Abstract
Aim: The objectives of this study are to assess the frequency of human bocavirus (HBoV) infection in hospitalised children and to study the clinical symptoms associated with the detection of HBoV.
Methods: Two groups of hospitalised children were included in this study: group 1 consisted of 1946 children hospitalised from 1st September 2004 to 30th May 2005, and group 2 consisted of 448 children hospitalised from 1st November 2003 to 30th March 2004. The respiratory specimens were tested by polymerase chain reaction.
Results: In the first group, HBoV was detected by polymerise chain reaction in 11/828 (1.3%) of nasal specimens that tested negative for other respiratory viruses. One child tested positive for HBoV in both a nasal aspirate and stool sample. In the second group, nasal specimens were tested for all respiratory viruses, including HBoV. The presence of HBoV infection was detected in seven children (1.6%). Detection of a mixed viral population was observed in four of these children. The main symptoms in children infected with HBoV were rhinitis (50%), cough (45%), dyspnoea (28%), wheezing (28%), fever (23%) and diarrhoea (22%). The final clinical diagnoses were bronchiolitis (seven children), rhinopharyngitis (five children), the exacerbation of asthma (two children) and pneumonia (one child). Moreover, four children have associated gastroenteritis.
Conclusion: These results contribute to the interest in the HBoV detection in children. HBoV detection in hospitalised children with or without any other respiratory virus detection was essentially associated with lower respiratory tract infection and in a lower score with upper respiratory tract infection and gastroenteritis.
Keywords: bocavirus, molecular detection, respiratory illness, respiratory virus
Infection of the respiratory tract is a major health problem in humans particularly in children. The human bocavirus (HBoV) is a recently identified parvovirus detected in respiratory secretions of children who had an infection of the lower respiratory tract. First reported in Sweden in September 2005, it is the second human pathogen member of the family Parvoviridae. 1
The objectives of this study are to assess the frequency of HBoV infection in hospitalised children, to study the clinical symptoms associated with the detection of HBoV and to describe the clinical course of this illness.
Key Points
-
1
Human bocavirus was detected in 1.4% and 1.6% of respiratory specimens sampled from two groups of hospitalized children.
-
2
The clinical characteristics associated with HBoV infection are predominantly bronchiolitis and exacerbation of asthma. Moreover, 22% of the HBoV infected children had a gastroenteritis associated with the respiratory symptoms.
-
3
The phylogenetic analysis based on the NP‐1 and VP partial genes showed that there is a high percentage of homology in these regions.
Material and Methods
Patients and samples
Two groups of hospitalised children were used in the study. The first group included 1946 children who were less than 15 years old. Nasal aspirations were sampled from these children following admission to the Pediatric Department of Caen University Hospital or Flers Hospital from 1st September 2004 to 30th May 2005. The children suffered from acute respiratory tract infections with suspicion of a viral aetiology.
The second group included children who were admitted to the emergency unit during 1st November 2003 to 30th March 2004. Likewise, these children suffered from an infection of the lower respiratory tract. The study population included nasal aspirates collected during a previous study. 2 In total, samples for analysis were obtained from 448 children.
Methods
Conventional techniques and molecular methods
The selection of negative samples setting up the first study population (group 1) was made using routine screening tests as described previously. 2 The tests used were, immunofluorescence assay (DFA, Imagen, Dako, Cambridgeshire, UK), culture isolation on human hepatocarcinoma (HUH7) cell culture and molecular methods. Such tests allowed one to detect the presence of the following viruses: influenza virus A, B and C, respiratory syncytial viruses A and B, parainfluenza viruses 1, 2 and 3, adenovirus, enterovirus and rhinovirus. Subsequently, a polymerase chain reaction (PCR) was carried out for the detection of HBoV on all remaining negative samples as described below.
