Abstract
Acute respiratory infections are responsible for high morbi–mortality in Peruvian children. However, the etiological agents are poorly identified. This study, conducted during the pandemic outbreak of H1N1 influenza in 2009, aims to determine the main etiological agents responsible for acute respiratory infections in children from Lima, Peru. Nasopharyngeal swabs collected from 717 children with acute respiratory infections between January 2009 and December 2010 were analyzed by multiplex RT‐PCR for 13 respiratory viruses: influenza A, B, and C virus; parainfluenza virus (PIV) 1, 2, 3, and 4; and human respiratory syncytial virus (RSV) A and B, among others. Samples were also tested with direct fluorescent‐antibodies (DFA) for six respiratory viruses. RT‐PCR and DFA detected respiratory viruses in 240 (33.5%) and 85 (11.9%) cases, respectively. The most common etiological agents were RSV‐A (15.3%), followed by influenza A (4.6%), PIV‐1 (3.6%), and PIV‐2 (1.8%). The viruses identified by DFA corresponded to RSV (5.9%) and influenza A (1.8%). Therefore, respiratory syncytial viruses (RSV) were found to be the most common etiology of acute respiratory infections. The authors suggest that active surveillance be conducted to identify the causative agents and improve clinical management, especially in the context of possible circulation of pandemic viruses. J. Med. Virol. 87:917–924, 2015. © 2015 Wiley Periodicals, Inc.
Keywords: respiratory viruses, respiratory infection, acute respiratory infections, virus detection, respiratory syncytial viruses, influenza viruses
INTRODUCTION
Acute respiratory infections are the leading cause of pediatric morbidity and mortality worldwide [Scheltinga et al., 2005; Grassi et al., 2014]. In Peru, respiratory infections are a major healthcare problem, especially in children under 5 years of age, with over 2.2 million infants being treated for this group of infectious diseases annually. The most affected areas are Lima East, Callao, Moquegua, Arequipa, and Ucayali, where a cumulative incidence as high as 19,783, and eight episodes per 10,000 children, was reported in 2013 [Gomez et al., 2013].
In 2009, Peru recorded one of the highest incidences of acute respiratory infections, with almost 2.8 million cases in children under 5 years of age [Gomez et al., 2009]. In May that year, the first influenza A (H1N1) case was confirmed. The pandemic rapidly spread throughout the country and affected primarily the 5–14 age group, with more than 80% of the cases being reported in Lima [Munayco et al., 2009]. Surveillance of the pandemic in Peru provided valuable information about its behavior in a developing southern hemisphere country, with over 8,000 confirmed cases until September 2009. This figure represents a 4% increase in acute respiratory infections [Gomez et al., 2009]. However, information about etiological agents of viral respiratory infections in Peru is still limited. Active surveillance is therefore essential to identify the causative agents and improve clinical management, especially in the context of possible circulation of pandemic viruses [Schoub, 2006; Ren et al., 2009].
Many viruses, including influenza virus, parainfluenza virus (PIV), human respiratory syncytial virus (RSV), human metapneumovirus (hMPV), human coronavirus (CoV), human rhinovirus (HRV), adenovirus (ADV), and human enterovirus (HEV), are known as common pathogens causing acute respiratory infections in children [Gerna et al., 2006]. Respiratory viruses can cause a wide spectrum of illnesses in children, including pharyngitis, otitis media, laryngitis, bronchitis, bronchiolitis, and pneumonia. This broad presentation of viral respiratory infections makes it difficult to clinically identify their etiology [Van Woensel et al., 2003]. Therefore, a rapid, sensitive, and specific diagnosis tool is essential for suitable therapeutic interventions [Barenfanger et al., 2000].
Viral diagnosis of respiratory tract infections is mainly done by direct antigen testing, virus isolation, or nucleic acid amplification using polymerase chain reaction (PCR). Viral culture and isolation is considered the gold standard, but this high‐cost, elaborated technique is usually performed for research purposes. Antigen detection tests, such as direct fluorescent antibody (DFA) test, are more commonly used because of their lower cost and rapid results, but often lack sensitivity or specificity. Furthermore, these tests produce false negatives in a high percentage of cases [Bellau‐Pujol et al., 2005]. Molecular techniques have revolutionized the diagnosis of respiratory viruses. Among them, PCR has shown to be the most sensitive and specific in detection of respiratory infectious agents, including samples with low copy number [van Elden et al., 2002; Freymuth et al., 2006; Van de Pol et al., 2007].
The aim of this study is to identify the etiological agents responsible for acute respiratory infections in children from January 2009 to December 2010 in Lima, Peru. For this purpose, the study was conducted according to the methodological guidance of National Reference Center for Influenza (WHO Centre for Influenza, National Center for Microbiology, ISCIII, Spain). A total number of 13 and 6 viruses were detected by multiplex RT‐PCR and DFA, respectively.
MATERIALS AND METHODS
Patients
A group of 717 patients under 18 years old with diagnosis of acute respiratory infection were recruited from the Hospital Nacional Cayetano Heredia, Lima, Peru, for prospective surveillance between January 2009 and December 2010. Epidemiological and clinical features were registered on a database. Those included age, symptoms (fever, defined as temperature higher than 38°C, rhinorrhea, cough, respiratory difficulty, sore throat, wheezing, discomfort, pharyngeal congestion, expectoration, vomiting, diarrhea, etc.), symptom duration, and time between sample taking and arrival in laboratory.
