Abstract
Background
Delivery of genes to airway mucosa would be a very valuable method for gene therapy and vaccination. However, there have been few reports on suitable gene delivery systems for administration. In this study, we use a cationic emulsion system, which is physically stable and facilitates the transfer of genes in the presence of up to 90% serum, as a mucosal gene carrier.
Methods and results
Cationic lipid emulsion was formulated with squalene and 1,2‐dioleoyl‐sn‐glycero‐3‐trimethylammoniumpropane (DOTAP) as major components. Emulsions formed stable complexes with DNA and protected and transferred DNA to target cells against DNase I digestion in the presence of mucosal destabilizers such as heparin sulfate (a polysaccharide of the glycosaminoglycan family in mucosa) and Newfectan (a natural lung extract of bovine) in an in vitro system. In contrast, commercial liposomes and counter liposomes, made with an identical lipid composition of emulsions, failed. After in vivo intranasal instillation, the cationic emulsion showed at least 200 times better transfection activity than the liposomal carriers in both nasal tissue and lung.
Conclusions
These findings show that cationic emulsions can mediate gene transfection into airway epithelium, making it a good choice for transferring therapeutic genes and for genetic vaccination against an pathogenic infection via an airway route. Copyright © 2005 John Wiley & Sons, Ltd.
Keywords: transfection, emulsion, liposome, heparin, lung surfactant, nasal mucosa
Introduction
Mucosal gene transfer with the aid of viral or non‐viral vectors is of great interest in the fields of gene therapy and DNA vaccine development 1, 2, 3. Since the early days of gene therapy, delivery of one therapeutic gene, cystic fibrosis transmembrane conductance regulator (CFTR) cDNA, to airway epithelia has been a long‐lasting attempt to correct the defects associated with cystic fibrosis. To induce mucosal as well as systemic immunities, this site also presents an attractive, non‐invasive target for the delivery of DNA vaccines against mucosal pathogens, especially infectious viruses such as severe acute respiratory syndrome coronavirus (SARS Co‐V), human papilloma virus (HPV) and hepatitis B virus (HBV) 4, 5, 6, 7, 8, 9, 10, 11.
In the case of viral vectors, however, the host immune system generates a series of immune reactions against the vector that creates potential concerns, such as vector‐induced hypersensitivity and inflammation 1, 2. To overcome these limitations, many non‐viral vectors have been developed and used for in vivo gene delivery to the respiratory system. Cationic liposomes, one of the most potent non‐viral vectors, however, still have low gene transfer efficiency in the respiratory system 1, 2.
There are many barriers that prevent a successful gene transfer via the airway route 1, 2, 3, 12, 13, 14, 15, 16, 17, 18. The thick secretions of anionic mucus and glycosaminoglycan are likely to prevent access of carrier/DNA complexes to the epithelial cell surface, and may dilute or alter the composition of the complexes. There are other barriers including mechanical hindrance by cilia and the glycocalyx. In the presence of many mucosal barriers, liposome/DNA complexes can lose their physical integrity and can be destabilized by negatively charged materials or lung surfactants 15, 19. The latter causes DNA to dissociate from the complex. DNA released from the complex is vulnerable to nuclease attack. Because of these problems, the use of liposomes for in vivo mucosal gene transfer has been limited.
In a previous study, we developed a gene carrier in the form of an oil‐in‐water (o/w) type cationic lipid emulsion 15, 20, 21, 22, 23, 24. This cationic emulsion successfully delivered a reporter gene into cells in vitro in the presence of up to 90% serum. It is well known that a small amount of serum (∼10%) can dramatically reduce the transfection activity of liposome/DNA complexes because it contains anionic materials 25. Unlike liposomal carriers, the cationic emulsion that we developed retained its physical integrity at high concentrations of serum (up to 90%) when complexed with DNA. In vitro DNA release tests showed that the physical integrity of our emulsion/DNA complex is strong, and is maintained even in the presence of anionic polymers, such as poly‐l‐aspartic acid and heparin 15, 20.