The 448 nasal aspirates obtained from group 2 were analysed by four multiplex reverse transcriptase‐polymerase chain reaction (RT‐PCR) recently set up in the laboratory 3 and by the bocavirus PCR. The multiplex PCRs allowed one to detect 15 different viruses: influenza viruses A, B and C, the human respiratory syncytial virus, the human metapneumovirus, parainfluenza virus types 1–4, human coronaviruses OC43, 229E, NL63 and HKU1, rhinovirus and enterovirus. PCR bocavirus was performed on all 448 samples even if the initial findings were negative or positive for other viruses, as described below.
Bocavirus detection and sequence analyses
Viral DNA was extracted from 200 µL of nasophavingael aspirate (NPA) samples using CHELEX 100 chelating resin as described by Zandotti. 4
Amplification of HBoV was performed with specific primers of predicted NP‐1 gene 188FHBoV and 542RHBoV as previously described. 1 Based on the published HBoV sequences (DQ000495‐6), new PCR primers was designed in the viral protein 1/viral protein 2 (VP1/VP2) gene, VPS (viral protein sens; 5′CGGATGTCTGGCACCAACTAA3′) and VPAS (viral protein antisens; 5′TGTAGGCGTGTAGTTGCTC3′), that were used for confirmation and for sequencing.
The PCR products of 354 bp from the NP‐1 gene were completely sequenced in both directions to confirm the sequence specificity. Similarly, the fragment of 483 bp from the VP gene was sequenced in order to analyse the variability of the bocaviruses as described below. The PCR products were purified through the use of ExoSAP‐IT (USB Corporation, Cleveland, OH, USA) and sequenced by using the CEQ Dye Terminator Cycle Sequencing Quick Start Kit (Beckeman Coulter, Fullerton, CA, USA). These latter steps were performed in the laboratory or by the Biofidal Company. The DNA sequences were assembled and analysed with the SEQMAN, EDITSEQ programs in the Lasergene program (DNASTAR, Madison, WI, USA) and BIOEDIT program (Ibis Biosciences, Carlsbad, CA, USA). Phylogenetic trees were generated by the neighbour‐joining method using the ClustalX program version 1.83 (National Center of Biotechnology Information, Bethesola, MD, USA). The nucleotide sequences were compared with the prototype sequences available in GenBank. The nucleotide sequence data reported in this paper were deposited in GenBank under accession numbers DQ924489–DQ924527.
Results
HBoV detection
In group 1, 1946 specimens were submitted for virus identification, of which 893 (45.9%) tested positive for a respiratory virus, as detected by reverse transcriptase‐polymerase chain reaction or PCR. The viral distribution in the 893 samples was as follows: respiratory syncitial virus in 259 (29%) specimens, influenza virus in 97 (10.9%), parainfluenza virus in 79 (8.8%), adenovirus in 42 (4.7%), enterovirus in 46 (5.2%), rhinovirus in 248 (27.8%), metapneumovirus in 49 (5.5%) and coronavirus in 4 (0.4%). Of the remaining 1053 specimens that tested negative, 828 were screened for the presence of HBoV.
HBoV was detected by PCR in 11/828 (1.4%) of the nasal specimens that tested negative with conventional methods. Interestingly, a stool specimen obtained from one child tested positive for the presence of HBoV. Most of the HBoV positive specimens were detected during winter as follows: in January, two cases (16.7%), February, four cases (33.3%) and March, five cases (41.7%) HBoV positive specimens were found, respectively. One positive result was detected in April and one in May, but HBoV was not found during the months of September through to December 2004. Children infected with HBoV aged from 4 months to 3 years of age, with the majority being less than 1 year old (n= 10; 56%). The gender distribution was 83% (n= 15) male and 17% (n= 3) female (Table 1).
Table 1.
