Abstract
Pandemic influenza A/H1N1 2009 (A/H1N1pdm) virus caused significant outbreaks worldwide last year (2009). A number of oseltamivir‐resistant A/H1N1pdm viruses possessing an H275Y substitution in the neuraminidase (NA) protein were reported sporadically in several countries, including Japan, but they were sensitive to zanamivir and did not spread in the community. In this study, to monitor rapidly and simply oseltamivir‐resistant A/H1N1pdm viruses possessing H275Y, a duplex one‐step RT‐PCR assay (H275Y RT‐PCR assay) was developed based on an endpoint genotyping analysis method. H275Y RT‐PCR assay evaluated using several subtypes/types of influenza A and B viruses and other respiratory pathogenic viruses and shown to have high sensitivity and high specificity. Forty‐four clinical specimens were tested after RNA purification using the H275Y RT‐PCR assay, resulting in one clinical specimen being found to contain a virus possessing the H275Y mutation. Seventy‐three clinical isolates were then tested with the H275Y assay by using clinical isolates in the cultured supernatants of cells directly, without RNA purification, and the results were consistent with the NA sequencing. Since the H275Y RT‐PCR assay could detect the H275Y mutation in clinical isolates without RNA purification, as well as a H275Y mutated virus in clinical specimens after RNA purification, the assay was considered a powerful tool for surveillance screening of oseltamivir‐resistant A/H1N1pdm virus activity. J. Med. Virol. 83:1121–1127, 2011. © 2011 Wiley‐Liss, Inc.
Keywords: oseltamivir resistant, pandemic influenza A/H1N1 2009 virus, rapid discrimination
INTRODUCTION
The novel H1N1 subtype influenza A virus (pandemic influenza A/H1N1 2009 virus) has been spreading worldwide since it was identified in Mexico and the United States in March and April 2009 [Garten et al., 2009; Smith et al., 2009; WHO, 2009a,b]. The initial characterization of pandemic influenza A/H1N1 2009 (A/H1N1pdm) virus isolates showed that they were susceptible to the neuraminidase inhibitors (NAIs) zanamivir and oseltamivir, but resistant to the ion channel M2 blocker adamantane [Smith et al., 2009]. Therefore, NAIs, especially oseltamivir, are pharmaceutical options for the prophylactic drugs and therapeutic drugs administered to A/H1N1pdm virus‐infected individuals.
After the introduction of two NAIs into clinical use, a low rate of resistant viruses for any subtypes and strains was observed until the 2006/2007 influenza season [Monto et al., 2006; Kramarz et al., 2009; Tashiro et al., 2009]. However, in the 2007/2008 influenza season, from November 2007, oseltamivir‐resistant seasonal influenza A/H1N1 viruses possessing a substitution in the neuraminidase (NA) protein from the histidine to tyrosine residue at position 275 (H275Y) were isolated predominantly in EU countries. In the 2008/2009 influenza season, oseltamivir‐resistant viruses spread dramatically worldwide, including Japan [Lackenby et al., 2008; Meijer et al., 2009; Ujike et al., 2010] (http://www.who.int/csr/disease/influenza/h1n1_table/en/index.html).
In the 2009/2010 influenza season, A/H1N1pdm viruses spread worldwide, replacing seasonal influenza A/H1N1 viruses, but only 285 oseltamivir‐resistant A/H1N1pdm viruses possessing H275Y substitution in the NA protein were reported sporadically in several countries, including Japan, as of April 14, 2010 (http://www.who.int/csr/disease/swineflu/laboratory07_05_2010/en/index.html). In the NAI surveillance in Japan, 5422 A/H1N1pdm clinical isolates were sequenced for identification of the H275Y substitution marker in the NA gene, and only 67 oseltamivir‐resistant viruses possessing H275Y substitution were found, as of April 5, 2010 (http://idsc.nih.go.jp/iasr/graph/tamiful09-10.gif). Those oseltamivir‐resistant viruses did not cause localized epidemics in Japan. Most oseltamivir‐resistant viruses were isolated from cases administered prophylactics or therapeutics, and at this point oseltamivir use was associated with the emergence of oseltamivir‐resistant A/H1N1pdm viruses [Baz et al., 2009; WHO, 2009c].
