Abstract
Viruses play a significant part in children's respiratory infections, sometimes leading to hospitalization in cases of severe respiratory distress. The aim of this study was to investigate respiratory infections in children treated in a hospital intensive care unit (ICU). Assays were performed using the CLART® Pneumovir DNA array assay (Genomica, Coslada, Madrid, Spain), which makes it possible to detect 11 genus of respiratory viruses simultaneously. During the winter of 2008–2009, 73 respiratory specimens collected from 53 children under 2 years of age and admitted to an ICU were tested. At least one virus was detected in 78% (57/73) of the samples. The virological diagnosis was based on single infections in 65% (37/57) and on multiple infections in 35% (20/57) of cases. The array assay revealed respiratory syncytial virus (RSV) in 73.6% (42/57) of the samples and rhinovirus in 24.6% (14/57), either on their own or in co‐infections. All viruses identified in single and multiple infections were tested, taking into account clinical features, risk factors, and severity criteria. Children with no risk factors presented more multiple infections, up to 42% of cases, than children with at least one risk factor. RSV seemed to induce severe symptoms by itself as no difference in intubation needs was observed when RSV was detected on its own or in co‐infection. The CLART® Pneumovir DNA array was useful for examining severe viral respiratory infections, when other viruses than those detected by conventional methods could be involved, particularly in an ICU. J. Med. Virol. 83:150–155, 2011. © 2010 Wiley‐Liss, Inc.
Keywords: respiratory viruses, DNA array, pediatric intensive care unit
INTRODUCTION
Acute respiratory diseases are very common in children and a viral etiology can be identified in up to 80% of cases [Mahony, 2008]. These infections can lead to damage to the lower respiratory tract, including bronchitis, bronchiolitis, pneumonia, and bacterial superinfections. In some cases, particularly for children under 2 years of age, hospitalization in an intensive care unit (ICU) may be required in severe cases.
Respiratory syncytial virus (RSV) is the most frequently reported virus in infants admitted to hospital, but many other viral agents can be associated with acute respiratory infections [Mentel et al., 2005; Freymuth et al., 2006]. Rhinovirus (hRV), influenza (Flu) virus, and metapneumovirus (hMPV) are also now well known to be involved in such pathologies, with prevalence of around 30%, 10%, and 5%, respectively [Bouscambert‐Duchamp et al., 2005; Freymuth et al., 2006]. In pediatric ICU, the prevalence of viruses may differ, with a larger proportion of dual or mixed infections [Paranhos‐Baccala et al., 2008].
Viral respiratory diagnosis has traditionally relied on antigen detection and virus isolation. But the lack of sensitivity of antigen detection, the delay in the results with virus isolation and the limitations of detecting only cultivable viruses make the diagnosis of respiratory viruses incomplete. As a consequence, some clinical cases remain virologically negative.
Molecular technology has better sensitivity and the development of multiplex amplifications makes it possible to detect a broader panel of viruses. Adenoviruses (ADV), parainfluenza viruses (PIV), enteroviruses (EV) but also coronaviruses (HCoV) and bocaviruses (HBoV) can now be detected by multiplex assays. These assays are based on different types of technology, such as ligation‐dependent probe amplification (MLPA®), dual priming oligonucleotide (DPO) technology, target‐specific primer extension (TSPE), or target‐specific extension (TSE) [Chun et al., 2007; Mahony et al., 2007; Marshall et al., 2007; Reijans et al., 2008]. Other techniques are based on RT‐PCR amplifications followed by microarray analysis, as performed in the present study [Li et al., 2007]. Broadening viral diagnosis in this way may help clinicians to decrease prescriptions of antibiotics, implement early antiviral treatments when available, and prevent virus transmission.
The aim of the present study was to investigate respiratory infections in children admitted to a hospital pediatric ICU. Assays were performed with the CLART® Pneumovir DNA array assay (Genomica, Coslada, Madrid, Spain) which makes it possible to detect 11 genus of respiratory viruses simultaneously. Technical aspects, virological, and clinical data were analyzed.