Clinical characteristics of children infected with HBoV
| Patient no. | Specimen no. | Sex | Age (months) | Diagnosis | Symptoms |
|---|---|---|---|---|---|
| 1 | 2117‐05 | M | 15 | Asthma | Dyspnoea, SaO2 < 94%, wheezing |
| 2 | 2141‐05 | M | 11 | Asthma | Dyspnoea, fever (T > 38°C), SaO2 < 94% |
| 3 | 1092‐05 | M | 14 | Rhinopharyngitis | Rhinitis, diarrhoea, cough |
| 4 | 1074‐05 | M | 10 | Rhinopharyngitis GE | Diarrhoea, rhinitis, cough |
| 5 | 1054‐05 | F | 16 | Rhinopharyngitis GE | Diarrhoea, pharyngitis, cough, expectoration |
| 6 | 1043‐05 | M | 37 | NA | NA |
| 7 | 6013‐05 | M | 19 | Rhinopharyngitis GE | Pharyngitis, otitis, rhinitis, cough |
| 8 | 3159‐05 | M | 7 | Bronchiolitis | Acute respiratory distress |
| 9 | 3168‐05 | M | 14 | Infant sudden death | – |
| 10 | 2087‐05 | M | 10 | Bronchiolitis GE | Cough, dyspnoea, SaO2 < 94%, wheezing |
| 11 | 1022‐05 | M | 15 | Pneumonia | Rhinitis, SaO2 < 94%, wheezing |
| 12 | 1087‐04 | F | 6 | Bronchiolitis | Dyspnoea, wheezing, SaO2 < 94% |
| 13 | 2021‐04 | M | 8 | NA | NA |
| 14 | 1164‐04 | M | 12 | Bronchiolitis | Cough, fever (T > 38°C), rhinitis, dyspnoea, wheezing SaO2 < 94% |
| 15 | 6013‐04 | F | 4 | Rhinopharyngitis Bronchitis | Cough, rhinitis |
| 16 | 4083‐04 | M | 10 | Rhinopharyngitis | Cough, rhinitis |
| 17 | 1058‐04 | M | 8 | Bronchiolitis | Cough, fever (T > 38°C), rhinitis |
| 18 | 3066‐04 | M | 8 | Bronchiolitis | Cough, fever (T > 38°C), rhinitis, dyspnoea, wheezing, SaO2 < 94% |
, no symptoms; GE, gastroenteritis; HBoV, human bocavirus; NA, not available.
In group 2, 448 nasal aspirates were collected and analysed during 1st November 2003 to 30th March 2004. The viruses most frequently detected in this population were respiratory syncytial virus (43.6%), rhinovirus (31.8%) and influenza virus (8.8%), and, to a lesser extent, human metapneumovirus (4.4%), coronavirus (3.4%), parainfluenza virus (3.2%), adenovirus (2.3%) and enterovirus (2.1%). In total, seven specimens (1.6%) tested positive for the presence of HBoV. Dual detection was noted in four, out of the seven, HBoV positive specimens: two associated to human metapneumovirus, one to respiratory syncytial virus and one to rhinovirus. One sample contained three viruses: HBoV, respiratory syncytial virus and rhinovirus. No differences were observed between the patients with unique or dual infections (data not shown).
Clinical data
Clinical data were obtained for 16/18 children that were HBoV positive in the two groups and are summarised in Table 1. The main clinical symptoms were rhinitis (50%), cough (45%), dyspnoea (28%), wheezing (28%), fever (23%) and diarrhoea (22%). The final clinical diagnoses were bronchiolitis (seven children), rhinopharyngitis (five children), the exacerbation of asthma (two children) and pneumonia (one child). Moreover, four children had associated gastroenteritis. The child who tested positive for HBoV in nasal aspirate and a stool specimen was subjected to a full investigation of sudden death syndrome. For this child, all other virological and bacteriological tests were negative.
Phylogenetic analyses
For all positive specimens, nucleotide sequences were determined for nucleotides 2376–2680 that encode the NP1 partial gene of HBoV (Genbank accession numbers DQ924489–DQ924508). In addition, for 19 of the 20 positive specimens, sequences were determined for nucleotides 4510–4946 that encode the VP partial gene of HBoV (Genbank accession numbers DQ924509–DQ924527). All molecular isolates shared 99% nucleotide identity in the same region of the NP‐1 gene and 97% nucleotide identity in the same region of the VP gene. Sequence comparison with previously characterised isolates revealed high identity (Fig. 1).
Figure 1.