In this study, we quantified the efficiency of cationic emulsions with different formulations in transferring genes to the airway cells after intranasal instillation. Transfection activity of emulsion in the nasal cavity and lung was at least 200 times higher than those of Lipofectamine, Lipofectin and DMRIE/c, which are commercially available liposomal carriers. We also investigated whether the cationic emulsion complexed with DNA maintained its integrity in the presence of mucosal destabilizers such as negatively charged heparin sulfate and a natural lung extract of bovine. In addition, the relationship between the transfection activity and the stability of the lipid carrier/DNA complexes in the presence of mucosal destabilizers was studied.
Materials and methods
Materials
Squalene, heparin sulfate and Tween 80 were purchased from Sigma Chemical Co. (St. Louis, MO, USA). DOTAP and 1,2‐dioleoyl‐sn‐glycero‐3‐phosphoethanolamine (DOPE) were purchased from Avanti Polar Lipids (Alabaster, AL, USA) and used without further purification. DMEM, MEM, RPMI 1640 and fetal bovine serum (FBS) were purchased from Gibco BRL/Life Technologies (New York, NY, USA). Newfectan was kindly supplied by Yuhan Pharmaceutical Company, Ltd. (Korea). Deionized‐distilled water was produced by using a Milli‐Q Plus water purification system (Millipore Corporation, Bedford, MA, USA). All other chemicals and reagents were of tissue‐culture grade.
Plasmid DNA
The plasmid pCMV‐Luc+ consists of the cytosolic form of Phontinus pyralis luciferase cDNA, which was obtained from the plasmid pGL3 (Promega, Madison, WI, USA) using XbaI L and HindIII and was subcloned into the plasmid pcDNA3.1 (Invitrogen) 21, 23. The plasmid was amplified in the E. coli DH5‐α strain and purified by using a Qiagen mega‐kit (Qiagen Inc., Chatsworth, CA, USA) according to the manufacturer's instructions. DNA purity was determined by agarose gel electrophoresis and by measuring optical density (OD). DNA having OD260/OD280 ≥1.8 was used in this study.
Preparation of lipid carriers
The emulsions contained 100 µl/ml of squalene to constitute the oil phase. Concentration and compositions of lipid emulsifiers were varied. Emulsions were prepared by following the procedure described previously 21. Briefly, lipid emulsifiers were weighed and dispersed in water. The emulsifier/water dispersion was sonicated until clear in an ice/water bath by using a probe type sonicator (High intensity ultrasonic processor, 600W model; Sonics and Materials, Danbury, CT, USA). The lipid solution was added to the oil and in turn sonicated in an ice/water bath for ca. 4 min to form emulsions. To prepare liposome carriers, the lipid solutions were further sonicated for ca. 4 min after the solution became clear. The prepared lipid carriers were kept at 4 °C for further experiments. The lipid compositions of prepared lipid carriers, which have been optimized for transfection in our previous study 21, 23, are summarized in Table 1. Throughout this paper, we will compare the transfection activity of emulsions with liposomes made with an identical lipid composition. We will refer to this liposome as ‘counterpart liposome’ in this paper. The counterpart liposome is defined as the liposome that has identical lipid compositions to the emulsion, but only lacking the oil component. The following terms will be used in the paper. C/D: the weight ratio between the cationic lipid in the lipid formulation and DNA in the complex. C/Dmin: the minimum C/D ratio to form a carrier/DNA complex without free DNA on agarose gel retardation assay. C/Dopt: the C/D ratio that shows the highest in vitro transfection activity.
Table 1.