Currently, the detection of oseltamivir‐resistant A/H1N1pdm viruses in Japan is performed by partial sequencing of the NA gene of the clinical isolates to identify the H275Y substitution marker, followed by NA inhibition assay using a chemiluminescence system. Those methods, however, require skill and involve laborious procedures.
In this study, to monitor rapidly and simply oseltamivir‐resistant A/H1N1pdm viruses possessing H275Y, a duplex RT‐PCR assay (H275Y RT‐PCR assay) was developed using an endpoint genotyping analysis method. This assay could discriminate an oseltamivir‐resistant A/H1N1pdm virus possessing the H275Y mutation in clinical specimens. RNA purification is required for using the assay to detect the H275Y mutation in clinical specimens, since specimens from nasal or nasopharyngeal swabs may contain some components that inhibit the RT‐PCR reaction. The assay also could detect the H275Y mutation of the clinical isolates in cultured supernatants directly without RNA purification. Since the RNA purification procedure for each sample increased the risk of cross‐contamination and was time‐consuming, the ability to perform the assay without RNA purification is advantageous in situations where a large number of clinical isolates are handled. The assay developed in this study is a useful tool for monitoring oseltamivir‐resistant viruses.
MATERIALS AND METHODS
Primer and Probe Design
The nucleotide sequences of the NA gene from A/H1N1pdm viruses posted on the Global Initiative on Sharing Avian Influenza Data (GISAID) database between August and October 2009 were aligned using the Clustal W software [Larkin et al., 2007]. Based on these sequences, two probes for detecting oseltamivir‐susceptible H275 and oseltamivir‐resistant Y275 were designed to discriminate C and T nucleotides at position 823 of the NA gene, respectively. The primer and probe sequences are listed in Table I.
Table I.
Primers and Probes
| Names | Sequences (5′–3′) | Orientation | Positiona |
|---|---|---|---|
| Primers for RT‐PCR | |||
| H1N1NA‐F690‐719 | ATGTGCATGTGTAAATGGTTCTTGCTTTAC | + | 690–719 |
| H1N1NA‐R847‐872 | ACACATGTGATTTCACTAGAATCAGG | − | 847–872 |
| Probes for RT‐PCR | |||
| FAM‐274Ya‐swH1N1‐F823‐835 (for detecting resistant virus) | (FAM)TACTATGAGGAAT(MGB) | + | 823–835 |
| VIC‐H274a‐swH1N1‐F823‐835 (for detecting susceptible virus) | (VIC)CACTATGAGGAAT(MGB) | + | 823–835 |
| Primers for sequencing | |||
| N1‐676‐694F | ACACAAGAGTCTGAATGTG | + | 676–694 |
| N1‐1130‐1111R | GGATCCCAAATCATCTCAAA | − | 1130–1111 |
The nucleotide positions of NA genes are based on cRNA sequences obtained from GISAID database. Isolate ID numbers for NA genes of A/Denmark/524/2009 and A/Denmark/528/2009 are EPI_ISL_33836 and EPI_ISL_33837.
Viruses
Influenza viruses were isolated from clinical specimens using Madin–Darby Canine Kidney (MDCK) cells or Caco 2 cells (human colonic carcinoma cells), and typing and subtyping of the clinical isolates were determined by a hemagglutination inhibition (HI) test. All clinical isolates of influenza viruses included in this study were cultured by passage through MDCK cells. The MDCK cells were cultured in Opti‐MEM (Invitrogen, Carlsbad, CA) containing 5 µg/ml acetylated trypsin (Sigma–Aldrich Corp., St. Louis, MO) and 200 µg/ml penicillin/streptomycin (Invitrogen) until a cytopathic effect was observed, and hemagglutination (HA) assays were performed using 0.5% turkey red blood cells so that the HA titer was confirmed to be higher than 8. As a control for the assay, A/Denmark/524/2009 (susceptible to NAI, possessing H275) and A/Denmark/528/2009 (resistant to NAI, possessing Y275) were obtained from the European Centre for Disease Prevention and Control (ECDC). A/Denmark/524/2009 and A/Denmark/528/2009 were isolated from the same patient before and after treatment with oseltamivir, respectively. These two isolates were plaque‐purified and used as controls for the H275Y RT‐PCR assay.