This study was approved by the Research Ethics Board of the Hospital Nacional Cayetano Heredia and Instituto de Investigación Nutricional (IIN). An informed consent was signed by parents or children's caregivers before enrollment.
Samples
Two nasopharyngeal samples were obtained per patient by inserting a swab into both nostrils parallel to the palate (Mini‐Tip Culture Direct, Becton‐Dickinson Microbiology System, MD) and a second swab from the posterior pharyngeal and tonsillar areas (Viral Culturette, Becton‐Dickinson Microbiology Systems). Both nasal and pharyngeal swabs were placed into the same tube containing viral transport medium (a minimal essential medium buffered with NaHCO3 and supplemented with 2% fetal bovine serum, penicillin and streptomycin 100 U/ml, amphotericin B 20 μg/ml, neomycin 40 μg/ml). Samples were then stored at 4°C until arrival in the laboratory of molecular biology at the Universidad Peruana de Ciencias Aplicadas (UPC)—Instituto de Investigación Nutricional (IIN). On receipt of the samples, the swabs were discarded and the tubes were centrifuged to pellet the cells, which were re‐suspended in 1.5 ml of PBS (Phosphate Buffered Saline). Antigen detection was performed by direct immunofluorescence assay (DFA), and two aliquots of each fresh specimen were stored at −20°C for reverse transcription PCR (RT‐PCR) analysis.
Immunofluorescence Detection
For direct immunofluorescence assay (DFA), samples were stained with fluorescein‐conjugated antibody for influenza A and B virus, human parainfluenza virus 1–3 (hPIV1–3), adenovirus (ADV), and respiratory syncytial virus (RSV) (Respiratory Panel 1, Viral Screening and Identification Kit; Light Diagnostic, CHEMICON International Temecula, USA). The presence of viral antigen in respiratory cells was indicated by the appearance of characteristic intracellular green fluorescence in ≥1 cell.
Reverse Transcription Polymerase Chain Reaction (RT‐PCR)
For multiplex RT‐PCR, viral genomic RNA and DNA were extracted from a total volume of 200 μl of sample by the guanidinium thiocyanate extraction method [Coiras et al., 2003]. The lysis buffer included 500 molecules of the cloned amplified product used as internal control in each reaction tube and; false negative results due to non‐specific inhibitors or extraction failure were discarded. Two independent multiplex reverse transcription RT‐PCR assays able to detect from 1 to 10 copies of viral genomes were performed [Van Woensel et al., 2003; Coiras et al., 2004]. One used specific primers for influenza A, B, and C virus, respiratory syncytial virus (RSV‐A and RSV‐B), and adenovirus (ADV); the other used specific primers for detection of human parainfluenza virus (PIV‐1, PIV‐2, PIV‐3, and PIV‐4), coronavirus (CoV‐229E and CoV‐OC43), human rhinovirus (HRV), and enterovirus (HEV). Negative (viral transport medium) and positive (cDNA viral) controls were prepared by the same procedure for both tests. Amplified products were recovered from the gel, purified (SpinPrep Gel DNA Kit; San Diego, CA) and sent for commercial sequencing (Macrogen, Korea). Finally, viral etiology was determined when RT‐PCR and/or DFA samples were positive for at least one of these viruses.
A viral etiology was considered, if RT‐PCR and/or DFA samples were positive for a respiratory virus.
Statistical Analysis
Quantitative variables were presented as an average interquartile range (IQR) in two groups (virus infection and viral co‐infection). Qualitative variables were reported as frequencies and percentages. Comparison of proportions was determined by χ2 or Fisher's exact test. Probability values of P < 0.05 were considered significant. Analysis was performed using SPSS Statistics 20.0.0 software (IBM Corporation, NY).
RESULTS
The study was conducted on 717 patients under 18 years old with diagnosis of acute respiratory infections between January 2009 and December 2010 in Lima, Peru. The most common age groups were infants under 1 year old (60.2%) and 2–5 (21.6%), followed by 6–10 (7.8%) and >10 (3.5%). Most were hospitalized, 80.9% (580) whereas 19.1% (137) were ambulatory patients. The most frequent symptoms were fever (67.8%), rhinorrhea (61.4%), cough (63.7%), and respiratory difficulty (54.1%) (Table I).
Table I.