Mean size and polydispersity of lipid carriers with different lipid compositions
| Lipid carrier | Oil | Lipid formulation | Size (nm) | Poly‐dispersity | |
|---|---|---|---|---|---|
| Lipid composition | Conc. (mg/ml) | ||||
| LE1 | squalene | DOTAP | 24 | 143.5 | 0.154 |
| LE2 | squalene | DOTAP/Tween 80 | 20/4.5 | 154.7 | 0.163 |
| LE3 | squalene | DOTAP/DOPE | 20/4.0 | 164.5 | 0.156 |
| LE4 | squalene | DOTAP/DOPE/Tween 80 | 20/4/4.5 | 156.4 | 0.145 |
| LP1 | — | DOTAP | 24 | 115.2 | 0.258 |
| LP2 | — | DOTAP/Tween 80 | 20/4.5 | 145.6 | 0.234 |
| LP3 | — | DOTAP/DOPE | 20/4.0 | 141.5 | 0.312 |
| LP4 | — | DOTAP/DOPE/Tween 80 | 20/4/4.5 | 120.4 | 0.239 |
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
Size of lipid carriers and DNA/lipid carrier complexes
The average particle size and the polydispersity of the lipid carriers (liposome and emulsions) or the carrier/DNA complexes were measured by using photon correlation spectroscopy (Malvern Zetasizer; Malvern Instruments Ltd., UK) as described previously 20, 21. The emulsion and liposomes were diluted 300 and 3 times with water, respectively, for the measurements. The mixtures containing 5 µg of plasmid DNA and an appropriate amount of the carrier were used to measure the size of the emulsion/DNA complexes in DMEM.
Plasmid DNA release from the lipid/DNA complexes by adding heparin sulfate or lung surfactant
DOTAP/DOPE emulsion (LE3, see Table 1 for the notation)/pCMV‐Luc+ and DOTAP/DOPE liposome (LP3)/pCMV‐Luc+ complexes were prepared by mixing 1 µg of DNA with 4 µg of the lipid carriers in 10 µl of DMEM. The complexes were incubated for 10 min at room temperature. Heparin sulfate corresponding to 50 equivalency of DNA and 5 µg of Newfectan were added to the complex solutions, where equivalency is defined as the negative charge ratio between heparin and DNA at pH 7.4. After incubation at room temperature for 10 min, the samples were analyzed by gel electrophoresis in a 1% agarose gel including 0.5 µg/ml ethidium bromide. To test the protection of DNA in the complexes against nuclease digestion, plasmid DNA, emulsion/DNA complex and Lipofectamine/DNA complex were incubated with or without the destabilizers in the presence of 0.5 U/ml of DNase I for 10 min at 37 °C. DNA from the samples was extracted twice with phenol/chloroform/isoamyl alcohol (50 : 49 : 1 by volume) prior to gel electrophoresis to remove lipids and oil.
In vitro transfection assay
A human squamous cell carcinoma, RPMI 2650, and a human large cell lung carcinoma, H1299, were cultured in MEM and RPMI 1640, respectively, supplemented with 10% FBS at 37 °C in a humidified 5% carbon dioxide incubator. RPMI 2650 cells have never shown cell polarization under our cultural conditions. Cells were seeded at 2 × 104 cells per well onto 96‐well plates 12 h before transfection. Cells were ca. 70–80% confluent at the time of transfection. pCMV‐Luc+ (500 ng) was complexed with an appropriate amount of the carrier in 40 µl of corresponding serum‐free media. In turn, 160 µl serum‐free media were added. To test the effect of mucosal destabilizers on the transfection efficiency of lipid formulations, 25 equivalency of heparin sulfate or 2.5 µg of Newfectan was added to 160 µl media after washing the cells with serum‐free media. To test the protection of DNA in the complexes against nuclease digestion in the presence of heparin and lung surfactant, 0.5 U/ml of DNase I was added to media. After 1 h incubation, the cells were washed with serum‐free media to remove the remaining carrier/DNA complexes. The cells were fed again with media containing 10% (v/v) FBS and cultured for 24 h after transfection. To evaluate the expression of luciferase, the transfected cells were lysed by adding 100 µl of lysis buffer (0.1 M Tris‐HCl, 2 mM EDTA, and 0.1% Triton X‐100, pH 7.8) per well. After one freeze/thaw cycle, 10 µl of the supernatants were assayed for luciferase activity using a kit purchased from Promega with a luminometer (Turner Designs luminometer model TD‐20/20, Promega). Peak light emission was measured for 20 s at room temperature. The luciferase content was calculated from relative light units (RLU) using a standard curve obtained from the purified firefly luciferase (Sigma). The formula for the conversion of RLU to picograms was 619.3 × RLU − 0.8254. Luciferase values were expressed as nanogram per well. Background of luciferase activity in each organ was measured from the lysates of untreated cells and was negligible (RLU <0.005).