MATERIALS AND METHODS
Patients and Samples
Seventy‐three respiratory samples from 53 children under 2 years of age and admitted to the hospital pediatric ICU in Lyon were collected between December 1, 2008 and March 30, 2009. Some children were sampled more than once but the samples corresponded to distinct episodes of respiratory disease. In most cases, the samples were nasopharyngeal or tracheo‐bronchic aspirates. Samples were collected in different clinical contexts: bronchiolitis (30 samples), pneumonia (14 samples), bronchiolitis associated with pneumonia (3 samples). Twenty samples were collected in other clinical contexts including isolated apnea (10 samples), sepsis (1 sample), apnea associated with sepsis (3), convulsions associated with fever (5), and asthma (1). No clinical information was available for 6 samples. Risk factors were considered for each patient. Of the 53 children, 12 were <42 days old (22.6%), 4 were born prematurely (7.5%), 2 had a cardiopathy (3.8%), 1 child had chronic respiratory insufficiency, and 1 child was immunodepressed. Twenty‐one children presented more than one risk factor (39.6%) and 12 children had no risk factors at all (22.6%). The severity of the respiratory disease was related to intubation requirements.
Nucleic Acid Extraction
Two hundred microliters of samples were treated with 10 µl of proteinase K at 20 mg/ml (Proteinase K PCR grade, Roche, Diagnostics GmbH, Mannheim, Germany). RNA was then extracted using the automated NucliSens easyMAG system (Biomerieux, SA, Mercy l'Etoiles,, France). Elution of the extracted nucleic acids was performed in 70 µl.
DNA Array Essay
The CLART® Pneumovir DNA array assay (Genomica, Coslada, Madrid, Spain) detects and characterizes the most frequent human viruses causing respiratory symptoms in a total of 8 hr after nucleic acid extraction. The viruses analyzed include: RSV A and B; Flu virus A, B, C; PIV 1, 2, 3, 4A, 4B; hMPV A and B; ADV (33 subtypes), EV (50 subtypes), hRV (65 subtypes); HCoV (subtype 229E); HBoV. This kit is based on the amplification of specific fragments of the viral genome by means of two multiplex RT‐PCR or PCR and a subsequent detection by hybridization with specific binding probes. During the 5‐hr RT‐PCR/PCR amplification, the amplified products were labeled with biotin. Following amplification, hybridization with specific probes immobilized in sites of the micro‐array was performed in a second step lasting 1 hr:30 min. After incubation with a streptavidin‐peroxidase conjugate, the addition of tetramethylbenzidine (TMB) induced the appearance of an insoluble product which precipitated at the hybridization sites on the micro‐array.
Statistical Analysis
Student's t‐test and Person's chi‐squared test were used to assess intergroup differences. Statistical analyses were performed on EpiInfo software (V 3.5.1 CDC). Odds ratio (OR) and 95% confidence interval (CI) were calculated for the likelihood of co‐detection and to test the association between multiple infections and risk factors with distress severity. A test was considered to be significant when the P‐value was <0.05.
RESULTS
Virological Data Obtained With CLART® Pneumovir in a Pediatric Intensive Care Unit
Of the 73 samples collected in this study, 57 samples contained at least one virus (78%; Table I). The virological diagnosis revealed single infections in 65% (37/57) and multiple infections in 35% (20/57) of the cases (Table I). Of the single infections, RSV was found in 49% (28/37) of cases and hRV in 7% (4/37). Other viruses, such as Flu B, ADV, PIV, hMPV, and HBoV were also detected. In multiple infections, RSV was found in co‐infection with hRV in 7% of cases, RSV A with RSV B in 5.3% of cases, hRV with HBoV in 5.3% of cases, and RSV A associated with both hRV and ADV in 3.5% of cases. Eight other viral co‐infections were also detected as reported in Figure 1. In total, RSV was detected with at least one other virus in 24.3% of cases, hRV in 17.5% of cases, and HBoV in 12% of cases.
Table I.