Clinical Summary of Pediatric Patients With Respiratory Disease
| Number (%) | |
|---|---|
| Children | |
| Age (range years) | |
| 0–1 | 432 (60.2) |
| 2–5 | 155 (21.6) |
| 6–10 | 56 (7.8) |
| >10 | 25 (3.5) |
| NR * | 49 (6.8) |
| Gender | |
| Male | 405 (56.5) |
| Women | 285 (39.7) |
| NR * | 27 (3.8) |
| Hospitalized | 580 (80.9) |
| Ambulatory | 137 (19.1) |
| Sample | |
| Nasopharyngeal swab | 706 (98.5) |
| Pharyngeal swab | 8 (1.1) |
| Nasopharyngeal aspired | 3 (0.4) |
| Clinical Symptoms | |
| Fever | 486 (67.8) |
| Cough | 457 (63.7) |
| Rhinorrhea | 440 (61.4) |
| Respiratory Difficulty | 388 (54.1) |
| Sore Throat | 121 (16.9) |
| Wheezing | 255 (35.6) |
| Discomfort | 52 (7.2) |
| Pharyngeal Congestion | 173 (24.1) |
| Expectoration | 165 (23.0) |
| Vomiting | 87 (12.1) |
| Diarrhea | 71 (9.9) |
| Others (<10% of cases): Ear pain, photophobia, conjunctival congestion, abdominal pain, lymphadenopathy, fatigue, headache, myalgia, skin rash. | |
| Clinical Diagnosis | |
| Viral Pneumonía | 106 (14.8) |
| Viral Pharyngitis/Rhinopharyngitis | 28 (3.9) |
| Bronchiolitis | 56 (7.8) |
| Influenza Infection | 47 (6.6) |
| Feverishness | 30 (4.2) |
| EDA | 7 (0.9) |
| Others (<2% of cases): Obstructive syndrome (bronchial, bronchial acute), sinusitis, respiratory distress syndrome, sepsis late atypical febrile seizure status epilepticus, atypical febrile seizure, gastroenteritis. | |
NR, not registered.
Regarding the 717 nasopharyngeal specimens tested, the presence of at least one viral respiratory pathogen was detected in 33.5% (240) of cases by PCR and in only 11.9% (85) by DFA (Table II). Human respiratory syncytial A (RSV‐A) was the most prevalent virus detected in 15.3% (110) by PCR, followed by influenza A in 4.6% (33) and parainfluenza‐1 (PIV‐1) in 3.6% (26). Their probability of occurrence (odds) was 18% for RSV‐A, 4.8% for influenza A and 3.8% for PIV‐1. As for DFA detection, the same pathogens were the most predominant, but the human respiratory syncytial was detected in only 6.2% (42) of patients, influenza A in 1.8% (13) and PIV‐1 in 0.8% (6) (Table III). In addition, PCR allowed the identification of viral co‐infections. Thus, a total of 13 cases of viral co‐infections, with influenza A and RSV‐A being the most common pathogens found in association with other respiratory viruses, were reported (Table IV).
Table II.
Comparison of the Diagnostic by PCR versus IFD
| Assay * | |||
|---|---|---|---|
| Cases (%) | PCR | IFD | P‐value ** |
| Positive | 240 (33.5) | 85 (11.9) | <0.001 |
| Negative | 477 (66.5) | 599 (83.5) | <0.001 |
| Unspecific | 0 (0.0) | 33 (4.6) | |
| Total | 717 (100.0) | 717 (100.0) | |
χ2‐test, P < 0.001.
z‐test.
Table III.
Prevalence of Pathogens Among Patients. Pathogens Agent Identified in the Diagnostic by PCR and IFD
| PCR | IFD | |||||
|---|---|---|---|---|---|---|
| Pathogen | Frequency | Prevalence (%) | Odds | Frequency | Prevalence (%) | Odds |
| RSV | — | — | — | 42 | 5.9 | 0.062 * |
| RSV‐A | 110 | 15.3 | 0.181 * | — | — | — |
| RSV‐B | 6 | 0.8 | 0.008 | — | — | — |
| Influenza A | 33 | 4.6 | 0.048 | 13 | 1.8 | 0.018 |
| Influenza B | 5 | 0.7 | 0.007 | 4 | 0.6 | 0.006 |
| Influenza C | 1 | 0.1 | 0.001 | — | — | — |
| Adenovirus | 1 | 0.1 | 0.001 | 1 | 0.1 | 0.001 |
| Rhinovirus | 5 | 0.7 | 0.007 | — | — | — |
| Coronavirus | 8 | 1.1 | 0.011 | — | — | — |
| Parainfluenza‐1 | 26 | 3.6 | 0.038 | 6 | 0.8 | 0.008 |
| Parainfluenza‐2 | 13 | 1.8 | 0.018 | 5 | 0.7 | 0.007 |
| Parainfluenza‐3 | 7 | 1.0 | 0.010 | 1 | 0.1 | 0.001 |
| Parainfluenza‐4 | 7 | 1.0 | 0.010 | — | — | — |
| Enterovirus | 18 | 2.5 | 0.026 | — | — | — |
z‐test, P < 0.05.
Table IV.
Viral Co‐Infections
| Pathogens: Virus‐Virus | Cases (%) |
|---|---|
| Influenza A & RSV‐A | 1 (0.14) |
| Influenza A & Rhinovirus | 1 (0.14) |
| Influenza A & RSV‐B | 2 (0.28) |
| Influenza A & Parainfluenza‐2 | 1 (0.14) |
| RSV‐A & Adenovirus | 1 (0.14) |
| RSV‐A & Enterovirus | 2 (0.28) |
| RSV‐A & Parainfluenza‐1 | 3 (0.41) |
| RSV‐A & Parainfluenza‐2 | 1 (0.14) |
| Parainfluenza‐1 & Enterovirus | 1 (0.14) |
| Total cases | 13 (1.81) |
*z‐test, P < 0.05.