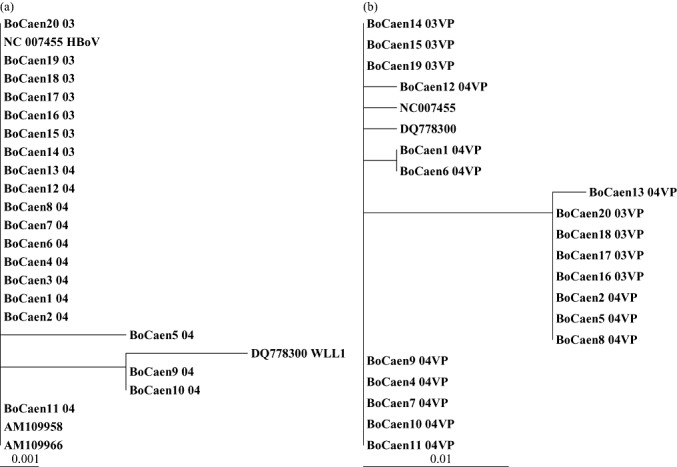
Phylogenetic analysis of the amplified products of the NP‐1 gene (a) and VP gene (b) presented on a topology tree prepared by the neighbour‐joining method using the ClustalX program and compared with strains obtained from GenBank. Specimens are labelled with the name BoCaen, number of isolate (1–20) followed by the year of isolation (03 for 2003 and 04 for 2004). HBoV, human bocavirus; VP, viral protein.
It is interesting to note the presence of one non‐silent recurrent transition in the NP‐1 gene of BoCaen9 and BoCaen10 at the 2645 nucleotide position. The mutation was present in both HBoV strains isolated from nasal and stool specimens of the child investigated for sudden death.
Discussion
HBoV was first detected in Sweden in September 2005 in the respiratory tract samples of patients suffering from respiratory disease. 1 The populations studied in different countries include primarily hospitalised children. 1 , 2 , 5 , 6 , 7 , 8 , 9 Even in studies that encompass a wide range of age groups, 10 , 11 the findings show that mostly children are involved with this disease. The present study population consisted of children less than 15 years old (group 1) and less than 2 years old (group 2). This study did not include patients suffering from whooping cough and thus differs from a recent American study. 8
The detection of HBoV requires molecular techniques because this virus has not yet been isolated in culture. One PCR positive nasal aspirate sample was inoculated on LLC‐MK2 and human rectum adenocarcinoma (HRT18) cells after two passages on primary kidney cells from a Cercopithecus monkey (data not shown). An attempt at isolation was also performed in Japan. They inoculated five patients' nasopharyngeal samples on LLC‐MK2 cells with unsuccessful results.
The prevalence of HBoV in children and its morbidity are not fully described. In the present study, HBoV was detected in 1.3% and 1.6% in groups 1 and 2, respectively. Published studies report detection rates in nasal aspirates that range from 1.4% to 10%. 1 , 5 , 6 , 7 , 8 , 10 , 11 There are several hypotheses to explain the low frequency of HBoV infection in patients such as: the prevalence of HBoV infection in the region (Normandy, France) might be low, and the patients inclusion criteria are different from other studies.
The first cohort (group 1), included samples that tested negative for other respiratory viruses. Consequently, one could not identify the presence of a co‐infection. However, this process allowed one to associate the described clinical symptoms with HBoV infection per se. In the second cohort (group 2), the samples were tested for 15 respiratory viruses and in some cases, a co‐detection with HBoV was identified. The high frequency of co‐detections in HBoV positive samples has been reported in many studies. 7 , 9 , 10 It makes it difficult to define HBoV induced symptoms.
The clinical symptoms associated with HBoV infection are similar to those reported previously with a predominance of bronchiolitis and asthma exacerbation in hospitalised patients. However, it is noteworthy that 22% of the children had a gastroenteritis associated with the respiratory symptoms. Indeed, in this study, we report the presence of HBoV in a stool specimen sampled from a child subjected to a full investigation of sudden death syndrome. It could not be established that there was a cause–effect link between the detection of the virus and the death of the child by overwhelming bocavirus infection. Nevertheless, as only one specimen was tested, the interpretation of this finding remains speculative. There are two studies that have recently been published in association with the detection of the bocavirus in stool specimens. 11 , 12 The authors suggest that the HBoV could be an enteric, as well as a respiratory, pathogen.