All A/H1N1pdm viruses used in this study are listed in Supplemental Table. The several subtypes/types of seasonal influenza viruses used for validating the specificity of the assay were as follows: A/Yamaguchi/26/2009 (H1N1), A/Oita/64/2009 (H1N1), A/Wakayama/198/2009 (H1N1), A/Sendai/85/2008 (H1N1), A/Toyama/104/2008 (H1N1), A/Kobe/41/2008 (H1N1), A/Uruguay/716/2007 (H3N2), A/Toyama/123/2008 (H3N2), A/Yamanashi/135/2008 (H3N2), A/Hiroshima‐C/41/2008 (H3N2), B/Sakai/41/2008, and B/Mie/1/2009.
The viral respiratory pathogens used for validating the specificity of the assay were as follows: Respiratory syncytial viruses A and B, human parainfluenza viruses 1–4, human rhinoviruses 1B, 2, 14, 36, and 89, human metapneumovirus, and human coronaviruses OC43, 229E, and NL63. Human metapneumovirus and human parainfluenza viruses 2 and 4 were obtained from Sendai Medical Center. Human rhinoviruses 1B, 2, 14, 36, and 89 were obtained from Nagasaki Prefectural Institute for Environmental Research and Public Health. Respiratory syncytial viruses A and B, human parainfluenza viruses 1 and 3, and human coronaviruses were stored in the National Institute of Infectious Diseases (NIID).
Clinical Specimens
Nasal or pharyngeal swabs collected from suspected and contact cases of influenza suspended in virus transport medium were obtained from hospitals and clinics in Japan. All specimens were collected between September 2009 and February 2010. The study protocol was approved by the Ethics Committee at NIID, and the study was performed in compliance with the declaration of Helsinki. Informed consent was obtained from all patients.
RNA Preparation
Supernatants of the cultured MDCK cells inoculated with influenza viruses were clarified by centrifugation at 10,000g for 10 min. Viral RNA was prepared from 140 µl of the supernatant using the QIAamp® Viral RNA kit (Qiagen, Duesseldorf, Germany) according to the manufacturer's instructions, except that the viral RNA was eluted in 70 µl of AVE (Qiagen).
Total RNA was prepared from clinical specimens using the QIAamp® Viral RNA kit (Qiagen) (using 140 µl of clinical specimen) or MagMAX™ 96 Viral Isolation Kit (Ambion, Austin, TX) (using 50 µl of clinical specimen) with KingFisher Flex (Thermo Fisher Scientific, Waltham, MA) according to each manufacturer's instructions. Total RNA from the 140 µl clinical specimen was eluted with 60 µl of AVE (Qiagen), whereas total RNA from the 50 µl clinical specimen was eluted with 30 µl of elution buffer (Ambion).
One‐Step Duplex RT‐PCR Assay
The reaction was performed using a QuantiTect® Virus + ROX Vial Kit (Qiagen) according to the manufacturer's instructions. Briefly, for testing clinical isolates using cultured medium, the 20 µl assay contained 4 µl of 5× QuantiTect Virus NR Master Mix, 0.2 µl of QuantiTect Virus RT Mix, 1.2 µl of 10 µM forward primer, 1.2 µl of 10 µM reverse primer, 0.4 µl for each of two 5 µM probes for H275Y RT‐PCR or 0.4 µl of 5 µM probe for Type A rRT‐PCR, 7.6 µl or 8 µl of water, and 2 µl of culture medium. For testing clinical specimens using extracted RNA, 5 µl of RNA template was used. Cycling was performed as follows: 20 min at 50°C to activate RT, followed by initial denaturation for 5 min at 95°C, with a subsequent 45 cycles of amplification (denaturation at 95°C for 15 sec and annealing as well as extension at 56°C for 45 sec) using LightCycler® 480 (Roche, Basel, Switzerland). Fluorescent signals were collected during annealing and extension steps, and amplification data and endpoint data were analyzed using the Light Cycler® 480 SW1.5 software according to the manufacturer's instructions.