Respiratory viruses were analyzed according to age distribution. The under 1‐year‐old group was the most affected by viral infections, 44% (190/432), followed by the under 10‐year‐old old group, 24% (6/25), the 6–10 group, 23.2% (13/56), and the 2–5 group, 18.7% (29/155). Incidence of RSV‐A was very high in the 1‐year old‐group (21.8%) whereas incidence of PIV‐1 (4.6%), influenza A (4.4%), enterovirus (3.5%), and PIV‐2 (2.5%), among others, were low. On the other hand, fewer cases of RSV‐A were reported in children aged 2–5, 6–10, and under 10 years old, with an incidence of 6.5%, 5.4%, and 8%, respectively (Table V).
Table V.
Virus Distribution According to Age Range
| Age Range (years) | ||||||
|---|---|---|---|---|---|---|
| Pathogens | 0–1 (n=432) | 2–5 (n=155) | 6–10 (n=56) | >10 (n=25) | * NR (n=49) | Total patients (n=717) |
| RSV‐A | 94 (21.8%) | 10 (6.5%) | 3 (5.4%) | 2 (8.0%) | 1 (2.0%) | 110 (15.3%) |
| RSV‐B | 6 (1.4%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 6 (0.8%) |
| Influenza A | 19 (4.4%) | 6 (3.9%) | 4 (7.1%) | 4 (16%) | 0 (0.0%) | 33 (4.6%) |
| Influenza B | 3 (0.7%) | 0 (0.0%) | 2 (3.6%) | 0 (0.0%) | 0 (0.0%) | 5 (0.7%) |
| Influenza C | 1 (0.2%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 1 (0.1%) |
| Adenovirus | 1 (0.2%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 1 (0.1%) |
| Rhinovirus | 4 (0.9%) | 1 (0.6%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 5 (0.7%) |
| Coronavirus | 5 (1.2%) | 3 (1.9%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 8 (1.1%) |
| Parainfluenza‐1 | 20 (4.6%) | 3 (1.9%) | 3 (5.4%) | 0 (0.0%) | 0 (0.0%) | 26 (3.6%) |
| Parainfluenza ‐2 | 11 (2.5%) | 0 (0.0%) | 1 (1.8%) | 0 (0.0%) | 1 (2.0%) | 13 (1.8%) |
| Parainfluenza ‐3 | 7 (1.6%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 7 (1.0%) |
| Parainfluenza ‐4 | 4 (0.9%) | 3 (1.9%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 7 (1.0%) |
| Enterovirus | 15 (3.5%) | 3 (1.9%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 18 (2.5%) |
| Total cases positives | 190 (44.0%) | 29 (18.7%) | 13 (23.2%) | 6 (24%) | 2 (4.1%) | 240 (33.5%) |
NR, not registered.
Finally, a seasonal distribution of all respiratory viruses detected between 2009 and 2010 was generally described (Fig. 1). Incidence of infections was higher during autumn and early winter in both years. The same distribution was reported for RSV‐A, with more than 50% of cases being detected during the periods of May–June in 2009 and March–April in 2010. Similarly, all enterovirus cases were found during May–June 2009. As for influenza A, one third of cases were detected during June–July 2009 whereas in 2010, only four cases were found. On the other hand, PIV‐1 exhibited a constant rather than a seasonal distribution.
Figure 1.
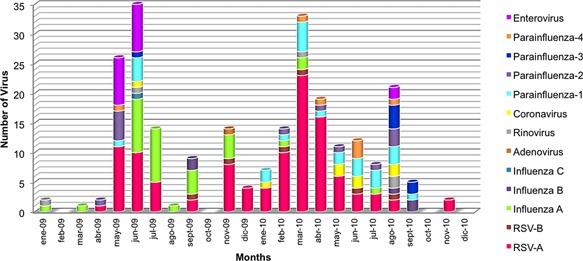
Respiratory virus seasonal distribution (2009–2010).
DISCUSSION
Acute respiratory infections are a major public health problem in children worldwide [Van Woensel et al., 2003; Scheltinga et al., 2005; Grassi et al., 2014], but most particularly in developing countries, where it is estimated that over 1.5 million children die every year, especially in the under 5‐year‐old age group [Alonso et al., 2012; Kwon et al., 2014]. Our study population was composed of 60.2% of infants under 1 year old and 21.6% of children between 2–5 years old. The World Health Organization (WHO) reports that, in Peru, less than 60% of these children have access to an appropriate health care provider [The United Nations Children's Fund (UNICEF), 2014UNICEF, 2014; World Health Organization (WHO), 2014WHO, 2014].
Viruses can cause a broad spectrum of respiratory illness in children. Etiological diagnosis of respiratory viral infections remains a clinical and laboratory challenge [Van de Pol et al., 2007; Alonso et al., 2012]. A 2012 study conducted with children from Rio de Janeiro, Brazil, with acute respiratory infections showed that the most frequent symptoms were cough (54%), fever (53.6%), rhinorrhea (47.8%), and nasal congestion (27.3%). Also, pneumonia was diagnosed in 24.4% of cases [Albuquerque et al., 2012]. The results of our study were similar since the most common symptoms were fever (67.8%), cough (63.7%), and rhinorrhea (61.4%). However, among 288 (40%) patients with a specific clinical diagnosis, 106 (14.8%) were diagnosed with pneumonia, probably because most samples were from hospitalized children, 580 (80.9%) (Table I). In addition, viruses were mostly identified in patients diagnosed with pneumonia, 63%, by far the major cause of respiratory‐related deaths [Alonso et al., 2012].