Virological Results on 73 Respiratory Samples
| Virus | Number of positive samples (%) detected by CLART® Pneumovir | ||
|---|---|---|---|
| Number | % | ||
| Positive samples | 57 | 78 | |
| Negative samples | 16 | 22 | |
| Single infections | RSV | 28 | 49 |
| RSV A | 20 | 35 | |
| RSV B | 8 | 14 | |
| hRV | 4 | 7 | |
| Flu A | 0 | 0 | |
| Flu B | 1 | 1.7 | |
| ADV | 1 | 1.7 | |
| PIV | 1 | 1.7 | |
| hMPV | 1 | 1.7 | |
| HBoV | 1 | 1.7 | |
| Total | 37 | 65 | |
| Multiple infections | RSV A + hRV | 4 | 7 |
| RSV A + RSV B | 3 | 5.3 | |
| hRV + HBoV | 3 | 5.3 | |
| RSV A + hRV + ADV | 2 | 3.5 | |
| RSVA + ADV | 1 | 1.7 | |
| RSV A + EV | 1 | 1.7 | |
| RSVA + HBoV | 1 | 1.7 | |
| RSV A + RSV B + ADV | 1 | 1.7 | |
| RSVA + hRV + FluA | 1 | 1.7 | |
| FluA + HBoV | 1 | 1.7 | |
| PIV + ADV + HBoV | 1 | 1.7 | |
| HBoV + hMPV | 1 | 1.7 | |
| Total | 20 | 35 | |
Figure 1.
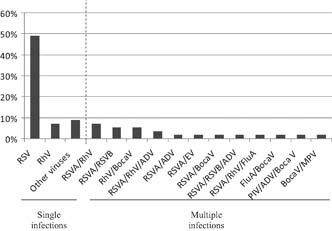
Distribution of virus detected in single and multiple infections by CLART®pneumovir in pediatric intensive care unit.
Clinical Data
Respiratory syndrome
CLART® Pneumovir DNA array made it possible to diagnose at least one virus in 85% of the bronchiolitis cases (Table II). In 57.6% of the cases, RSV was detected as a single infection. Multiple infections were also detected in around 27% of the bronchiolitis cases. They involved mostly RSV A associated with other viruses (8/33, 24.2%). hRV and HBoV were also detected in co‐infections in a proportion of 15% (5/33) and 6% (2/33), respectively.
Table II.
Viral Distribution Correlated to Respiratory Syndrome
| Number of positive samples (%) detected by CLART® Pneumovir | |||
|---|---|---|---|
| Virus | Number | % | |
| Bronchiolitis a n = 33 | |||
| Single infections | RSV | 19 | 57.6 |
| RSV A | 13 | ||
| RSV B | 6 | ||
| hRV | 0 | ||
| Total | 19 | 57.6 | |
| Multiple infections | RSV A + hRV | 3 | 9 |
| RSV A + RSV B | 2 | 6 | |
| RSV A + EV | 1 | 3 | |
| RSV A + HBoV | 1 | 3 | |
| hRV + HBoV | 1 | 3 | |
| RSV A + hRV + ADV | 1 | 3 | |
| Total | 9 | 27.3 | |
| Global positivity rate | 85 | ||
| Pneumonia a n = 17 | |||
| Single infections | RSV | 4 | 23.5 |
| RSV A | 4 | ||
| RSV B | 0 | ||
| hRV | 1 | 5.9 | |
| Flu B | 1 | 5.9 | |
| ADV | 1 | 5.9 | |
| PIV | 1 | 5.9 | |
| Total | 8 | 47.1 | |
| Multiple infections | RSV A + hRV | 1 | 5.9 |
| RSV A + RSV B | 1 | 5.9 | |
| hRV + HBoV | 2 | 11.8 | |
| RSV A + hRV + ADV | 1 | 5.9 | |
| Total | 5 | 29.5 | |
| Global positivity rate | 76.4 | ||
Three samples were collected in case of bronchiolitis associated to pneumonia.