In vivo gene transfer and luciferase assay
To prepare DNA/carrier complexes, DNA solution containing 6.7–40 µg of pCMV‐Luc+ and the carrier solution each diluted with DMEM were mixed by inversion. The amount of the carrier used to form the complex was set at C/Dopt. The complex solution was allowed to incubate at room temperature for 10 min and was administrated into female Balb/c mice (6–8 week old) by intranasal instillation under general anesthesia using Ketamine. Twenty‐four hours later, the mice were sacrificed, and nose and organs were removed and homogenized in lysis buffer. The volume‐to‐weight ratio of lysis buffer was 5 µl/mg for each collected organ. After two freeze/thaw cycles, the homogenized organ extracts were centrifuged at 4 °C for 10 min at 12 000 rpm. A portion of the supernatants was assayed for protein concentration by using a Bio‐Rad protein assay kit (Bio‐Rad Laboratories, Hercules, CA, USA) based on the Bradford method 26. Luciferase activity was expressed as picograms per milligram of tissue protein. Background of luciferase activity in each organ was measured from the organs of untreated mice and was negligible.
Immunohistochemistry
Mice (n = 3) were anesthetized with ether, and then perfused with chilled phosphate‐buffered saline (PBS) and 4% paraformaldehyde in PBS. After perfusion, the nose of the mouse and the lung were cut off. Before freezing nasal samples, the isolated nasal fragments were further incubated in neutral buffered EDTA for 10 days to decalcificate the nasal bone. The isolated lung and the nose were mounted in OCT freezing compound (Miles, Tarrytown, NY, USA) and frozen in a deep freezer at −70 °C. The tissues were cut into 6 µm serial cryosections. More than 100 sections per group were selected for immunostaining to detect the expression of luciferase. Sections were washed twice with PBS containing 0.05% Tween 20 (PBST) and blocked with 5% skim milk in PBST for 1 h at room temperature. Sections were then washed and incubated for 1 h at room temperature with a goat polyclonal anti‐luciferase antibody (Ab) used in 1 : 100 dilution (Promega). Following additional washes, donkey anti‐goat IgG conjugated with peroxidase (Promega) was applied for 1 h. Ab binding was detected by adding 3,3′‐diaminobenzidine tetrahydrochloride (DAB, Dako, Carpenteria, CA, USA). Hematoxylin was used for counterstaining. No staining was detected in sequential tissue sections treated with a normal goat serum, demonstrating negligible background level of this experiment. Cells having higher than three‐fold above the background were scored as positive.
Results
Resistance of the emulsion/DNA complex against mucosal destabilizers
When the complex moves along the intranasal tract, the barriers that the emulsion/DNA complex faces include mucosal destabilizers such as negative charged polymers and nucleases. Therefore, the stability of emulsion/DNA complexes was monitored by observing a competitive exchange between the mucosal destabilizers and DNA in the emulsion/DNA complexes. Heparin sulfate, a polysaccharide of the glycosaminoglycan family, or Newfectan, a naturally derived pulmonary surfactant preparation obtained from bovine lung homogenate, was selected as a nasal or a lung destabilizer, respectively. The study was performed with DOTAP emulsion (LE1, see Table 1) at C/Dmin = 4. DOTAP liposome (LP1) was used as a control. The complexes were incubated for 10 min at room temperature after adding heparin or Newfectan, and then applied to an agarose gel for electrophoresis (Figure 1A). DNA complexed with LP1 was completely released after the addition of heparin (lane 4) or the lung surfactant (lane 5). In distinct contrast, DNA in the LE1/DNA complex was not released when both of the mucosal destabilizers (lanes 7 and 8) were present. The pale fluorescent band, however, was observed in the well occasionally by adding the lung surfactant into the LE1/DNA complex (lane 8).