To evaluate the sensitivity of the H275Y RT‐PCR assay, the influenza A virus detection assay (simplex real‐time RT‐PCR assay designated as Type A rRT‐PCR assay) was performed using previously designed primers and probe to correspond to a conserved region of the matrix gene segment as a control [Nakauchi et al., 2010].
To confirm the subtype of influenza virus, all clinical specimens were tested by simplex real‐time RT‐PCR assays for detecting specifically A/H1N1pdm virus (H1pdm rRT‐PCR assay) as described previously [Nakauchi et al., 2010].
Sequence
To amplify the partial NA gene between 676 and 1111 (from the start codon) using extracted RNA from the isolated virus, RT‐PCR was carried out with the following paired primers: swine N1‐676‐694F and swine N1‐1130‐1111R (Table I) using the One‐Step RNA PCR Kit (AMV) (TaKaRa, Tokyo, Japan) according to the manufacturer's instructions. The RT‐PCR conditions were as follows: 30 min at 50°C to activate RT, followed by initial denaturation for 2 min at 94°C, with a subsequent 30 cycles of amplification (denaturation at 94°C for 30 sec, annealing at 45°C for 30 sec, and extension at 72°C for 2 min). The PCR products were purified, and then sequenced using swine N1‐676‐694F primer with BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Carlsbad, CA) by Applied Biosystems DNA Analyzer. Total RNA was prepared as described in the RNA Preparation Section.
RESULTS
Development of Duplex Real‐Time RT‐PCR Assay for Detecting Oseltamivir‐Resistant Pandemic Influenza A/H1N1 2009 Virus
The specificity of the duplex endpoint genotyping RT‐PCR assay for detecting oseltamivir‐resistant A/H1N1pdm virus (H275Y RT‐PCR) was evaluated using human seasonal influenza A/H1N1 and A/H3N2 viruses, influenza B viruses, and other viral respiratory pathogens. The H275Y RT‐PCR assay reacted specifically to A/H1N1pdm viruses but showed no cross‐reactivity against other subtypes/types of influenza A and B viruses or other viral respiratory pathogens (data not shown).
The sensitivity of the H275Y RT‐PCR assay was compared with that of the Type A rRT‐PCR assay using serial dilutions of A/Denmark/524/2009 (H1N1)pdm and A/Denmark/528/2009 (H1N1)pdm viruses in six replicates for each assay. As shown in Table II, the minimum viral titer for 100% detection of A/Denmark/524/2009 (H1N1)pdm and A/Denmark/528/2009 (H1N1)pdm using the Type A rRT‐PCR and H275Y RT‐PCR assays was 3.16 × 10−2 TCID50/reaction, respectively.
Table II.
Detection Limits of RT‐PCR Assays Using Serial Dilutions of Viral RNA
| Viral titer (TCID50/reaction) | A/Denmark/524/2009 | A/Denmark/528/2009 | ||||
|---|---|---|---|---|---|---|
| No. of positive replicates/No. of tests for each assay (positive %) | No. of positive replicates/No. of tests for each assay (positive %) | |||||
| Type A rRT‐PCR | H275Y RT‐PCR | Type A rRT‐PCR | H275Y RT‐PCR | |||
| H275 | Y275 | H275 | Y275 | |||
| 3.16 × 10−1 | 6/6 (100) | 6/6 (100) | 0/6 (0) | 6/6 (100) | 0/6 (0) | 6/6 (100) |
| 3.16 × 10−2 | 6/6 (100) | 6/6 (100) | 0/6 (0) | 6/6 (100) | 0/6 (0) | 6/6 (100) |
| 3.16 × 10−3 | 2/6 (33.3) | 2/6 (33.3) | 0/6 (0) | 3/6 (50) | 0/6 (0) | 3/6 (50) |
| 3.16 × 10−4 | 0/6 (0) | 0/6 (0) | 0/6 (0) | 0/6 (0) | 0/6 (0) | 0/6 (0) |
Endpoint fluorescence plots of the H275Y RT‐PCR (Fig. 1A) and the standard curve of the Type A rRT‐PCR assay were also generated (Fig. 1B,C). The standard curve showed a linear relationship between the log of viral titer and the crossing point (Cp) value (Fig. 1B for A/Denmark/524/2009 and Fig. 1C for A/Denmark/528/2009). The correlation coefficient of the standard curve was 0.99, indicating a precise log‐linear relationship between the viral titer and Cp value (Fig. 1B,C).