In the pneumonia cases (17 samples), CLART® Pneumovir DNA array detected viruses in 76.4% of cases (Table II). The implication of RSV in 41.2% (7/17) of the cases was less than in bronchiolitis, with 23.5% (4/17) in single infections and 17.6% (3/17) in multiple infections. Other viruses were involved, such as hRV, Flu B, ADV, and PIV in single infections. Two cases of hRV associated with HBoV were also reported.
In total, RSV was implicated in 81.8% (27/33) of the bronchiolitis cases and in 41.2% (7/17) of the pneumonia cases, either in single or multiple infections. hRV was implicated in 15.2% (5/33) of the bronchiolitis cases and in 29.5% (5/17) of the pneumonia cases.
Risk factors
Of the 53 children, 20 presented one risk factor (37.8%), 21 presented more than one risk factor (39.6%), and 12 children had no risk factors at all (22.6%). For the children with no risk factors, viral factors that could have led to the severe respiratory infection were investigated. Children with no risk factors presented more multiple infections, up to 42% of cases, than children with at least one risk factor, even if the difference was not significant, probably because of the low number of patients (n = 12, P = 0.23; Table III).
Table III.
Risk Factors Associated to Single or Multiple Infections
| Risk factors | Single infection | Multiple infection | No virus detected |
|---|---|---|---|
| Age <42 years | 67% | 20% | 13% |
| Prematurity | 48% | 15% | 27% |
| Chronical respiratory disease | 42% | 25% | 33% |
| Cardiopathy | 38% | 25% | 37% |
| No risk factor | 42% | 42% | 16% |
Severity
Need for intubation was used as a surrogate for the severity of the respiratory distress. Nineteen episodes of respiratory disease required intubation and 52 did not. Information regarding intubation was missing in two cases. There was no difference in the severity of the distress between single and multiple infections (P = 0.62). In addition, the detection of RSV in a co‐infection did not worsen the respiratory distress, as intubation was required in 23.1% of the cases when RSV was detected alone and in 14.3% of the cases when detected in co‐infection with another viral agent. No association was observed between RSV in either single or multiple infections and distress severity (P = 0.4). RSV seemed to induce severe symptoms on its own. In RSV negative samples, no difference was observed in the distress severity between single and multiple infections (P = 0.35), probably because of the low number of RSV negative samples.
HBoV was detected in 8 clinical cases (1 in single, 7 in multiple infections) with intubation required in 3 cases (Table IV). Interestingly, detection of HBoV resulted in an increased likelihood of co‐detecting another virus as reflected by an OR of >1 (95% CI: 3.15–248) P = 0.0003. But there was no difference in distress severity between single and multiple infections when HBoV was detected (P = 0.5), probably because of the low number of cases (n = 8).
Table IV.
Clinical Cases Involving HBoVirus
| Virus | Clinical data | Ventilation | Risk factor |
|---|---|---|---|
| HBoV | Apnea, convulsions | IV | Age <42 days |
| HBoV + hRV | Bronchiolitis | NIV | None |
| HBoV + hRV (n = 2) | Pneumonia | IV | CRD, cardiopathy |
| HBoV + RSV A | Bronchiolitis, apnea | NIV | Age <42 days |
| HBoV + influenza A | Apnea | NIV | Prematurity, CRD |
| HBoV + hMPV | Apnea | NIV | None |
| HBoV + ADV + PIV | Apnea, convulsions | IV | Age <42 days |
CRD, chronical respiratory disease; IV, invasive ventilation; NIV, non‐invasive ventilation.