Figure 1.
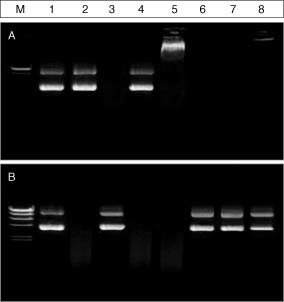
Gel electrophoresis of pCMV‐Luc+ after an exchange reaction of the plasmid DNA complexed with liposome or emulsion by heparin or lung surfactant in the absence (A) and presence of DNase I (B). Lipid carrier/DNA complexes were prepared by mixing pCMV‐Luc+ with LP1 (lanes 3–5) or LE1 (lanes 6–8) at C/D = 4 in 20 µl PBS. The formed complexes were incubated for 1 h at room temperature with 50 equivalency of heparin solutions (lanes 4 and 7) or 5 µg of lung surfactant (lanes 5 and 8). After the incubation, samples were analyzed by gel electrophoresis (A). The heparin or lung surfactant treated carrier/DNA complexes as well as naked DNA were incubated with 0.5 U/ml of DNase I for 10 min at 37 °C. DNAs in the complexes were extracted with phenol/chloroform/isoamyl alcohol (50 : 49 : 1 by volume) and were analyzed by gel electrophoresis. M: DNA molecular marker (λ Hind III); lane 1: 1 µg of DNA (no treatment); lane 2: 1 µg of DNA (treated with DNase I); lanes 3–5: LP1/DNA complexes; lanes 6–8: LE1/DNA complexes
We also examined whether the complexed DNA could be protected from enzymatic digestion by DNase I in the presence of the destabilizers. The naked DNA, liposome/DNA complex and emulsion/DNA complex were incubated first with heparin or Newfectan for 10 min, and then further incubated with 0.5 U/ml of DNase I for 10 min at 37 °C. DNA was retrieved and visualized on agarose gel (Figure 1B). Naked DNA was completely digested by DNase I (lane 2). DNA in the LP1 complex was protected from the enzymatic digestion under destabilizer‐free conditions (lane 3). The released DNAs from LP1/DNA complexes by adding heparin sulfate (lane 4) or Newfectan (lane 5), however, were degraded completely by enzymatic digestion. In contrast, DNAs in the emulsion/DNA complexes were not degraded in the presence of destabilizers, but a topological change occurred from super‐coiled to nicked DNA (lanes 7 and 8). These results indicate that DNA in the emulsion/DNA complex is more protected than that in the liposome/DNA complex against the destructive effects of mucosal destabilizers.
