Abstract
To explore the epidemiological and clinical features of different human metapneumovirus (hMPV) genotypes in hospitalized children. Reverse transcription polymerase chain reaction (RT‐PCR) or PCR was employed to screen for both hMPV and other common respiratory viruses in 2613 nasopharyngeal aspirate specimens collected from children with lower respiratory tract infections from September 2007 to February 2011 (a period of 3.5 years). The demographics and clinical presentations of patients infected with different genotypes of hMPV were compared. A total of 135 samples were positive for hMPV (positive detection rate: 5.2%). Co‐infection with other viruses was observed in 45.9% (62/135) of cases, and human bocavirus was the most common additional respiratory virus. The most common symptoms included cough, fever, and wheezing. The M gene was sequenced for 135 isolates; of these, genotype A was identified in 72.6% (98/135) of patients, and genotype B was identified in 27.4% (37/135) of patients. The predominant genotype of hMPV changed over the 3.5‐year study period from genotype A2b to A2b or B1 and then to predominantly B1. Most of clinical features were similar between patients infected with different hMPV genotypes. These results suggested that hMPV is an important viral pathogen in pediatric patients with acute lower respiratory tract infection in Changsha. The hMPV subtypes A2b and B1 were found to co‐circulate. The different hMPV genotypes exhibit similar clinical characteristics. J. Med. Virol. 87:1839–1845, 2015. © 2015 Wiley Periodicals, Inc.
Keywords: human metapneumovirus, genotype, acute lower respiratory tract infections, clinical feature, children
INTRODUCTION
Human metapneumovirus (hMPV) is a new respiratory viral pathogen identified in the Netherlands in 2001 [van den Hoogen et al., 2001]. hMPV is an RNA virus with a negative‐sense, single‐stranded, non‐segmental genome. hMPV belongs to the Metapneumovirus genus within the Pneumovirinae subfamily of the Paramyxoviridae family and is currently the only member of the genus Metapneumovirus that is known to cause disease in humans.
hMPV is widespread throughout the world [Feuillet et al., 2012] and infects human populations of different age groups, especially children younger than 5 years of age [McAdam et al., 2004; Vicente et al., 2006; Matsuzaki et al., 2008; Zhu et al., 2011; Zhang,,2012t al., 2012; Lu et al., 2013; Wei et al., 2013; Xiao et al., 2013]. hMPV causes acute upper and lower respiratory tract infections. The clinical manifestations of hMPV infection are similar to those of respiratory syncytial virus infection [van den Hoogen et al., 2003; van den Hoogen et al., 2004].
Based on the coding genes, hMPV has been classified into genotypes A and B, which are divided into 4 sub‐genotypes: A1, A2, B1, and B2. The sub‐genotype A2 is divided further into sub‐genotypes A2a and A2b. The new sub‐genotype A2b was discovered in children with respiratory tract infections by German researchers in 2006 [Huck et al., 2006]. The previous studies have identified distinct predominant prevalences of hMPV genotypes in different regions and different years [Matsuzaki et al., 2008; Pitoiset et al., 2010; Apostoli et al., 2012; Kim et al., 2012; Lu et al., 2013; Wei et al., 2013]. However, the data on the epidemiological and clinical characteristics of various genotypes of hMPV are currently limited.
Xiao et al in our research group has reported that hMPV is an important pathogen causing acute lower respiratory tract infections in children of the Hunan Province [Xiao et al., 2013]. To further explore the epidemiological and clinical features of various genotypes of hMPV, we summarized the data on the hMPV positive patients hospitalized for acute lower respiratory tract infections in Hunan Province over a period of 3.5 years (Sep. 2007–Feb. 2011)
MATERIALS AND METHODS
Patients and Specimens
Nasopharyngeal aspirates samples were collected from 2613 children hospitalized for acute lower respiratory tract infection at Hunan provincial People's Hospital, China, during fixed 2 days of each week between September 2007 and February 2011. All enrolled hospitalized patients were 14 years of age or younger and were admitted for acute lower respiratory tract infection (pneumonia, bronchitis, bronchiolitis, and asthma complicate pulmonary infection). Acute lower respiratory tract infection was diagnosed on the basis of clinical and radiologic findings. The enrolled children were consulting with an illness where an acute or worsened cough was the main or dominant symptom, or had a clinical presentation that suggested a lower respiratory tract infection, with a duration of ≤ 28 days. All nasopharyngeal aspirates were collected within 1–3 days of admission. Demographic data and details of the clinical findings were recorded from case history. Informed consent was obtained from the parents of all children who provided specimens. The study protocol was approved by the hospital ethics committee.