DISCUSSION
Microarray assay was found to be highly sensitive and made it possible to detect viruses that are not detected otherwise, either with the standard multiplex RT‐PCR (PIV, ADV, EV, HBoV, hMPV, HCoV) or in culture (HBoV, hMPV, HCoV) and which may be involved in multiple infections. From a practical point of view, the CLART® Pneumovir array assay is easy to use even if technical training is necessary to avoid false positives because of insufficient washing or overexposure of the arrays. Nevertheless, after nucleic acid extraction, the array assay takes 8 hr and is divided into two steps: multiplex RT‐PCR followed by hybridization. The main advantages are its high sensitivity and broader detection of respiratory viruses, improving viral diagnosis. However, the detection of multiple infections raises certain questions regarding the pathogenicity of the different viruses. Are all the viruses detected responsible for the clinical signs presented by the patient? These multiple viruses can be successive infections and molecular tests could detect a persistent genome in the absence of virus activity. This array assay is only a qualitative technique. However, quantitation should help to identify the predominant viral agent believed to be responsible for an acute respiratory syndrome.
Different studies have described previously the clinical characteristics of multiple viral infections versus RSV single infection. Multiple infections varied from 17.4% to 30% in children under 2 years of age admitted to hospital [Calvo et al., 2008; Miron et al., 2010] and were associated with higher fever, longer hospitalization, and more frequent use of antibiotics than in the case of infection with RSV alone [Calvo et al., 2008]. Infants with multiple infections are at higher risk of being admitted to a pediatric ICU [Paranhos‐Baccala et al., 2008; Richard et al., 2008]. In the present study, multiple infections were detected by arrays in 35% of the positive cases. In bronchiolitis, viral co‐infection was found in 27.3% of the cases with a predominance of the RSV and hRV co‐infection, as described previously [Richard et al., 2008]. RSV seems to induce severe symptoms on its own. Viral co‐infections with RSV effectively did not increase the need for intubation in comparison to infections with RSV as a single agent, as recently reported in a study considering duration of ventilation and length of hospitalization [Marguet et al., 2009]. Interestingly, children admitted to a pediatric ICU with no risk factor were found to present more co‐infections than children with risk factors, even if the difference was not significant.
RSV prevalence has previously been detected at around 40% using multiplex RT‐PCR in children admitted to the emergency room during winter [Freymuth et al., 2006]. In the population studied, the array assay revealed RSV in 73% of the samples either alone or in co‐infection. This prevalence of RSV may be explained by a higher sensitivity of array assay versus RT‐PCR, but this point needs to be evaluated in more detail. It may also suggest that RSV may be responsible for a higher risk of hospitalization in a pediatric ICU. The detection of hRV has been greatly improved by molecular techniques [Mahony, 2008]. The prevalence of hRV was previously reported as being around 30–35% with multiplex RT‐PCR [Freymuth et al., 2006]. This study reported hRV in 24% of the samples. This could suggest that infections caused by hRV are less represented in pediatric ICU than in other pediatric units.
hMPV is an RNA virus discovered in 2001 and is known to induce symptoms very similar to those caused by RSV [van den Hoogen et al., 2001, 2003]. hMPV outbreaks occur predominantly in the winter and spring months, overlapping or following RSV outbreaks [Mahony, 2008]. However, there can be significant differences in the detection of hMPV across seasons from year to year. Overall prevalence is around 7%, ranging from 2% to 25% [Mahony, 2008; Gaunt et al., 2009]. Only two hMPV infections were detected in this study, probably because the study period missed potential hMPV circulation during the spring season.
HBoV was discovered in 2005 using large‐scale molecular viral techniques [Allander et al., 2005]. Prevalence, ranging from 2% to 11% in respiratory samples from symptomatic patients, has been reported worldwide. In most cases, as in this study, HBoV was detected in co‐infections with other known viral agents raising the question of its actual pathogenic role [Mahony, 2008]. However, HBoV has been associated with low respiratory infections [Kesebir et al., 2006; Manning et al., 2006; Fry et al., 2007] and acute wheezing [Allander et al., 2007]. Even if HBoV can be found in asymptomatic patients, HBoV genome quantitation could help to identify the implication of HBoV in lower respiratory tract infections, as a high‐HBoV load has been reported in patients with lower respiratory tract infections [van de Pol et al., 2009]. In this study, HBoV prevalence was around 14%, higher than the prevalence usually reported [van de Pol et al., 2009], almost certainly because of the differences in detection techniques, patient recruitment and the seasonal study period. Nevertheless, Fry et al. [2007] have previously reported up to 12% of HBoV in children under 5 years of age with pneumonia.