In vitro transfection activity of emulsion/DNA complexes in the presence of mucosal destabilizers
The activity of emulsion/DNA complexes can be diminished greatly in the airway tract due to mucosal destabilizers since they can dissociate DNA from the complex. Subsequently, the released DNA is vulnerable to nuclease attack. Therefore, the transfection activity of the emulsion/DNA complex against DNase I digestion was evaluated with or without destabilizers using a human nasal squamous cell carcinoma (RPMI 2650) and a human large cell lung carcinoma (H1299) to mimic the in vivo airway situations. Heparin sulfate and Newfectan were added as destabilizers to the RPMI 2650 and H1299, respectively. Before this experiment, the transfection efficiency was evaluated at various C/D ratios to find C/Dopt. C/Dopt values of LE1, LE3, LP1, and LP3 were 4. The C/Dopt values, however, increased by adding Tween 80 into the lipid formulations to 8 for DOTAP/Tween 80 emulsion (LE2) and DOTAP/DOPE/Tween 80 emulsion (LE4) and 6 for DOTAP/Tween 80 liposome (LP2) and DOTAP/DOPE/Tween 80 liposome (LP4), respectively 21, 23. The transfection activities of different formulations at C/Dopt are shown in Figure 2. Naked DNA was also used as a control. As shown in Figures 2A and 2B, although the expression level of luciferase between the cell types varied, the transfection efficiency of liposomes and emulsions increased by adding the other lipid components referred to as the helper lipids such as DOPE and Tween 80 23. Although DNase I alone could not significantly affect the transfection activity of the liposome/DNA complexes in both cell lines, the addition of heparin sulfate and a lung surfactant dramatically inhibited the transfection activity of the liposome/DNA complexes. This decrease was accelerated to an undetectable level by adding DNase I. The inhibition might be not due to the toxic effect of destabilizers, as judged by the number and morphology of living cells (data not shown). In the emulsion system, however, these destabilizers attenuated only slightly the transfection efficiency of the complexes. Over 40% of the original efficiency was maintained even in the mixed media containing heparin or a lung surfactant in the presence of DNase I in both cell systems.
Figure 2.
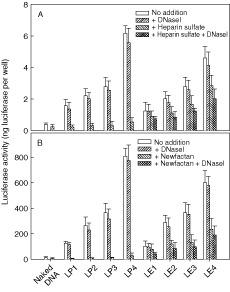
In vitro transfection efficiency of lipid formulations with different lipids in the presence of mucosal destabilizers for RPMI 2650 (A) and H1299 (B) cells. Emulsions and liposomes were complexed with 0.5 µg of pCMV‐Luc+ at C/Dopt, and then applied to cells in the presence of heparin sulfate (25 equivalency), lung surfactant (2.5 µg), or DNase I (0.5 U/ml). Naked DNA was used as a negative control. Twenty‐four hours after transfection, the cell lysates were prepared in lysis buffer and analyzed for luciferase activity by luminescence assay. Data are expressed as the mean ± SEM (n = 3) in terms of luciferase content (nanograms of luciferase/well)
Expression of luciferase in the nasal cavity and lung using different lipid carriers
We evaluated the efficiency of in vivo transgene expression after intranasal instillation of naked DNA or its complex with various lipid carriers. To prepare DNA/carrier complexes, 6.7 µg of pCMV‐Luc+ were complexed with different lipid carriers in 50 µl of DMEM at C/Dopt of each carrier. The transfection efficiencies for different carriers are shown in Figures 3B and 3C. Lipofectin, Lipofectamine and DMRIE/c, which are commercially available liposomal carriers, were also used for comparison. These liposomal carriers were complexed with DNA at the charge ratio, cationic lipid (+)/DNA (−), of 1.54, corresponding to C/D = 4 in LP3 (Figure 3A). Naked DNA was used as a control. Luciferase activity was determined at 24 h post‐administration in the nasal cavity and lung. The commercialized liposome/DNA complexes had higher transfection efficiency than naked DNA. Especially, Lipofectamine and DMRIE/c yielded the maximum expression in the nasal cavity and lung, respectively (Figures 3A and 3B). Compared with Lipofectamine and DMRIE/c, the emulsion system showed a robust luciferase expression in both nasal cavity and lung (Figure 3C). The highest transfection activity was obtained with LE4 for the nasal cavity, and LE3 for lung (Figure 3C). For counterpart liposomes, however, low luciferase activity was observed in both of the tissue lysates (<2 pg luciferase/mg tissue protein).
Figure 3.