Collection and Processing of Nasopharyngeal Aspirates Samples
All nasopharyngeal aspirates were collected and transported immediately to the laboratory at the National Institute for Viral Disease Control and Prevention, China CDC, and stored at –80°C until required for further analysis. Viral DNA and RNA were extracted from 140 µl of each nasopharyngeal aspirates specimen using the QIAamp viral DNA and the QIAamp viral RNA Mini Kits (Qiagen, Shanghai, China) according to the manufacturer's instructions. cDNA was synthesized using random hexamer primers with the Superscript II RH– reverse transcriptase (Invitrogen, Carlsbad, CA).
Detection of hMPV
Screening for hMPV was conducted using traditional PCR methods. hMPV forward (5′‐CCC TTT GTT TCA GGC CAA‐3′) and reverse (5′‐GCA GCT TCA ACA GTA GCT G‐3′) primers, which target the M gene and generate a 416‐bp product, were used as described previously [Bellau‐Pujol et al., 2005]. All PCR products were purified using the QIAquick PCR purification kit (Qiagen). The reaction mix contained 10 pmol of each primer and 1.25 units of EXTaq DNA polymerase (Takara Bio Inc., Tokyo, Japan). Reactions were incubated at 94°C for 8 min, followed by 35 cycles at 94°C for 30 s, 55°C for 30 s, and 72°C for 45 s, followed by a final extension at 72°C for 10 min.
Nucleotide Sequence Analysis
All PCR products were purified using the QIAquick PCR purification kit (Qiagen) and cloned into the pGEM‐T Easy Vector (Promega, Madison, WI) and then sequenced. Sequences were determined and analyzed using the DNASTAR software package. Phylogeneticanalysis was performed with Mega version 5.2 by using 1,000 bootstrapped replicates and the neighborjoining algorithm. The GenBank accession numbers of the previously published sequences are as follows: CAN98‐74(AY145258), CAN98‐75(AY145259), CAN99‐81(AY145264), CAN97‐82(AY145265), CAN00‐12(AY145267), CAN00‐14(AY145269), CAN00‐16(AY145271), BJ1887(DQ843659) and JPS03‐240(AY530095).
Screening for Other Respiratory Viruses
A standard reverse transcription‐PCR technique was used to screen for respiratory syncytial virus (RSV), human rhinovirus (HRV), influenza viruses (IFVA, IFVB), parainfluenza viruses (PIV types 1–3), human coronaviruses HKU1 (HCoV‐HKU1), and NL63 (HCoV‐NL63), and PCR for adenovirus (ADV) and human bocavirus (HBOV) [Hierholzer et al., 1993; Bellau‐Pujol et al., 2005].
Statistical Analysis
Inferential statistics performed included non‐parametric Mann‐Whitney U test for continuous variables, with data reported as medians and 25th and 75th interquartile ranges (IQR). Categorical variables were compared using the chi‐square test or Fisher's exact test. Multivariate logistic regression analysis was used to identify independent risk factors. Variables with a plausible relationship with P values less than 0.2 in the univariate analysis were entered into the multivariate analyses. Results are summarized as odds ratios (OR) and 95% confidence intervals (CI). All analyses were performed using SPSS version 20.0 software (SPSS Inc., Chicago, IL). P < 0.05 was considered statistically significant.
RESULTS
Patient Characteristics
In total, 2613 patients were included, and about 100 cases were excluded for samples missing when those were transporting from Changsha to Beijing (March–June 2009). The enrolled 2613 study samples represented 23.03% of 11348 admissions for acute lower respiratory tract infection to Hunan provincial People's Hospital, China. The ages of enrolled children ranged from 1 day to 166 months, with a median age of 11 (IQR 5–24) months. The majority of the specimens (2231/2613, 85.4%) were collected from patients under 36 months old (Figure 1), and the male to female ratio was 1.86:1 (1698:915).
Figure 1.