In conclusion, microarrays have a high level of sensitivity and make broader viral detection possible. However, the relevance of the positive results obtained with this highly sensitive array is not easy to determine, particularly in cases of multiple infections caused by the lack of quantitation. DNA array may be useful regarding severe viral respiratory infections, particularly in pediatric ICU when no viral diagnosis has been established using conventional techniques that detect the most common viral respiratory pathogens.
Emilie Frobert and V. Escuret contributed equally to this study.
Conflict of interest: B.L. declares conflicts of interest with Argène, Biocryst, bioMérieux, GSK, MedImmune, Merck, Novartis, Plasmair, Roche, Sanofi‐Pasteur, and Wittycell. E.F., V.E., M.B.D. and F.M. declare conflicts of interest with Argène, bioMérieux, Cepheid, and R‐Biopharm. D.F. received research grants from Biomerieux and Merieux Foundation. E.Y. and Y.G. declare no conflict of interest.
Ethics: This study was conducted during the standard diagnosis of virological parameters and did not require any additional samples. For standard diagnoses, typically no approval by the Ethics Committee or informed consent from the parents is required. Nevertheless, at the Hospices Civils de Lyon, diagnosis is performed in compliance with French laws and HCL guidelines and in accordance with the ethical standards of the Declaration of Helsinki.
References
- Allander T, Tammi MT, Eriksson M, Bjerkner A, Tiveljung‐Lindell A, Andersson B. 2005. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc Natl Acad Sci USA 102: 12891–12896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allander T, Jartti T, Gupta S, Niesters HG, Lehtinen P, Osterback R, Vuorinen T, Waris M, Bjerkner A, Tiveljung‐Lindell A, van den Hoogen BG, Hyypia T, Ruuskanen O. 2007. Human bocavirus and acute wheezing in children. Clin Infect Dis 44: 904–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouscambert‐Duchamp M, Lina B, Trompette A, Moret H, Motte J, Andreoletti L. 2005. Detection of human metapneumovirus RNA sequences in nasopharyngeal aspirates of young French children with acute bronchiolitis by real‐time reverse transcriptase PCR and phylogenetic analysis. J Clin Microbiol 43: 1411–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo C, Garcia‐Garcia ML, Blanco C, Vazquez MC, Frias ME, Perez‐Brena P, Casas I. 2008. Multiple simultaneous viral infections in infants with acute respiratory tract infections. J Clin Virol 42: 268–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun JY, Kim KJ, Hwang IT, Kim YJ, Lee DH, Lee IK, Kim JK. 2007. Dual priming oligonucleotide system for the multiplex detection of respiratory viruses and SNP genotyping of CYP2C19 gene. Nucleic Acids Res 35: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freymuth F, Vabret A, Cuvillon‐Nimal D, Simon S, Dina J, Legrand L, Gouarin S, Petitjean J, Eckart P, Brouard J. 2006. Comparison of multiplex PCR assays and conventional techniques for the diagnostic of respiratory virus infections in children admitted to hospital with an acute respiratory illness. J Med Virol 78: 1498–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry AM, Lu X, Chittaganpitch M, Peret T, Fischer J, Dowell SF, Anderson LJ, Erdman D, Olsen SJ. 2007. Human bocavirus: A novel parvovirus epidemiologically associated with pneumonia requiring hospitalization in Thailand. J Infect Dis 195: 1038–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaunt E, McWilliam‐Leitch EC, Templeton K, Simmonds P. 2009. Incidence, molecular epidemiology and clinical presentations of human metapneumovirus; Assessment of its importance as a diagnostic screening target. J Clin Virol 46: 318–324. [DOI] [PubMed] [Google Scholar]