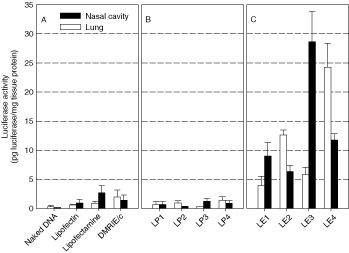
In vivo transfection efficiency of commercial liposomes (A) and the liposomes (B) and emulsions (C). pCMV‐Luc+ (6.7 µg) was complexed with various lipid formulations at C/Dopt. Naked DNA was used as a control. Twenty‐four hours after intranasal instillation, nasal and lung lysates were prepared in lysis buffer and analyzed for luciferase activity by luminescence assay. Data are expressed as the mean ± SEM (n = 3) in terms of luciferase concentration (picograms luciferase per milligram total protein)
Particle size of the DOTAP/DOPE emulsion/DNA complexes affects the distribution and the level of luciferase expression
We tried to explain the reason for differential expression levels in emulsion‐mediated transfection after intranasal instillation. It is well known that in vivo deposition of inhaled particles in the respiratory tract is determined mainly by the surface charge and particle size 27, 28. Therefore, in this study, we further investigated whether the particle size of the emulsion/DNA complex affects the differential expression of transgene in the nasal cavity and lung. To this end, LE3 was complexed at C/D = 4. The volume and DNA dose were changed for intranasal administration. By fixing the C/D value, the surface charge effect was excluded under this experimental condition. The average particle size of LE3/DNA complexes was measured by using photon correlation spectroscopy. The mean particle size of the complex was dependent on the volume of the complex solution (Figure 4A). As the volume of complex solution increased, the mean particle size decreased gradually; the expression of transgene increased in the lung, but decreased in the nasal cavity. In 25 µl, the expression level dramatically decreased in both of the tissues due to inactive large aggregates (>450 nm) between DNA and the emulsion during complexation.
Figure 4.
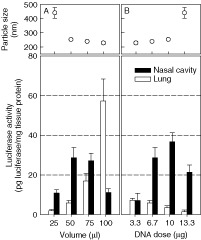
In vivo transfection efficiency of LE3/DNA complexes in the nasal cavity and lung as a function of volume (A) or DNA dose (B). Various amounts of pCMV‐Luc+ were complexed with LE3 at C/Dopt, and administered via nares. Twenty‐four hours after intranasal instillation, the luciferase activity in the lysates from the tissues was measured (n = 3)
The DNA dose was increased with the volume fixed (Figure 4B). Since the administered volume was fixed, the concentration became higher with an increase in DNA dose. As the DNA dose increased, the mean particle size increased, and the expression of transgene in the lung decreased gradually. It is worthy of note that the smallest DNA dose (3.3 µg in 50 µl) produced the maximum luciferase activity in lung lysates. In contrast, the luciferase activity in a mouse lung decreased when the largest DNA dose (13.3 µg in 50 µl) was administered. These results showed clearly that the particle size of the emulsion/DNA complexes affects the distribution and the level of luciferase expression via intranasal instillation.
DNA dose response for transgene expression in the nasal cavity and lung
To determine the optimal amount of DNA for transgene expression, dose‐response experiments were carried out. The complex was prepared by mixing 6.7 µg of pCMV‐Luc+ and LP3, LE3 or LE4 in 50 µl of DMEM at C/Dopt, as described in Figure 3, and administered topically to the nares of BALB/c mice. Naked DNA was also inhaled as a control. Luciferase activity was determined at 24 h post‐administration in the nasal cavity and lung (Figure 5). In the nasal cavity, the naked DNA and their lipid formulation complexes followed a dose‐dependent expression. At 40 µg of DNA, LE3 yielded more than 1000 times higher luciferase activity than naked DNA. In lungs of mice administered with 40 µg of DNA, the expression level of LE4/DNA increased more than 500‐fold than that of the corresponding naked DNA controls, which showed higher expression levels than the LP3/DNA complex. No signs of toxicity were noticed when the DNA dose was below 40 µg per mouse.
Figure 5.