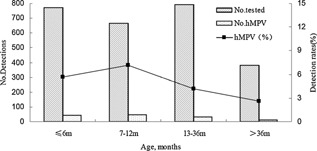
Age distribution of hMPV in children with acute lower respiratory tract infections during 3.5‐years study period.
Detection of hMPV and Other Viral Agents
Of 2613 respiratory specimens tested in the study period, at least one respiratory virus was detected in 1629 (62.34%). The most commonly detected virus was RSV (503, 19.25%), followed by HRV (475, 18.18%), PIV‐3 (317, 12.13%), HBOV (224, 8.57%), ADV (153, 5.86%), hMPV (135, 5.2%), IFVB (62, 2.37%), IFVA (51, 1.95%), HCoV‐HKU1 (20, 0.77%), PIV‐1 (15, 0.57%), PIV‐2 (11, 0.42%), HCoV‐NL63 (11, 0.42%). Sixty‐two of 135 (45.9%) hMPV‐positive samples were found to be coinfected with other respiratory viruses, including twenty‐two with HBOV, seventeen with RSV, fifteen with HRV, thirteen with PIV3, six with ADV, three with IFVB, two with PIV4, two with HCoV‐HKU1, and one with HCoV‐NL63. HBOV was the most common coinfecting virus, accounting for 22/62 (35.5%) coinfections. Among the patients with co‐detections, 36.3% (49/135) were positive for hMPV plus one additional viral agent and 9.6% (13/135) were positive for hMPV plus two or three additional viruses; of these 9.6%, seven patients were positive for three and six patients were positive for four viral agents. The epidemiological and clinical characteristics of patients infected with hMPV single infection and patients with hMPV co‐detections were similar. However, rhonchus were more common in patients with hMPV co‐detections (χ2 = 8.427, P = 0.004) (Table I). The univariates of cyanosis, crackles, and rhonchus were entered into the multivariate analyses, and rhonchus was associated with hMPV co‐infection (OR = 3.057; 95%CI 1.463–6.385; P = 0.003).
Table I.
Clinical Characteristics of Patients Infected With Human Metapneumovirus (hmpv) From September 2007 Through August 2008
| hMPV‐A (n = 98) | hMPV‐B (n = 37) | P‐value | single hMPV‐A (n = 51) | single hMPV‐B (n = 22) | P‐value | single infection hMPV (n = 73) | co‐infection hMPV (n = 62) | P‐value | |
|---|---|---|---|---|---|---|---|---|---|
| No. (%) | No. (%) | No. (%) | No. (%) | No. (%) | No. (%) | ||||
| Male gender | 75(76.5) | 28(75.7) | 0.917 | 39(76.5) | 18(81.8) | 0.762 a | 57(78.1) | 46(74.2) | 0.596 |
| Age, months, median(IQR) b | 10 (6–23.25) | 10 (5.5–15.5) | 0.501 | 11 (6–24) | 7.5 (5.0–11.25) | 0.098 | 10 (6–20) | 8 (5–16) | 0.599 |
| Age in months | 0.598 | 0.349 a | 0.958 | ||||||
| ≤6 | 32(32.7) | 12(32.4) | 16(31.4) | 8(36.4) | 24(32.9) | 20(32.3) | |||
| 6.1 to≤12 | 33(33.7) | 15(40.5) | 15(29.4) | 10(45.5) | 25(34.2) | 23(37.1) | |||
| 12.1 to≤36 | 24(24.5) | 9(24.3) | 16(31.4) | 3(13.6) | 19(26.0) | 14(22.6) | |||
| >36 | 9(9.2) | 1(2.7) | 4(7.8) | 1(4.5) | 5(6.8) | 5(8.1) | |||
| LOS, days, median (IQR) b | 7 (6–10) | 8 (7–9) | 0.322 | 7 (6–9) | 8 (6.75–9) | 0.296 | 7 (6–9) | 8 (6–10) | 0.326 |
| Symptoms and signs | |||||||||