Figure 1.
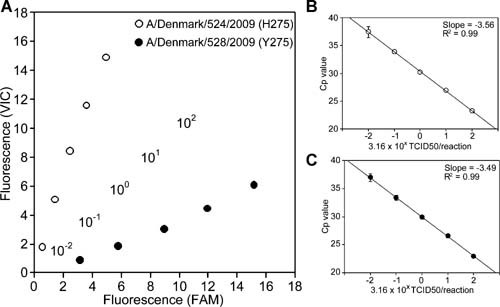
A: Endpoint fluorescence plot. Serial dilutions of A/Denmark/524/2009 (open circle) and A/Denmark/528/2009 (filled circle) were analyzed by H275Y RT‐PCR assay. Viral titers of these two isolates were 3.16 × 102, 3.16 × 101, 3.16 × 100, 3.16 × 10−1, and 3.16 × 10−2 TCID50/reaction and are represented in the centerline of the plot as 102, 101, 100, 10−1, and 10−2, respectively. The plot was generated using the average of the results obtained from the assay performed in six replicates. Relative H275 (VIC) and Y275 (FAM) fluorescence are plotted on the y‐axis and x‐axis, respectively. B,C: Serial dilutions of A/Denmark/524/2009 (B) and A/Denmark/528/2009 (C) were analyzed by Type A rRT‐PCR assay. The standard curves were generated using average crossing point (Cp) values obtained from the assay performed in six replicates. The correlation coefficient and slope of the standard curve are represented in the graphs.
Validation of H275Y RT‐PCR Assay Performed Using Clinical Specimens
From October 2009 to February 2010, clinical specimens (nasal or pharyngeal swabs, or nasal aspirate) from suspected cases of A/H1N1pdm virus infection (n = 44) were obtained from hospitals and clinics in Japan. The clinical specimens were tested by Type A and H1pdm rRT‐PCR assays after RNA purification. Of 44 clinical specimens, 39 clinical specimens were confirmed to be positive for Type A and H1pdm, and 5 clinical specimens were confirmed to be negative for Type A and H1pdm (Table III). Of 39 Type A‐ and H1pdm‐positive clinical specimens tested by H275Y RT‐PCR assay, 38 specimens were determined as H275 (oseltamivir‐susceptible) and 1 specimen was determined as Y275 (oseltamivir‐resistant) (Table III). All five Type A‐ and H1pdm‐negative clinical specimens were confirmed as negative by H275Y RT‐PCR assay (Table III). Some of these results are also shown as endpoint fluorescence plots (Fig. 2).
Table III.
Detection of Pandemic Influenza A/H1N1 2009 Virus Having H275Y in Clinical Specimens by H257Y RT‐PCR Assay
| Results of Type A and H1pdm rRT‐PCR assays | Results of H275Y RT‐PCR assay | ||
|---|---|---|---|
| H275 | Y275 | Negative | |
| Type A‐ and H1pdm‐positive (n = 39) | 38 | 1 | 0 |
| Type A‐ and H1pdm‐negative (n = 5) | 0 | 0 | 5 |
Figure 2.
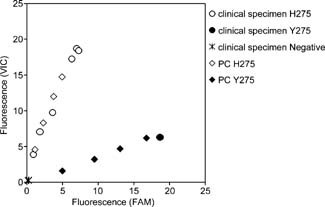
Endpoint fluorescence plot of H275Y RT‐PCR assay using clinical specimens. Relative H275 (VIC) and Y275 (FAM) fluorescence are plotted on the y‐axis and x‐axis, respectively. Clinical specimens discriminated as H275 or Y275 are represented as open circles or filled circles, respectively. Viral titers of 3.16 × 101, 3.16 × 100, 3.16 × 10−1, and 3.16 × 10−2 TCID50/reaction of A/Denmark/524/2009 (PC H275, open diamond) and A/Denmark/528/2009 (PC Y275, filled diamond) were used as positive controls for the assay.