- Kesebir D, Vazquez M, Weibel CS, Shapiro ED, Ferguson D, Landry ML, Kahn JS. 2006. Human bocavirus infection in young children in the United States: Molecular epidemiological profile and clinical characteristics of a newly emerging respiratory virus. J Infect Dis 194: 1276–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, McCormac MA, Estes RW, Sefers SE, Dare RK, Chappell JD, Erdman DD, Wright PF, Tang YW. 2007. Simultaneous detection and high‐throughput identification of a panel of RNA viruses causing respiratory tract infections. J Clin Microbiol 45: 2105–2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahony JB. 2008. Detection of respiratory viruses by molecular methods. Clin Microbiol Rev 21: 716–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahony J, Chong S, Merante F, Yaghoubian S, Sinha T, Lisle C, Janeczko R. 2007. Development of a respiratory virus panel test for detection of twenty human respiratory viruses by use of multiplex PCR and a fluid microbead‐based assay. J Clin Microbiol 45: 2965–2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning A, Russell V, Eastick K, Leadbetter GH, Hallam N, Templeton K, Simmonds P. 2006. Epidemiological profile and clinical associations of human bocavirus and other human parvoviruses. J Infect Dis 194: 1283–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marguet C, Lubrano M, Gueudin M, Le Roux P, Deschildre A, Forget C, Couderc L, Siret D, Donnou MD, Bubenheim M, Vabret A, Freymuth F. 2009. In very young infants severity of acute bronchiolitis depends on carried viruses. PLoS ONE 4: 4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall DJ, Reisdorf E, Harms G, Beaty E, Moser MJ, Lee WM, Gern JE, Nolte FS, Shult P, Prudent JR. 2007. Evaluation of a multiplexed PCR assay for detection of respiratory viral pathogens in a public health laboratory setting. J Clin Microbiol 45: 3875–3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mentel R, Ilgert U, Wegner U, Zimmerman K, Bruns R, Gurtler L. 2005. Molecular and clinical characteristics of respiratory syncytial virus infections in hospitalized children. Med Microbiol Immunol 194: 67–71. [DOI] [PubMed] [Google Scholar]
- Miron D, Srugo I, Kra‐Oz Z, Keness Y, Wolf D, Amirav I, Kassis I. 2010. Sole pathogen in acute bronchiolitis: Is there a role for other organisms apart from respiratory syncytial virus? Pediatr Infect Dis J 29: 7–10. [DOI] [PubMed] [Google Scholar]
- Paranhos‐Baccala G, Komurian‐Pradel F, Richard N, Vernet G, Lina B, Floret D. 2008. Mixed respiratory virus infections. J Clin Virol 43: 407–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijans M, Dingemans G, Klaassen CH, Meis JF, Keijdener J, Mulders B, Eadie K, van Leeuwen W, van Belkum A, Horrevorts AM, Simons G. 2008. RespiFinder: A new multiparameter test to differentially identify fifteen respiratory viruses. J Clin Microbiol 46: 1232–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard N, Komurian‐Pradel F, Javouhey E, Perret M, Rajoharison A, Bagnaud A, Billaud G, Vernet G, Lina B, Floret D, Paranhos‐Baccala G. 2008. The impact of dual viral infection in infants admitted to a pediatric intensive care unit associated with severe bronchiolitis. Pediatr Infect Dis J 27: 213–217. [DOI] [PubMed] [Google Scholar]
- van de Pol AC, Wolfs TF, Jansen NJ, Kimpen JL, van Loon AM, Rossen JW. 2009. Human bocavirus and KI/WU polyomaviruses in pediatric intensive care patients. Emerg Infect Dis 15: 454–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Hoogen BG, de Jong JC, Groen J, Kuiken T, de Groot R, Fouchier RA, Osterhaus AD. 2001. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat Med 7: 719–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Hoogen BG, van Doornum GJ, Fockens JC, Cornelissen JJ, Beyer WE, de Groot R, Osterhaus AD, Fouchier RA. 2003. Prevalence and clinical symptoms of human metapneumovirus infection in hospitalized patients. J Infect Dis 188: 1571–1577. [DOI] [PubMed] [Google Scholar]