Appropriate viral etiology diagnosis in children with acute respiratory infections is decisive to proper care and treatment of this group of patients, as well as to proper choice of preventive strategies [Barenfanger et al., 2000; Van Woensel et al., 2003; Nasreen et al., 2014]. Surveillance of influenza and other respiratory infections is crucial to identifying the distribution, trend and impact of viruses circulating in a population, even in non‐pandemic periods [Watanabe et al., 2011; Zhao et al., 2014]. Using routinely available hospital laboratory data at minimum extra cost is an important addition to the current respiratory virus surveillance system [Zhao et al., 2014].
Polymerase Chain Reaction (PCR) is a highly sensitive, specific method for the detection of a broad spectrum of respiratory viruses [van Elden et al., 2002; Mahony et al., 2007; Lamarão et al., 2012]. Compared to conventional diagnostic tests, like Direct Immunofluorescence Assay (DFA), PCR increases diagnostic yields of respiratory virus detection from 24% to 43% of cases [Turner et al., 2013]. Therefore, PCR is the laboratory surveillance method of choice [Barenfanger et al., 2000; van Elden et al., 2002; Mahony et al., 2007; Van de Pol et al., 2007; Zhao et al., 2014]. The above figures are consistent with our study results, which show that respiratory viral detection increases from 11.9%, by DFA to 33.5%, by PCR (Table II). However, limited data are available regarding clinical presentation and strains of these viruses in Latin America [Sovero et al., 2011; Garcia et al., 2013; Turner et al., 2013; Grassi et al., 2014]. The prevalence of these pathogens is underestimated due to lack of rapid and proper virus identification tools in many areas [Alonso et al., 2012]. In Peru, respiratory virus surveillance has been conducted in 28 hospitals around the country since 2012. Samples of acute respiratory infection patients are analyzed by DFA and only influenza positive cases are subjected to PCR detection [Tejada, [Link]]. Furthermore, collected data remain unpublished and few studies have been undertaken about circulating respiratory viruses in the last 5 years, especially concerning specific viral etiology detection (although a few more on viral surveillance can be found) [Forshey et al., 2010; Padilla‐Ygreda et al., 2010; Salmón‐Mulanovich et al., 2011; Budge et al., 2014a].
A household‐based study conducted in high‐altitude rural Peruvian settings reported an adjusted incidence of 6.2 respiratory infection/child‐year, and the virus‐specific incidences per 100 child‐years were rhinovirus, 236; adenovirus, 73; parainfluenza, 46; influenza, 37; respiratory syncytial virus, 30; and human metapneumovirus, 17 [Budge et al., 2014b]. In our study, human respiratory syncytial virus A (RSV‐A) was the most prevalent virus in 15.3% of patients (110), followed by influenza A in 4.6% (33) and parainfluenza‐1 (PIV‐1) in 3.6% (26) (Table III). This was probably due to differences in respiratory virus circulation patterns among the regions and the large number of hospitalized patients in our study population.
Respiratory syncytial virus (RSV) has frequently been identified as the most common viral etiology in infants with respiratory infections [Van Woensel et al., 2003; Turner et al., 2013]. Incidence rates as high as 53.2% are typically reported for RSV, a virus strongly associated with severe respiratory infections, including pneumonia [Alonso et al., 2012; Lamarão et al., 2012; Garcia et al., 2013; Tejada, [Link]; Turner et al., 2013; Kwon et al., 2014; Zhao et al., 2014]. In the present study, RSV was also the predominant viral etiology (42%), with a very high incidence in under 1‐year‐old infants (21.8%), whereas lower incidences were found in other age groups (Table V).
A study performed in Ecuador described that influenza viruses may be the second most common cause of acute respiratory infections in childhood [Alonso et al., 2012]. Another finding was that influenza viruses are more likely to be detected in under 5‐year‐old patients, with an incidence of 9.4–9.6% in the last 2 years [Watanabe et al., 2011; Turner et al., 2013; Kwon et al., 2014]. Nonetheless, a recent work showed that infants and young children have a 12‐fold increased risk of admission to hospital for respiratory tract infection caused by influenza virus compared with children aged 5–17 years [Van Woensel et al., 2003]. Influenza virus was also the second most frequent etiology detected in our samples: influenza A in 4.6% and influenza B and C in only 0.7% and 0.1%, respectively (Table V). Influenza A was more common in children under 10 years old, with 16% of cases. However, an influenza H1N1v outbreak occurred during our study period, which may explain an abrupt increase in Influenza A cases during June to September in 2009.