| Fever | 50(51.0) | 20(54.1) | 0.753 | 31(60.8) | 9(40.9) | 0.084 | 40(54.8) | 30(48.4) | 0.458 |
| Cough2 | 95(96.9) | 36(97.3) | 1 | 49(96.1) | 21(95.5) | 1 | 70(95.9) | 61(98.4) | 0.624 a |
| Wheezing | 54(55.1) | 25(67.6) | 0.190 | 28(54.9) | 16(72.7) | 0.153 | 44(60.3) | 35(56.5) | 0.653 |
| Shortness of breath | 11(11.2) | 5(13.5) | 0.767 a | 7(13.7) | 4(18.2) | 0.724 a | 11(15.1) | 5(8.1) | 0.210 |
| Cyanosis | 2(2.0) | 3(8.1) | 0.126 a | 2(3.9) | 3(13.6) | 0.157 a | 5(6.8) | 0 | 0.062 a |
| Crackles | 70(71.4) | 29(78.4) | 0.415 | 39(76.5) | 18(81.8) | 0.762 a | 57(78.1) | 42(67.7) | 0.176 |
| Rhonchus | 45(45.9) | 11(29.7) | 0.089 | 19(37.3) | 3(13.6) | 0.044 | 22(30.1) | 34(54.8) | 0.004 |
| Diarrhea2 | 10(10.2) | 9(24.3) | 0.035 | 6(11.8) | 5(22.7) | 0.289 a | 11(15.1) | 8(12.9) | 0.718 |
| supplemental oxygen | 8(8.2) | 7(18.9) | 0.120 a | 4(7.8) | 3(13.6) | 0.424 | 7(9.6) | 8(12.9) | 0.541 |
| Coinfection | 47(48.0) | 15(40.5) | 0.440 | / | / | / | / | / | / |
| Underlying disease | 22(22.4) | 13(35.1) | 0.134 | 11(21.6) | 5(22.7) | 1 | 16(21.9) | 19(30.6) | 0.249 |
| Seasonal distribute | 0.366 a | 0.570 a | 0.368 | ||||||
| Spring | 49(50.0) | 16(43.2) | 30(58.8) | 10(45.5) | 40(54.8) | 25(40.3) | |||
| Summer | 8(8.2) | 6(16.2) | 3(5.9) | 3(13.6) | 6(8.2) | 8(12.9) | |||
| Autumn | 8(8.2) | 5(13.5) | 5(9.8) | 2(9.1) | 7(9.6) | 6(9.7) | |||
| Winter | 33(33.7) | 10(27.0) | 13(25.5) | 7(31.8) | 20(27.4) | 23(37.1) |
Fisher's exact test,
Mann‐Whitney U test, all the other: Chi‐square test.
hMPV‐A indicate type A hMPV, hMPV‐B indicate type B hMPV, single hMPV‐A indicate single type A hMPV infection, single hMPV‐B indicate single type B hMPV infection.
Seasonal distribution: Spring (March‐May), Summer (June‐August), Autumn (September‐ November), Winter (December‐February).
IQR, interquartile range; LOS, length of hospital stay.
Epidemiology of hMPV
Approximately 6.1% (103/1698) of the males and 3.5% (32/915) of the females tested positive for hMPV in this study. The male‐to‐female ratio in the hMPV‐infected group was 3.2:1 and differed significantly between the patients with and without hMPV infections (χ2 = 8.007, P = 0.005). The age of patients infected with hMPV varied from 20 days to 12 years of age (median (IQR), 10 (6–16) months), and 68.1% (92/135) were ≤ 12 months of age. Children 7–12 months of age exhibited the highest infection rate (7.2%). The hMPV infection rates among four age groups differed significantly (χ2 = 12.808, P = 0.005; Fig. 1). hMPV infections occurred most frequently in spring. The positive rate of hMPV in spring reached 10.7% (65/607). The seasonal distribution exhibited statistically significant differences in the positive rates of hMPV among four different seasons (χ2 = 57.483, P = 0.000; Fig. 2).
Figure 2.
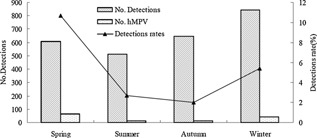
Seasonal distribution of hMPV infections in children with acute lower respiratory tract infections, September 2007–February 2011.
