inhibitors and the protein conserved active site pocket amino acids, the metal interactions and other interactions that strengthen the binding of the drugs to the protein active site. It would be useful to further examine the binding through molecular dynamics simulations.
IDX‐184 has the advantage of low toxicity due to its low concentration in blood. This emerges from its nature as a prodrug which converts to the active form inside its target cell, as reported for Hepatitis C Virus (HCV).44 Further optimization may be required to increase the potency of IDX‐184 and other drugs to fit the active site pockets of the polymerases more precisely. A modification in carbon 2′ of the ribose ring may result in more potent inhibitors probably able to stop the replication of HCoV, HCV, ZIKV, and other viruses.45, 46
4. CONCLUSION
QSAR and docking studies of the investigated nucleotides, their analogues and Ribavirin unfold that IDX‐184 and MK‐0608 are two promising keystones for the treatment of a wide variety of HCoV strains including the most recent MERS CoV. The results of docking to both HCV and HCoV suggest that both H‐bonds and metal interactions strongly contribute to the binding of nucleotide inhibitors to the polymerase active site and hence protein inhibition.
For MERS CoV models, IDX‐184 yields better docking scores compared to Ribavirin. It would be suggested to lunch an experimental study on the potency of IDX‐184 on MERS CoV. A number of possible modifications and optimizations to the lead compound (IDX‐184) may be useful for further increasing the potency of the drug against MERS CoV polymerase.
5. AUTHORS' CONTRIBUTION
AAE suggested the research plan, wrote the manuscript and constructed tables and figures. SMM made the core calculations while WME shared in scientific discussions, revised the whole manuscript and gave ideas on presenting the tables and data statistics.
Supporting information
Additional Supporting Information may be found online in the supporting information tab for this article.
Structure files hosted here: https://drive.google.com/drive/folders/0B3TVdGWxKpKuMDMyOUN3U3hsSXc?usp=sharing
Supporting Figure S1.
ACKNOWLEDGMENT
Dr Wael A. Mohamed is appreciated for helpful discussions and suggestions.
Elfiky AA, Mahdy SM, and Elshemey WM. Quantitative structure‐activity relationship and molecular docking revealed a potency of anti‐hepatitis C virus drugs against human corona viruses. J Med Virol. 2017; 89:1040–1047. 10.1002/jmv.24736
REFERENCES
- 1. Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus ADME, Fouchier RAM. 2012. Isolation of a novel Coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 367:1814–1820. [DOI] [PubMed] [Google Scholar]
- 2. Zumla AC, Azhar JF, Hui EI, Yuen DS, KY. 2016. Coronaviruses—drug discovery and therapeutic options. Nat Rev Drug Discov. 15:327–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. van den Brand JMA, Smits SL, Haagmans BL. 2015. Pathogenesis of Middle East respiratory syndrome coronavirus. J Pathol. 235:175–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sharif‐Yakan A, Kanj SS. 2014. Emergence of MERS‐CoV in the Middle East: origins, transmission, treatment, and perspectives. PLoS Pathog. 10:e1004457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Guan Y, Zheng BJ, He YQ, Liu XL, Zhuang ZX, Cheung CL, Luo SW, Li PH, Zhang LJ, Guan YJ, Butt KM, Wong KL, Chan KW, Lim W, Shortridge KF, Yuen KY, Peiris JS, Poon LL. 2003. Isolation and characterization of viruses related to the SARS Coronavirus from animals in southern China. Science. 302:276–278. [DOI] [PubMed] [Google Scholar]
- 6.WHO, 2016. Middle East respiratory syndrome coronavirus (MERS‐CoV) WHO.