Co‐infection with other respiratory viruses was not rare and was described in 2.9–8% of samples [Mahony et al., 2007; Sovero et al., 2011; Watanabe et al., 2011; Alonso et al., 2012; Lamarão et al., 2012]. The most common co‐infections (67%) were RSV co‐occurrences with other viruses, chiefly influenza A, followed by parainfluenza‐3, adenovirus, and hMPV [Mahony et al., 2007; Alonso et al., 2012]. Other influenza A virus co‐infections have also been reported, especially with rhinovirus, parainfluenza virus, and hMPV [Mahony et al., 2007]. These data are compatible with our results, 13 cases of co‐infection of influenza A and RSV‐A, the most common pathogens associated with parainfluenza, rhinovirus, adenovirus, and enterovirus (Table IV).
Although most respiratory viral infections occur throughout the year, seasonal variation in a comparable pattern is obvious for certain viruses, such as respiratory syncytial virus, influenza virus and parainfluenza virus [Van Woensel et al., 2003; Watanabe et al., 2011; Alonso et al., 2012; Lamarão et al., 2012; Zhao et al., 2014]. On the contrary, no clear seasonal patterns have been described for adenovirus and rhinovirus, not even after long periods of study [Alonso et al., 2012; Ampuero et al., 2012; Zhao et al., 2014].
In the present study, a higher incidence of respiratory viral infections was observed during autumn and early winter in both years. Nevertheless, to establish seasonality for respiratory viruses can be challenging. A study aimed to provide epidemiologic information about human parainfluenza viruses in Latin American cities between 2006 and 2010 revealed differences in patterns depending on the climate, altitude, and latitude. Particularly, in Piura (Peru) PIV‐1 showed several primary peaks between June and July. However, comparison of PIV‐1 incidence between Piura (Peru) and Fortaleza (Brazil), located at only 2°N, showed differences in distribution, suggesting that seasonality may be associated with temperature rather than distance from the Equator [Villaran et al., 2014]. In contrast, a constant, rather than seasonal, incidence was observed for PIV‐1 in the Lima study whereas a higher incidence was reported during autumn for RSV‐A, with more than 50% of cases detected, especially during May–June in 2009 and March–April in 2010. A similar seasonal distribution had been reported in Brazil, Argentina, and Chile [Luchsinger et al., 2008; Vidaurreta et al., 2011; Albuquerque et al., 2012].
In conclusion, respiratory virus surveillance programs are essential in Peru, just like in other Latin American countries, since they would prevent inappropriate antibiotic prescription and resistance to other viral etiologies accounting for increased health care demands and hospitalizations.
ACKNOWLEDGMENTS
This work has been partially supported by Programa de Ciencia y Tecnología (Grant FINCyT‐ PIN‐2008), Peru. LJdV is in debt to support from the Generalitat de Catalunya (2009SGR1208).
Disclosure Statement: No author has conflicts of interest.
REFERENCES
- Albuquerque M, Varella R, Santos N. 2012. Acute respiratory viral infections in children in Rio de Janeiro and Teresópolis, Brazil. Rev Inst Med Trop Sao Paulo 54:249–255. [DOI] [PubMed] [Google Scholar]
- Alonso WJ, Laranjeira BJ, Pereira S, et al. 2012. Comparative dynamics, morbidity, mortality burden of pediatric viral respiratory infections in an equatorial city. Pediatr Infect Dis J 31:9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ampuero JS, Ocaña V, Gómez J, Gamero ME, Garcia J, Halsey ES, Laguna‐Torres VA. 2012. Adenovirus respiratory tract infections in Peru. PLoS ONE 7:e46898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barenfanger J, Drake C, Leon N, Mueller T, Troutt T. 2000. Clinical and financial benefits of rapid detection of respiratory viruses: An outcomes study. J Clin Microbiol 38:2824–2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellau‐Pujol S, Vabret A, Legrand L, et al. 2005. Development of three multiplex RT‐PCR assays for the detection of 12 respiratory RNA viruses. J Virol Methods 126:53–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budge P, Griffin M, Edwards K, Williams JV, Verastegui H, Hartinger S, Mäusezahl D, Johnson M, Klemenc J, Zhu Y, Gil A, Lanata CF, Grigalva CG, for the RESPIRA PERU Group. 2014a. Impact of home environment interventions on the risk of influenza‐associated ARI in Andean children: Observations from a prospective household‐based cohort study. PLoS ONE 9:e91247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budge P, Griffin M, Edwards K, Williams J, Verastegui H, Hartinger S, Johnson M, Klemenc J, Zhu Y, Gil A, Lanata CF, Grijalva C. 2014b. Acute viral respiratory illnesses in Andean children: a household‐based study. Pediatr Infect Dis J 33:443–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casas I, Powell L, Klapper P, Cleator GM. 1995. New method for the extraction of viral RNA and DNA from cerebrospinal fluid for use in the polymerase chain reaction assays. J Virol Methods 53:25–36. [DOI] [PubMed] [Google Scholar]