Clinical Characteristics of hMPV in Children
Among the 135 pediatric patients with acute lower respiratory tract infections who tested positive for hMPV, the majority had developed a cough (131 patients, 97.0%), 70 experienced a fever (51.9%), 79 suffered from wheezing (58.5%), 99 (73.3%) developed moist rales, and rhonchi were audible in 56 patients (41.5%). In addition, 16 patients experienced shortness of breath (11.9%), 5 developed cyanosis (3.7%), 19 had diarrhea (14.1%), and 15 required supplemental oxygen (11.1%). Thirty‐five patients developed underlying disorders or complications (25.9%), including 6 patients had thrush, 6 patients had granulocytopenia, 3 children had congenital heart defects, 3 children had cytomegao‐ virus infection, and some other diseases or complications. All hospitalized patients exhibited good prognoses and the median length of hospital stay was 7 (IQR 6–9) days.
Phylogenetic Analysis of hMPV
The sequence of positive products and standard sequences from GenBank exhibited high homology (96%–100%). Phylogenetic analyses indicated that the 135 hMPV specimens were classified into the two main genetic lineages, A and B. Ninety‐eight (98/135, 72.6%) hMPV strains were subgroup A2b, thirty‐six (26.6%) strains were subgroup B1, one was subgroup B2, and none were either subgroup A1 and A2a. The nucleotide and deduced amino acid sequences of the M gene of 135 hMPV specimens were compared with those of hMPV strains available at the GenBank site (Fig. 3).
Figure 3.
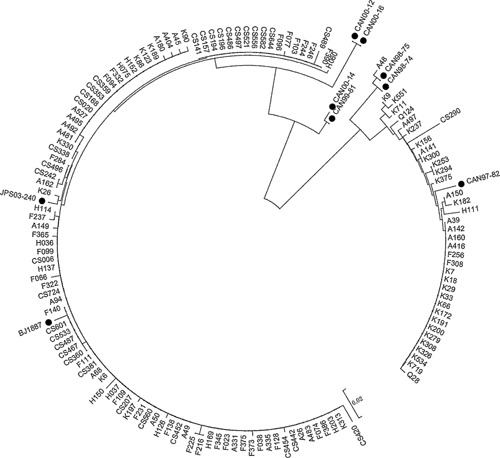
Phylogenetic analysis of nucleotide sequences from the partial M gene of the 135 hMPV strains from nasopharyngeal aspirates. Phylogenetic trees were constructed by the neighbor‐joining method using MEGA5.2. The viral sequences in black solid rhombus were generated from GenBank (GenBank accession no. CAN98‐74 (AY145258), CAN98‐75 (AY145259), CAN99‐81 (AY145264), CAN97‐82 (AY145265), CAN00‐ 12 (AY145267), CAN00‐14 (AY145269), CAN00‐16 (AY145271), BJ1887 (DQ843659) and JPS03‐240 (AY530095)); other reference sequences were obtained from the present study. Genotypes of the previously published hMPV sequences: CAN00‐12 and CAN00‐16 were subgroup A2a; BJ1887 and JPS03‐ 240 were subgroup A2b; CAN00‐14 and CAN99‐81 were subgroup A1; CAN97‐82 was subgroup B1; CAN98‐ 74 and CAN98‐75 were subgroup B2.
Epidemiological and Clinical Characteristics of hMPV Genotypes A and B
The epidemiological and clinical data of 135 pediatric patients infected with type A (98 cases) or type B hMPV (37 cases) were summarized and analyzed. The results of the distribution of epidemic seasons indicated that hMPV genotypes A and B prevailed alternately in different years. From the autumn of 2007 to the summer of 2008, hMPV genotype A2b was the predominant epidemic strain. From the autumn of 2008 to the winter of 2009, the predominant genotype of hMPV changed to combined A2b and B1 genotype. From the spring to the winter of 2010, hMPV genotype B1 became the predominant epidemic strain (Fig. 4). Comparison studies revealed that the epidemiological data and clinical characteristics were largely similar between the patients infected with genotype A hMPV and patients infected with genotype B hMPV. However, patients infected with genotype B hMPV developed diarrhea more frequently compared with pediatric patients infected with genotype A hMPV (χ2 = 4.328, P = 0.035). Excluding the factor of mixed infection, the epidemiological and clinical characteristics of patients infected with type A hMPV alone and patients infected with type B hMPV alone were also basically similar. However, rhonchus were more common in patients with type A hMPV infection (χ2 = 4.072, P = 0.044). The univariates of fever, wheezing, cyanosis, rhonchus were entered into the multivariate analyses, and rhonchus was associated with type A hMPV infection (OR = 0.164; 95%CI 0.036–0.751; P = 0.020).