Validation of H275Y RT‐PCR Assay Performed Using Clinical Isolates Without RNA Purification
To facilitate the assay scheme, the H275Y RT‐PCR assay was performed by using the cultured supernatants of infected MDCK cells directly, without RNA purification, as the template of the assay for screening H275Y mutation from a large number of clinical isolates. Twenty‐four A/H1N1pdm clinical isolates possessing H275, 45 A/H1N1pdm clinical isolates possessing Y275, and 4 A/H1N1pdm clinical isolates having a combination of viruses possessing H275 and Y275 were used directly for the validation of the H275Y RT‐PCR assay without RNA purification. Three isolated seasonal influenza A/H1N1 viruses possessing H275 and three isolated seasonal influenza A/H1N1 viruses possessing Y275 were also used directly as negative controls for the H275Y RT‐PCR assay without RNA purification. All the isolated seasonal influenza A/H1N1 viruses showed no cross‐reactivity and were negative in the assay (Fig. 3). In the assay, all 24 and 45 isolated A/H1N1pdm viruses possessing H275 and Y275 were confirmed as H275 (Fig. 3) and Y275 (the results of 5 clinical isolates are shown in Fig. 3), respectively. The four clinical isolates having a combination of viruses were confirmed as “mix” (Fig. 3).
Figure 3.
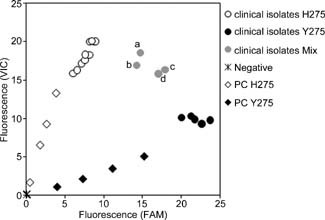
Endpoint fluorescence plot of H275Y RT‐PCR assay using clinical isolates. Relative H275 (VIC) and Y275 (FAM) fluorescence are plotted on the y‐axis and x‐axis, respectively. Clinical isolates discriminated as H275, Y275, or mix are represented as open circles, filled circles, or gray circles, respectively. Viral titers of 3.16 × 101, 3.16 × 100, 3.16 × 10−1, and 3.16 × 10−2 TCID50/reaction of A/Denmark/524/2009 (PC H275, open diamond) and A/Denmark/528/2009 (PC Y275, filled diamond) were used as positive controls for the assay.
DISCUSSION
In this study, a rapid and simple duplex one‐step endpoint genotyping RT‐PCR assay for discriminating A/H1N1pdm viruses possessing the H275Y mutation (H275Y RT‐PCR assay) was developed. The sensitivity of the H275Y RT‐PCR assay was comparable to that of the Type A rRT‐PCR assay (Table II), with a detection limit of 7.5 copies/reaction [Nakauchi et al., 2010]. No cross‐reactivity or nonspecific reactions were observed in the assay performed using other subtypes/type of influenza viruses (Fig. 3) or clinical specimens (Table III and Fig. 2). These results demonstrated that the H275Y RT‐PCR assay was highly specific and sensitive at detecting the H275Y mutation.
Using the H275Y RT‐PCR assay, purified RNA from 44 clinical specimens was tested (Table III and Fig. 2). Of 44 clinical specimens tested, 39 were positive for Type A and H1pdm (Table III and Fig. 2). Among 39 specimens, the H275Y RT‐PCR assay detected 38 oseltamivir‐susceptible viruses with H275 and 1 oseltamivir‐resistant virus with Y275. These results corresponded to those of NA sequencing. There was no discrepancy between the H275Y RT‐PCR assay and NA sequencing, suggesting that the H275Y RT‐PCR assay could discriminate A/H1N1pdm viruses possessing H275 and Y275 in clinical specimens.
The clinical isolates in the cultured supernatants of MDCK cells (Fig. 3) were used directly for the H275Y RT‐PCR assay, without RNA purification. The assay results were compared with the results of NA sequencing, and no discrepancies were found between the assays (data not shown, all sequence data of the NA gene of the clinical isolates used in this study have been deposited in the GISAID database). Four clinical isolates, A/Aichi/1210/2009, A/Mie/137/2009, A/Tochigi/373/2009, and A/Niigata/1459/2009 (Fig. 3a–d) were found to be a mixed population of the viruses possessing H275 and Y275 by sequencing of the NA gene (data not shown), and were detected as “mix” in the H275Y RT‐PCR assay (Fig. 3). These results suggested that the H275Y RT‐PCR assay could discriminate specifically A/H1N1pdm viruses possessing H275, Y275, or a mix of H275 and Y275.