A previous phylogenetic analysis based on the NP‐1 and VP partial genes showed that there is a high percentage of genetic homology in these regions. Nevertheless, some authors defined two closely related genotypes based on minor variations of the nucleotide sequence of VP1/VP2 coding gene. 13 , 14 , 15
In conclusion, prospective, population‐based and serological studies are required to provide a better understanding of the epidemiology and clinical involvement of HBoV infection. Moreover, the development of real‐time PCR will probably be also very helpful to improve the knowledge of this respiratory disease.
References
- 1. Allander T, Tammi MT, Eriksson M, Bjerkner A, Tiveljung‐Lindell A, Andersson B. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc. Natl. Acad. Sci. USA 2005; 102: 12891–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Freymuth F, Vabret A, Cuvillon‐Nimal D et al. Comparison of multiplex PCR assays and conventional techniques for the diagnostic of respiratory virus infections in children admitted to hospital with an acute respiratory illness. J. Med. Virol. 2006; 78: 1498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bellau‐Pujol S, Vabret A, Legrand L et al. Development of three multiplex RT‐PCR assays for the detection of 12 respiratory RNA viruses. J. Virol. Methods 2005; 126: 53–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zandotti C, De Lamballerie X, Guignole‐Vignoli C, Bollet C, De Micco P. A rapid DNA extraction method from culture and clinical samples. Suitable for the detection of human cytomegalovirus by the polymerase chain reaction. Acta Virol. 1993; 37: 106–8. [PubMed] [Google Scholar]
- 5. Weissbrich B, Neske F, Schubert J et al. Frequent detection of bocavirus DNA in German children with respiratory tract infections. BMC Infect. Dis. 2006; 6: 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Foulongne V, Rodiere M, Segondy M. Human bocavirus in children. Emerg. Infect. Dis. 2006; 12: 862–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Choi EH, Lee HJ, Kim SJ et al. The association of newly identified respiratory viruses with lower respiratory tract infections in Korean children, 2000–2005. Clin. Infect. Dis. 2006; 43: 585–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Arnold JC, Singh KK, Spector SA, Sawyer MH. Human bocavirus: prevalence and clinical spectrum at a children's hospital. Clin. Infect. Dis. 2006; 43: 283–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Arden KE, McErlean P, Nissen MD, Sloots TP, Mackay IM. Frequent detection of human rhinoviruses, paramyxoviruses, coronaviruses, and bocavirus during acute respiratory tract infections. J. Med. Virol. 2006; 78: 1232–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sloots TP, McErlean P, Speicher DJ, Arden KE, Nissen MD, Mackay IM. Evidence of human coronavirus HKU1 and human bocavirus in Australian children. J. Clin. Virol. 2006; 35: 99–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vicente D, Gustavo C, Montes M et al. Human bocavirus, a respiratory and enteric virus. Emerg. Infect. Dis. 2007; 13: 636–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lau S, Yip C, Que TI et al. Clinical and molecular epidemiology of human bocavirus in respiratory and fecal samples from children in Hong Kong. J. Infect. Dis. 2007; 196: 986–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bastien N, Brandt K, Dust K, Ward D, Human LIY. Bocavirus infection, Canada. Emerg. Infect. Dis. 2006; 12: 848–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kaplan NM, Dove W, Abu‐Zeid AF, Shamoon HE, Abd‐Eldayem SA, Hart CA. Human bocavirus infection among children, Jordan. Emerg. Infect. Dis. 2006; 12: 1418–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Smuts H, Hardie D. Human bocavirus in hospitalized children, South Africa. Emerg. Infect. Dis. 2006; 12: 1457–8. [DOI] [PMC free article] [PubMed] [Google Scholar]