Figure 4.
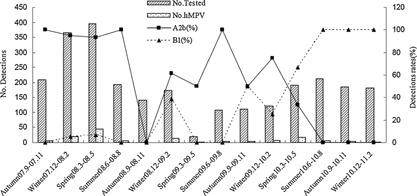
Seasonal distribution of hMPV genetypes from September 2007 to February 2011.
DISCUSSION
The present study summarized the prevalence and clinical features of hMPV infections in Hunan Province between September 2007 and February 2011, including the 76 hMPV cases (Sep. 2007–Aug. 2008) reported in the preliminary study [Xiao et al., 2013]. The present study revealed that the hMPV positive rate was 5.2%, in hospitalized children with acute lower respiratory tract infections in Hunan Province, that was similar to Beijing [Zhu et al., 2011; Lu et al., 2013], higher than Southern China (2.6%) [Cai et al., 2014], and lower than Chongqing (10.2%) [Zhang et al., 2012] and Taiwan (23%) [Wei et al., 2013]. Compared with studies in other countries, that was higher than the reported rate in Japan (2.5%) [Matsuzaki et al., 2008], lower than the reported rate in Germany (11.9%) [Reiche et al., 2014], and similar to the reported rates in the United States [van den Hoogen et al., 2003; Edwards et al., 2013] and South Korea [Kim et al., 2012] (6% and 7.1%, respectively). The variation in hMPV detection rates between various regions may be related to the differences in climate, geographic locations, year and human populations.
The present study found that the hMPV positive rate was higher in male pediatric patients, which is consistent with the most of the previous studies [Peiris et al., 2003; van den Hoogen et al., 2003; Wei et al., 2013], but different from a study in the United States which found hMPV infections were equally distributed between males and females [McAdam et al., 2004]. Furthermore most previous studies have indicated that the majority of hMPV‐positive patients are children of young ages [McAdam et al., 2004; Vicente et al., 2006; Lu et al., 2013; Wei et al., 2013; Reiche et al., 2014], which is similar to the findings of the present study. The peak of hMPV infections was found in the spring in the present study, which is similar to that of Beijing [Zhu et al., 2011], but different from that of Chongqing where the peak of hMPV epidemics are observed in the spring/summer and the winter/spring seasons [Zhang et al., 2012]. In the United States, hMPV infections have been reported to occur predominately in the winter and spring seasons [McAdam et al., 2004]. These unique characteristics of hMPV infections may be related to the geographic region and climate, and requires the further study for verification.
In the present study the most common clinical symptoms of hMPV‐positive patients included coughing, wheezing and fever, which in some pediatric patients, were accompanied by shortness of breath, cyanosis and moist rales heard in pulmonary auscultation, whcih were similar to those reported previously [Zhang et al., 2012; Lu et al., 2013; Wei et al., 2013]. The present study revealed a rather high mixed infection of hMPV with other viruses (45.9%) and HBOV was the most frequently detected virus with hMPV. The rates of hMPV coinfection of previous studies range from approximately 9.4% to 70.8% [Zhu et al., 2011; Fathima et al., 2012; Kim et al., 2012; Zhang et al., 2012; Lu et al., 2013; Wei et al., 2013; Cai et al., 2014], and the commonly co‐detected viruses include RSV, HRV and PIV‐3. These discrepancies are most likely due to the differences in the types of viruses examined and the test methods employed in the studies. However, the discrepancies may also be related to the selection of research objects, geographic regions and research periods. Previous studies have indicated that mixed infection may aggravate the diseases and affect prognosis [Greensill et al., 2003]. In the present study, lung auscultation did reveal that rhonchi were more common in patients with mixed hMPV infection compared with patients with single hMPV infection. However, no other significant differences were found between the patients, including the requirement of supplemental oxygen and duration of hospitalization.