- Coiras M, Aguilar J, García M, Casas I. 2004. Simultaneous detection of fourteen respiratory viruses in clinical specimens by two multiplex reverse transcription nested‐PCR assays. J Virol. 72:484–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coiras M, Pérez‐Breña P, García M, Casas I, Pérez‐Breña P. 2003. Simultaneous detection of influenza A, B, and C viruses, respiratory syncytial virus, and adenoviruses in clinical samples by multiplex reverse transcription nested‐PCR assay. J Med Virol 69:132–144. [DOI] [PubMed] [Google Scholar]
- Forshey B, Laguna‐Torres V, Vilcarromero S, Bazan I, Rocha C, Morrison AC, Stoddard ST, Alegre Y, Gomez J, Scott TW, Kochel TJ. 2010. Epidemiology of influenza‐like illness in the Amazon Basin of Peru, 2008–2009. Influenza Other Respir Viruses 4:235–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freymuth F, Vabret A, Cuvillon‐Nimal D, Simon S, Dina J, Legrand L, Gouarin S, Petitjean J, Eckart P, Brouard J. 2006. Comparison of multiplex PCR assays and conventional techniques for the diagnostic of respiratory virus infections in children admitted to hospital with an acute respiratory illness. J Med Virol 78:1498–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia J, Espejo V, Nelson M, Sovero M, Villaran MV, Gomez J, Barrantes M, Sanchez F, Comach G, Arango AE, Aguayo N, de Rivera IL, Chicaiza W, Jimenez M, Aleman W, Rodriguez F, Gonzales MS, Kochel TJ, Halsey ES. 2013. Human rhinoviruses,enteroviruses in influenza‐like illness in Latin America. Virol J 10:305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerna G, Campanini G, Rovida F, Percivalle E, Sarasini A, Marchi A, Baldanti F. 2006. Genetic variability of human coronavirus OC43‐, 229E‐, and NL63‐like strains and their association with lower respiratory tract infections of hospitalized infants and immunocompromised patients. J Med Virol 78:938–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez J. 2013. Situación epidemiológica de las infecciones respiratorias agudas (IRA) , neumonías y SOB (asma) en el Perú hasta la SE 39–2013 22:822–828. [Google Scholar]
- Gomez J, Munayco C, Arrasco J. 2009. Pandemic influenza in a southern hemisphere setting: The experience in Peru from May to September, 2009. Euro Surveill 14:pii 19371. [DOI] [PubMed] [Google Scholar]
- Grassi T, Mancini F, Ciervo A, Vescio MF, Ghazal A, Ashour H, Saleh E, El Zalabani M, Donatelli I, El Sawaf G, Rezza G. 2014. Chlamydophila pneumoniae, Mycoplasma pneumoniae, and influenza in children with respiratory infections in Alexandria. Egypt J Infect Dev Ctries 8:379–383. [DOI] [PubMed] [Google Scholar]
- Kwon J, Shim J, Kim D, Jung HL, Park MS, Shim JY. 2014. Prevalence of respiratory viral infection in children hospitalized for acute lower respiratory tract diseases, and association of rhinovirus and influenza virus with asthma exacerbations. Korean J Pediatr 57:29–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamarão LM, Ramos FL, Mello WA, Santos MC, Barbagelata LS, Justino MC, da Silva AF, Quaresma AJ, da Silva VB, Burbano RR, Linhares AC. 2012. Prevalence and clinical features of respiratory syncytial virus in children hospitalized for community‐acquired pneumonia in northern Brazil. BMC Infect Dis 12:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luchsinger V, Noy A, Avendaño L. 2008. Human respiratory syncytial virus genomic and antigenic variants isolated in two hospitals during one epidemic, in Santiago, Chile. J Clin Virol 42:260–263. [DOI] [PubMed] [Google Scholar]
- Mahony J, Chong S, Merante F, Yaghoubian S, Sinha T, Lisle C, Janeczko R. 2007. Devolopment of a respiratory virus panel test for detection of twenty human respiratory viruses by use of multiplex PCR and a fluid microbead‐based assay. J Clin Microbiol 45:2965–2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munayco C, Gomez J, Laguna‐Torres V. 2009. Epidemiological and transmissibility analysis of influenza A(H1N1)v in a southern hemisphere setting: Peru. Euro Surveill 14:pii 19299. [PubMed] [Google Scholar]
- Nasreen S, Luby SP, Brooks WA, Homaira N, Al Mamun A, Bhuiyan MU, Rahman M, Ahmed D, Abedin J, Rahman M, Alamgir AS, Fry AM, Streatfield PK, Rahman A, Bresee J, Widdowson MA, Azziz‐Baumgartner E. 2014. Population‐based incidence of severe acute respiratory virus infections among children aged <5 years in rural Bangladesh, June‐October 2010. PLoS ONE 9:e89978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padilla‐Ygreda J, Lindo‐Pérez F, Rojas‐Galarza R, Tantaleán Da Fieno J, Suárez Moreno V, Cabezas Sánchez C, Morales de Santa Gadea S, Hijar Guerra G. 2010. Etiology of community acquired pneumonia in children 2–59 months old in two ecologically different communities from Peru. Arch Argent Pediatr 108:516–523. [DOI] [PubMed] [Google Scholar]