- 7. Huynh J, Li S, Yount B, Smith A, Sturges L, Olsen JC, Nagel J, Johnson JB, Agnihothram S, Gates JE. 2012. Evidence supporting a zoonotic origin of human coronavirus strain NL63. J Virol. 86:12816–12825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vijgen L, Keyaerts E, Moes E, Thoelen I, Wollants E, Lemey P, Vandamme AM, Van Ranst M. 2005. Complete genomic sequence of human coronavirus OC43: molecular clock analysis suggests a relatively recent zoonotic coronavirus transmission event. J Virol. 79:1595–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Corman VM, Baldwin HJ, Tateno AF, Zerbinati RM, Annan A, Owusu M, Nkrumah EE, Maganga GD, Oppong S, Adu‐Sarkodie Y. 2015. Evidence for an ancestral association of human coronavirus 229E with bats. J Virol. 89:01755–01715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Han HJ, Yu H, Yu XJ. 2016. Evidence for zoonotic origins of Middle East respiratory syndrome coronavirus. J Gen Virol. 97:274–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sheahan T, Rockx B, Donaldson E, Sims A, Pickles R, Corti D, Baric R. 2008. Mechanisms of zoonotic severe acute respiratory syndrome Coronavirus host range expansion in human airway epithelium. J Virol. 82:2274–2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Belouzard S, Millet JK, Licitra BN, Whittaker GR. 2012. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 4:1011–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fouchier RA, Hartwig NG, Bestebroer TM, Niemeyer B, de Jong JC, Simon JH, Osterhaus AD. 2004. A previously undescribed coronavirus associated with respiratory disease in humans. Proc Natl Acad Sci USA. 101:6212–6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Coleman CM, Frieman MB. 2014. Coronaviruses: important emerging human pathogens. J Virol. 88:5209–5212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Azhar EI, El‐Kafrawy SA, Farraj SA, Hassan AM, Al‐Saeed MS, Hashem AM, Madani TA. 2014. Evidence for camel‐to‐human transmission of MERS coronavirus. N Engl J Med. 370:2499–2505. [DOI] [PubMed] [Google Scholar]
- 16. Reusken CB, Haagmans BL, Müller MA, Gutierrez C, Godeke GJ, Meyer B, Muth D, Raj VS, Smits‐De Vries L, Corman VM, Drexler JF, Smits SL, El Tahir YE, De Sousa R, van Beek J, Nowotny N, van Maanen K, Hidalgo‐Hermoso E, Bosch BJ, Rottier P, Osterhaus A, Gortázar‐Schmidt C, Drosten C, Koopmans MP. 2013. Middle East respiratory syndrome coronavirus neutralising serum antibodies in dromedary camels a comparative serological study. Lancet Infect Dis. 13:859–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hilgenfeld R, Peiris M. 2013. From SARS to MERS: 10 years of research on highly pathogenic human coronaviruses. Antiviral Res. 100:286–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Stadler K, Masignani V, Eickmann M, Becker S, Abrignani S, Klenk HD, Rappuoli R. 2003. SARS‐beginning to understand a new virus. Nat Rev Microbiol. 1:209–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chan JF, Lau SK, To KK, Cheng VC, Woo PC, Yuen KY. 2015. Middle East respiratory syndrome coronavirus: another zoonotic betacoronavirus causing SARS‐like disease. Clin Microbiol Rev. 28:465–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA, Somasundaran M, Sullivan JL, Luzuriaga K, Greenough TC, Choe H, Farzan M. 2003. Angiotensin‐converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 426:450–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yeager CL, Ashmun RA, Williams RK, Cardellichio CB, Shapiro LH, Look AT, Holmes KV. 1992. Human aminopeptidase N is a receptor for human coronavirus 229E. Nature. 357:420–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Raj VS, Mou H, Smits SL, Dekkers DH, Muller MA, Dijkman R, Muth D, Demmers JA, Zaki A, Fouchier RA, Thiel V, Drosten C, Rottier PJ, Osterhaus AD, Bosch BJ, Haagmans BL. 2013. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus‐EMC. Nature. 495:251–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Huang X, Dong W, Milewska A, Golda A, Qi Y, Zhu QK, Marasco WA, Baric RS, Sims AC, Pyrc K, Li W, Sui J. 2015. Human Coronavirus HKU1 spike protein uses O‐Acetylated sialic acid as an attachment receptor determinant and employs hemagglutinin‐Esterase protein as a receptor‐Destroying enzyme. J Virol. 89:7202–7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hofmann H, Pyrc K, van der Hoek L, Geier M, Berkhout B, Pohlmann S. 2005. Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc Natl Acad Sci USA. 102:7988–7993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. van Boheemen S, de Graaf M, Lauber C, Bestebroer TM, Raj VS, Zaki AM, Osterhaus AD, Haagmans BL, Gorbalenya AE, Snijder EJ, Fouchier RA. 2012. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. MBio. 3:e00473‐12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yang PL, Gao M, Lin K, Liu Q, Villareal VA. 2011. Anti‐HCV drugs in the pipeline. Curr Opin Virol. 1:607–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Elfiky AA, Elshemey WM, Gawad WA, Desoky OS. 2013. Molecular modeling comparison of the performance of NS5b polymerase inhibitor (PSI‐7977) on prevalent HCV genotypes. Protein J. 32:75–80. [DOI] [PubMed] [Google Scholar]