Consistent with the findings reported by Xiao et al [2013], the present study revealed that both hMPV genotype A and genotype B were prevalent in the Changsha area from September 2007 to February 2011. hMPV genotypes A2 and B1 were the most predominant genotypes. The annual distribution indicated that genotype A2 and genotype B1 prevailed alternately. Previous studies have reported the prevalence of certain predominant hMPV genotypes in a given year. hMPV sub‐genotypes may vary in different years and alternative predominance of hMPV sub‐genotypes has been observed [Matsuzaki et al., 2008; Pitoiset et al., 2010; Kim et al., 2012; Lu et al., 2013; Wei et al., 2013]. The present study found that the epidemiological characteristics and clinical features were fundamentally similar among patients infected with different genotypes of hMPV, which is consistent with the results of some studies [Bosis et al., 2008; Wei et al., 2013]. Diarrhea was a clinical manifestation that occurred more frequently in patients infected with type B hMPV compared with patients infected with type A hMPV in this study. However, there are different epidemiological and clinical findings in other two studies [Matsuzaki et al., 2008; Kim et al., 2012]. Furthermore, a study indicated that patients with type A hMPV infection are often associated with more severe symptoms [Vicente et al., 2006]. However, there is no apparent link between the genotype of hMPV and the severity factors of the disease in other studies [Agapov et al., 2006; Pitoiset et al., 2010]. Certainly, more data are needed to elucidate this issue.
In conclusion, the present study explores the epidemiological and clinical manifestations of infections with various genotypes of hMPV and especially the predominant prevalence of hMPV genotypes in different years. However, retrospective analysis was conducted in the present study without control samples, and the sample sizes of certain sub‐genotypes of hMPV were small. These are the major limitations. And a long period of data accumulation with a larger sample size and control is required in the future study.
Conflict of interest: None.
REFERENCES
- Agapov E, Sumino KC, Gaudreault‐Keener M, Storch GA, Holtzman MJ. 2006. Genetic variability of human metapneumovirus infection: Evidence of a shift in viral genotype without a change in illness. J Infect Dis 193:396–403. [DOI] [PubMed] [Google Scholar]
- Apostoli P, Zicari S, Lo Presti A, Ciccozzi M, Ciotti M, Caruso A, Fiorentini S. 2012. Human metapneumovirus‐associated hospital admissions over five consecutive epidemic seasons: evidence for alternating circulation of different genotypes. J Med Virol 84:511–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellau‐Pujol S, Vabret A, Legrand L, Dina J, Gouarin S, Petitjean‐Lecherbonnier J, Pozzetto B, Ginevra C, Freymuth F. 2005. Development of three multiplex RT‐PCR assays for the detection of 12 respiratory RNA viruses. J Virol Methods 126:53–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosis S, Esposito S, Osterhaus AD, Tremolati E, Begliatti E, Tagliabue C, Corti F, Principi N, Niesters HG. 2008. Association between high nasopharyngeal viral load and disease severity in children with human metapneumovirus infection. J Clin Virol 42:286–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai XY, Wang Q, Lin GY, Cai ZW, Lin CX, Chen PZ, Zhou XH, Xie JC, Lu XD. 2014. Respiratory virus infections among children in South China.J. Med Virol 86:1249–1255. [DOI] [PubMed] [Google Scholar]
- Edwards KM, Zhu Y, Griffin MR, Weinberg GA, Hall CB, Szilagyi PG, Staat MA, Iwane M, Prill MM, Williams JV. 2013. New Vaccine Surveillance Network. Burden of human metapneumovirus infection in young children. N Engl J Med 368:633–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fathima S, Lee BE, May‐Hadford J, Mukhi S, Drews SJ. 2012. Use of an innovative web‐based laboratory surveillance platform to analyze mixed infections between human metapneumovirus (hMPV) and other respiratory viruses circulating in Alberta (AB), Canada (2009–2012). Viruses 4:2754–2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuillet F, Lina B, Rosa‐Calatrava M, Boivin G. 2012. Ten years of human metapneumovirus research. J Clin Virol 53:97–105. [DOI] [PubMed] [Google Scholar]