- Ren L, Gonzalez R, Wang Z, Xiang Z, Wang Y, Zhou H, Li J, Xiao Y, Yang Q, Zhang J, Chen L, Wang W, Li Y, Li T, Meng X, Zhang Y, Vernet G, Paranhos‐Baccalà G, Chen J, Jin Q, Wang J. 2009. Prevalence of human respiratory viruses in adults with acute respiratory tract infections in Beijing, 2005–2007. Clin Microbiol Infect 15:1146–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmón‐Mulanovich G, Sovero M, Laguna‐Torres V, Kochel TJ, Lescano AG, Chauca G, Sanchez JF, Rodriguez F, Parrales E, Ocaña V, Barrantes M, Blazes DL, Montgomery JM. 2011. Frequency of human bocavirus (HBoV) infection among children with febrile respiratory symptoms in Argentina, Nicaragua and Peru. Influenza Other Respir Viruses 5:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheltinga SA, Templeton KE, Beersma MF, Claas EC. 2005. Diagnosis of human metapneumovirus and rhinovirus in patients with respiratory tract infections by an internally controlled multiplex real‐time RNA PCR. J Clin Virol 33:306–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoub B. 2006. Recommendations pertaining to the use of viral vaccines: Influenza 2006. S Afr Med J 96:103. [PubMed] [Google Scholar]
- Sovero M, Garcia J, Kochel T, Laguna‐Torres VA, Gomez J, Chicaiza W, Barrantes M, Sanchez F, Jimenez M, Comach G, de Rivera IL, Arango AE, Agudo R, Halsey ES. 2011. Circulating strains of human respiratory syncytial virus in central and South America. PLoS ONE 6:e22111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tejada C. Directiva Sanitaria para la Vigilancia Epidemiológica de Influenza, de otros virus respiratorios (OVR) e infecciones respiratorias agudas graves (IRAG) en el Perú [Internet]. Lima, Perú. Ministerio de Salud (MINSA). Available at: http://www.minsa.gob.pe/portada/Especiales/2013/h1n1/manejo_clinico/RM108-2012_DS%20p%20Vigilancia%20Epidemiologica%20de%20Influenza%20y%20otros%20virus%20respiratorios%20e%20IRAG.pdf. [Accessed May 15, 2014; Cited on June 14, 2013].
- The United Nations Children's Fund (UNICEF). Pneumonia: The forgotten killer of children [Internet] Geneva: WHO. Available at: http://unicefinnovation.org/sites/unicef.jjcdev2.com/files/Pneumonia_The_Forgotten_Killer_of_Children1.pdf. [Accessed May 28, 2014; Cited on June 15, 2014].
- Turner P, Turner C, Watthanaworawit W, Carrara V, Cicelia N, Deglise C, Phares C, Ortega L, Nosten F. et al. 2013. Respiratory virus surveillance in hospitalised pneumonia patients on the Thailand‐Myanmar border. BMC Infect Dis 13:434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Pol AC, Van Loon AM, Wolfs TF, Jansen NJ, Nijhuis M, Breteler EK, Schuurman R, Rossen JW. 2007. Increased detection of respiratory syncytial virus, influenza viruses, parainfluenza viruses, and adenovirus with real time PCR in samples from patients with respiratory symptoms. J Clin Microbiol 45:2260–2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Elden LJ, van Kraaij MG, Nijhuis M, Hendriksen KA, Dekker AW, Rozenberg‐Arska M, van Loon AM. 2002. Polymerase chain reaction is more sensitive than viral culture and antigen testing for the detection of respiratory viruses in adults with hematological cancer and pneumonia. Clin Infect Dis 34:177–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Woensel JB, van Aalderen WM, Kimpen JL. 2003. Viral lower respiratory tract infection in infants and young children. BMJ 327:36–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidaurreta SM, Marcone DN, Ellis A, Ekstrom J, Cukier D, Videla C, Carballal G, Echavarría M. 2011. Acute viral respiratory infection in children under 5 years: Epidemiological study in two centers in Buenos Aires, Argentina. Arch Argent Pediatr 109:296–304. [DOI] [PubMed] [Google Scholar]
- Villaran MV, García J, Gomez J, Arango AE, Gonzales M, Chicaiza W, Alemán W, Lorenzana de Rivera I, Sanchez F, Aguayo N, Kochel TJ, Halsey ES. 2014. Human parainfluenza virus in patients with influenza‐like illness from Central and South America during 2006‐2010. Influenza Other Respir Viruses 8:217–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe AS, Carraro E, Moreira L, Camargo C, Sinohara J, Puerari D, Guatura S, Granato C, Bellei N. 2011. Respiratory virus infections among hospitalized patients with suspected influenza A H1N1 2009 virus during the first pandemic wave in Brazil. Braz J Infect Dis 15:220–224. [DOI] [PubMed] [Google Scholar]
- World Health Organization (WHO). Care seeking for pneumonia: Situation and trends. [Internet] Geneva: WHO. Available at: http://www.who.int/gho/child_health/prevention/pneumonia/en/. [Accessed May 25, 2014; Cited on June 15, 2014].
- Zhao H, Green H, Lackenby A, Donati M, Ellis J, Thompson C, Bermingham A, Field J, Sebastianpillai P, Zambon M, Watson J, Pebody R. 2014. A new laboratory‐based surveillance system (Respiratory DataMart System) for influenza and other respiratory viruses in England: Results and experience from 2009 to 2012. Euro Surveill 19:pii 20680. [DOI] [PubMed] [Google Scholar]