It is noteworthy that the H275Y RT‐PCR assay could detect the H275Y mutation in clinical isolates without RNA purification, since a sufficient amount of viral RNA may be released from the virion under the RT conditions (50°C, 20 min). When considering the handling of a large number of clinical isolates, the H275Y RT‐PCR assay can be performed rapidly and simply with a lower risk of cross‐contamination, because RNA purification is unnecessary.
Recently, an assay was developed using probes containing LNA nucleotides for discriminating A/H1N1pdm possessing H275Y [van der Vries et al., 2010]. Their assay was highly specific, and the results were interpreted easily; however, the oseltamivir‐resistant A/H1N1pdm virus has not been evaluated with their assay. Two real‐time RT‐PCR assays were also developed: one for detecting A/H1N1pdm viruses possessing H275, and the other for detecting those possessing Y275 [Hindiyeh et al., 2010]. However, in the two independent assays described above for discriminating H275Y, it is not simple to interpret the results, and doing so is costly.
The H275 RT‐PCR assay developed in this study could be performed rapidly and simply, since the cultured supernatants of infected cells were used directly without RNA purification (Fig. 3), and the endpoint genotyping analysis program of the Light Cycler® 480 SW1.5 software interpreted the results of the assay automatically by using positive controls, like A/Denmark/524/2009 (H1N1)pdm and A/Denmark/528/2009 (H1N1)pdm, suggesting that this assay is useful for the screening of the H275Y mutation in NAI surveillance activity. Additionally, the H275Y RT‐PCR assay could detect whether patients were infected with oseltamivir‐susceptible or ‐resistant viruses by testing clinical specimens, suggesting that this assay is a powerful diagnostic method for determining how to manage patients.
Supporting information
Additional Supporting Information may be found in the online version of this article.
Supplemental Tables
Acknowledgements
We thank Dr. Lars Nielsen (European Centre for Disease Prevention and Control) for providing us with A/Denmark/524/2009 and A/Denmark/528/2009. We are greatly obliged to Dr. Hidekazu Nishimura (Sendai Medical Center), Dr. Masanobu Agoh (Nagasaki Prefectural Institute for Environmental Research and Public Health), and Dr. Fumihiro Taguchi (Department of Virology III, NIID and Nippon Veterinary and Life Science University) for providing us with the respiratory pathogens. Finally, we thank all the members of the Influenza Virus Research Center for their helpful technical advice.
The authors declare no potential conflict of interest relevant to this article.
REFERENCES
- Baz M, Abed Y, Papenburg J, Bouhy X, Hamelin ME, Boivin G. 2009. Emergence of oseltamivir‐resistant pandemic H1N1 virus during prophylaxis. N Engl J Med 361:2296–2297. [DOI] [PubMed] [Google Scholar]
- Garten RJ, Davis CT, Russell CA, Shu B, Lindstrom S, Balish A, Sessions WM, Xu X, Skepner E, Deyde V, Okomo‐Adhiambo M, Gubareva L, Barnes J, Smith CB, Emery SL, Hillman MJ, Rivailler P, Smagala J, de Graaf M, Burke DF, Fouchier RA, Pappas C, Alpuche‐Aranda CM, Lopez‐Gatell H, Olivera H, Lopez I, Myers CA, Faix D, Blair PJ, Yu C, Keene KM, Dotson PD Jr, Boxrud D, Sambol AR, Abid SH, St George K, Bannerman T, Moore AL, Stringer DJ, Blevins P, Demmler‐Harrison GJ, Ginsberg M, Kriner P, Waterman S, Smole S, Guevara HF, Belongia EA, Clark PA, Beatrice ST, Donis R, Katz J, Finelli L, Bridges CB, Shaw M, Jernigan DB, Uyeki TM, Smith DJ, Klimov AI, Cox NJ. 2009. Antigenic and genetic characteristics of swine‐origin 2009 A(H1N1) influenza viruses circulating in humans. Science 