- Greensill J, McNamara PS, Dove W, Flanagan B, Smyth RL, Hart CA. 2003. Human metapneumovirus in severe respiratory syncytial virus bronchiolitis. Emerg Infect Dis. 9:372–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hierholzer JC, Halonen PE, Dahlen PO, Bingham PG, McDonough MM. 1993. Detection of adenovirus in clinical specimens by polymerase chain reaction and liquid‐phase hybridization quantitated by time‐resolved fluorometry. J Clin Microbiol 31:1886–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huck B, Scharf G, Neumann‐Haefelin D, Puppe W, Weigl J, Falcone V. 2006. Novel human metapneumovirus sublineage. Emerg Infect Dis 12:147–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HR, Cho AR, Lee MK, Yun SW, Kim TH. 2012. Genotype variability and clinical features of human metapneumovirus isolated from korean children, 2007 to 2010. J Mol Diagn 14:61–64. [DOI] [PubMed] [Google Scholar]
- Lu G, Li J, Xie Z, Liu C, Guo L, Vernet G, Shen K, Wang J. 2013. Human metapneumovirus associated with community‐acquired pneumonia in children in Beijing. China. J Med Virol 85:138–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAdam AJ, Hasenbein ME, Feldman HA, Cole SE, Offermann JT, Riley AM, Lieu TA. 2004. Human metapneumovirus in children tested at a tertiary‐care hospital. J Infect Dis 190:20–26. [DOI] [PubMed] [Google Scholar]
- Peiris JS, Tang WH, Chan KH, Khong PL, Guan Y, Lau YL, Chiu SS. 2003. Children with respiratory disease associated with metapneumovirus in Hong Kong. Emerg Infect Dis 9:628–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitoiset C, Darniot M, Huet F, Aho SL, Pothier P, Manoha C. 2010. Human metapneumovirus genotypes and severity of disease in young children (n¼100) during a 7‐year study in dijon hospital, france. J Med Virol 82:1782–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiche J, Jacobsen S, Neubauer K, Hafemann S, Nitsche A, Milde J, Wolff T, Schweiger B. 2014. Human metapneumovirus: Insights from a ten‐year molecular and epidemiological analysis in Germany. PLoS One 9:E88342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Hoogen BG, de Jong JC, Groen J, Kuiken T, de Groot R, Fouchier RA, Osterhaus AD. 2001. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat Med 7:719–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Hoogen BG, van Doornum GJ, Fockens JC, Cornelissen JJ, Beyer WE, de Groot R, Osterhaus AD, Fouchier RA. 2003. Prevalence and clinical symptoms of human metapneumovirus infection in hospitalized patients. J Infect Dis 188:1571–1577. [DOI] [PubMed] [Google Scholar]
- van den Hoogen BG, Osterhaus DM, Fouchier RA. 2004. Clinical impact and diagnosis of human metapneumovirus infection. Pediatr Infect Dis J 23:S25–S32. [DOI] [PubMed] [Google Scholar]
- Vicente D, Montes M, Cilla G, Perez‐Yarza EG, Perez‐Trallero E. 2006. Differences in clinical severity between genotype a and genotype b human metapneumovirus infection in children. Clin Infect Dis 42:111–113. [DOI] [PubMed] [Google Scholar]
- Wei HY, Tsao KC, Huang CG, Huang YC, Lin TY. 2013. Clinical features of different genotypes/genogroups of human metapneumovirus in hospitalized children. J Microbiol Immunol Infect 46:352–357. [DOI] [PubMed] [Google Scholar]
- Xiao NG, Zhang B, Xie ZP, Zhou QH, Zhang RF, Zhong LL, Ding XF, Li J, Song JR, Gao HC, Hou YD, Duan ZJ. 2013. Prevalence of human metapneumovirus in children with acute lower respiratory infection in Changsha, China. J Med Virol 85:546–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoko Matsuzaki, Tsutomu Itagaki, Chieko Abiko, Yoko Aoki, Asuka Suto, Katsumi Mizuta. 2008. Clinical impact of human metapneumovirus genotypes and genotype‐specific seroprevalence in yamagata, Japan. J Med Virol 80:1084–1089. [DOI] [PubMed] [Google Scholar]
- Zhang C, Du LN, Zhang ZY, Qin X, Yang X, Liu P, Chen X, Zhao Y, Liu EM, Zhao XD. 2012. Detection and genetic diversity of human metapneumovirus in hospitalized children with acute respiratory infections in Southwest China. J Clin Microbiol 50:2714–2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu RN, Qian Y, Zhao LQ, Deng J, Sun Y, Wang F, Liao B, Li Y, Huang RY. 2011. Characterization of human metapneumovirus from pediatric patients with acute respiratory infections in a 4‐year period in Beijing. China. Chin Med J (Engl) 124:1623–1628. [PubMed] [Google Scholar]