- 28. Asselah T. 2014. Daclatasvir plus sofosbuvir for HCV infection: an oral combination therapy with high antiviral efficacy. J Hepatol. 61:435–438. [DOI] [PubMed] [Google Scholar]
- 29. Elfiky AA, Elshemey WM. 2016. IDX‐184 is a superior HCV direct‐acting antiviral drug: a QSAR study. Med Chem Res. 25:1005–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Saleh NA. 2014. The QSAR and docking calculations of fullerene derivatives as HIV‐1 protease inhibitors. Spectrochim Acta A Mol Biomol Spectrosc. 136PC:1523–1529. [DOI] [PubMed] [Google Scholar]
- 31. Rodrigues JP, Bonvin AM. 2014. Integrative computational modeling of protein interactions. FEBS J. 281:1988–2003. [DOI] [PubMed] [Google Scholar]
- 32. Elfiky AA, Elshemey WM, Wissam AG. 2015. 2′‐Methylguanosine prodrug (IDX‐184), phosphoramidate prodrug (Sofosbuvir), diisobutyryl prodrug (R7128) are better than their parent nucleotides and ribavirin in hepatitis C virus inhibition: a molecular modeling study. J Comput Theor Nanosci. 12:376–386. [Google Scholar]
- 33. Summers KL, Mahrok AK, Dryden MD, Stillman MJ. 2012. Structural properties of metal‐free apometallothioneins. Biochem Biophys Res Commun. 425:485–492. [DOI] [PubMed] [Google Scholar]
- 34. Squizzato S, Park YM, Buso N, Gur T, Cowley A, Li W, Uludag M, Pundir S, Cham JA, McWilliam H, Lopez R. 2015. The EBI Search engine: providing search and retrieval functionality for biological data from EMBL‐EBI. Nucleic Acids Res. 43:W585–W588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high‐quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 7:539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Robert X, Gouet P. 2014. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42:W320–W324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. 2015. The I‐TASSER Suite: protein structure and function prediction. Nat Methods. 12:7–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Colovos C, Yeates TO. 1993. Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci. 2:1511–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pontius J, Richelle J, Wodak SJ. 1996. Deviations from standard atomic volumes as a quality measure for protein crystal structures. J Mol Biol. 264:121–136. [DOI] [PubMed] [Google Scholar]
- 40. O'Farrell D, Trowbridge R, Rowlands D, Jäger J. 2003. Substrate complexes of hepatitis C virus RNA polymerase (HC‐J4): structural evidence for nucleotide import and de‐novo initiation. J Mol Biol. 326:1025–1035. [DOI] [PubMed] [Google Scholar]
- 41. Noolvi MN, Patel HM. 2013. A comparative QSAR analysis and molecular docking studies of quinazoline derivatives as tyrosine kinase (EGFR) inhibitors: a rational approach to anticancer drug design. J Saudi Chem Soc. 17:361–379. [Google Scholar]
- 42. Xu X, Liu Y, Weiss S, Arnold E, Sarafianos S G, Ding J. 2003. Molecular model of SARS coronavirus polymerase: implications for biochemical functions and drug design. Nucleic Acids Res. 31:7117–7130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.1.7.6, V., The PyMOL Molecular Graphics System, Version 1.7.6 Schrödinger, LLC.
- 44. Bellissent‐Funel M‐C, Hassanali A, Havenith M, Henchman R, Pohl P, Sterpone F, van der Spoel D, Xu Y, Garcia AE. 2016. Water determines the structure and dynamics of proteins. Chem Rev. 116:7673–7697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhou XJ, Pietropaolo K, Chen J, Khan S, Sullivan‐Bolyai J, Mayers D. 2011. Safety and pharmacokinetics of IDX184, a liver‐targeted nucleotide polymerase inhibitor of hepatitis C virus, in healthy subjects. Antimicrob Agents Chemother. 55:76–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Elfiky AA. 2016. Zika viral polymerase inhibition using anti‐HCV drugs both in market and under clinical trials. J Med Virol. 88:2044–2051. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found online in the supporting information tab for this article.
Structure files hosted here: https://drive.google.com/drive/folders/0B3TVdGWxKpKuMDMyOUN3U3hsSXc?usp=sharing
Supporting Figure S1.