325:197–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindiyeh M, Ram D, Mandelboim M, Meningher T, Hirsh S, Robinov J, Levy V, Orzitzer S, Azar R, Grossman Z, Mendelson E. 2010. Rapid detection of influenza A pandemic (H1N1) 2009 virus neuraminidase resistance mutation H275Y by real‐time RT‐PCR. J Clin Microbiol 48:1884–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramarz P, Monnet D, Nicoll A, Yilmaz C, Ciancio B. 2009. Use of oseltamivir in 12 European countries between 2002 and 2007—Lack of association with the appearance of oseltamivir‐resistant influenza A(H1N1) viruses. Euro Surveill 14: pii19112. [DOI] [PubMed] [Google Scholar]
- Lackenby A, Thompson CI, Democratis J. 2008. The potential impact of neuraminidase inhibitor resistant influenza. Curr Opin Infect Dis 21:626–638. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. [DOI] [PubMed] [Google Scholar]
- Meijer A, Lackenby A, Hungnes O, Lina B, van‐der‐Werf S, Schweiger B, Opp M, Paget J, van‐de‐Kassteele J, Hay A, Zambon M. 2009. Oseltamivir‐resistant influenza virus A (H1N1), Europe, 2007–08 season. Emerg Infect Dis 15:552–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monto AS, McKimm‐Breschkin JL, Macken C, Hampson AW, Hay A, Klimov A, Tashiro M, Webster RG, Aymard M, Hayden FG, Zambon M. 2006. Detection of influenza viruses resistant to neuraminidase inhibitors in global surveillance during the first 3 years of their use. Antimicrob Agents Chemother 50:2395–2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakauchi M, Yasui Y, Miyoshi T, Minagawa H, Tanaka T, Tashiro M, Kageyama T. 2010. Improving one‐step real‐time reverse transcription‐PCR assays for detecting and subtyping pandemic (H1N1) 2009, seasonal H1N1, and seasonal H3N2 influenza viruses. J Virol Methods 171:156–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GJ, Vijaykrishna D, Bahl J, Lycett SJ, Worobey M, Pybus OG, Ma SK, Cheung CL, Raghwani J, Bhatt S, Peiris JS, Guan Y, Rambaut A. 2009. Origins and evolutionary genomics of the 2009 swine‐origin H1N1 influenza A epidemic. Nature 459:1122–1125. [DOI] [PubMed] [Google Scholar]
- Tashiro M, McKimm‐Breschkin JL, Saito T, Klimov A, Macken C, Zambon M, Hayden FG. 2009. Surveillance for neuraminidase‐inhibitor‐resistant influenza viruses in Japan, 1996–2007. Antivir Ther 14:751–761. [DOI] [PubMed] [Google Scholar]
- Ujike M, Shimabukuro K, Mochizuki K, Obuchi M, Kageyama T, Shirakura M, Kishida N, Yamashita K, Horikawa H, Kato Y, Fujita N, Tashiro M, Odagiri T, Working Group for Influenza Virus Surveillance in Japan . 2010. Oseltamivir‐resistant influenza viruses A (H1N1) during 2007–2009 in fluenza seasons, Japan. Emerg Infect Dis 16:926–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vries E, Jonges M, Herfst S, Maaskant J, Van der Linden A, Guldemeester J, Aron GI, Bestebroer TM, Koopmans M, Meijer A, Fouchier RA, Osterhaus AD, Boucher CA, Schutten M. 2010. Evaluation of a rapid molecular algorithm for detection of pandemic influenza A (H1N1) 2009 virus and screening for a key oseltamivir resistance (H275Y) substitution in neuraminidase. J Clin Virol 47:34–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . 2009a. Global influenza surveillance network: Laboratory surveillance and response to pandemic H1N1 2009. Wkly Epidemiol Rec 84:361–365. [PubMed] [Google Scholar]
- WHO . 2009b. New influenza A(H1N1) virus infections: Global surveillance summary, May 2009. Wkly Epidemiol Rec 84:173–179. [PubMed] [Google Scholar]
- WHO . 2009c. Oseltamivir‐resistant pandemic (H1N1) 2009 influenza virus, October. Wkly Epidemiol Rec 84:453–468. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article.
Supplemental Tables