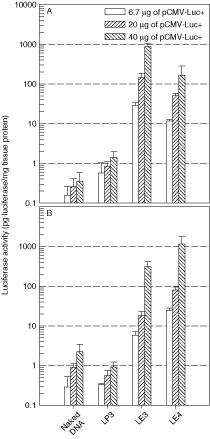
Dose dependence of gene expression in the nasal cavity (A) and lung (B) by intranasal instillation of naked DNA and their complexes with LP3, LE3, or LE4. Various amounts of pCMV‐Luc+ were complexed with LP3, LE3, or LE4 at C/Dopt, and administered via nares. Naked DNA was used as a control. Twenty‐four hours after intranasal instillation, the luciferase activity in the lysates from the tissues was measured (n = 3)
One of the main obstacles to the development of gene therapy for the airway is the inability of current viral vectors to direct sustained expression of a therapeutic transgene 14. Thus, the duration of protein expression after intranasal emulsion‐mediated transfection using the luciferase system was also evaluated. In all transfected groups, the highest level of expression was observed at day 1 in both the nasal cavity and lung. Luciferase expression persisted 24 days after transfection with DNA/emulsion complexes, although the degree of decline was steeper in lung than in the nasal cavity. About 1% of expression level by LE3/DNA and LE4/DNA complexes at day 1 remained in both tissues after 24 days (Figures 6A and 6C). In contrast, the transgene expression in the tissues of mice transfected by naked DNA and its complex with LP3 liposome almost disappeared in 24 days.
Figure 6.
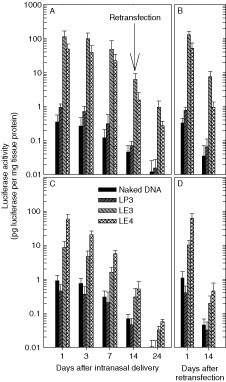
Time course of gene expression after the first and second intranasal instillation of naked DNA and their complexes with LP3, LE3, or LE4. Twenty micrograms of pCMV‐Luc+ were complexed with lipid carriers at each C/Dopt, and administered via nares. Naked DNA was used as a control. The luciferase activity in the lysates from the tissues was measured 24 days after transfection. Some of the mice were re‐transfected at day 14 by the same method as the initial administration. Each column represents the mean value from three BALB/c mice
For virus‐based vectors, there are many hurdles to be overcome for re‐administration 1, 2, 3, 12, 13. These include: (1) acute inflammation mediated by inflammatory cytokines; (2) induction of cytotoxic CD8 T cells leading to a decrease in persistent transgene expression; and (3) production of neutralizing antibodies against viral vectors that limit repeated viral vector administration. In contrast, non‐viral vectors are generally regarded as safer and less immunogenic alternatives to viral vectors. There, however, may be a potential concern of mucosal gene transfer using a cationic emulsion, induction of an immune response against the carrier/DNA complex resulting in lack of persistent transgene expression. To check this potential adversity, we re‐transfected respiratory tracts intranasally 14 days after the first transfection with DNA and the complexes (Figures 6B and 6D). One day after the re‐administration, a similar gene expression level to the level obtained after initial administration that was obtained at day 1 was observed in both tissues and the expression was maintained for 14 days. This result shows the possibility of repeat injections of the emulsion/DNA complexes.
Immunohistochemical analysis of gene expression in the nasal cavity and lung
To visualize the population and localization of transfected cells in the nasal cavity and lung, an immunohistochemical analysis was carried out after the intranasal instillation of emulsion/DNA complexes. Forty micrograms of pCMV‐Luc+ were complexed with LE3 or LE4 and administered to the nares as described in Figure 5 (Figure 7). The luciferase expression in the nasal cavity and lung was detected by immunoperoxidase staining. In situ immunostaining of nasal serial sections from the mice administered with LE3/DNA complexes was weak and rare in the epithelium on the nasal septum (Figure 7B). Although there were positive signals on monocyte‐like cells in nasal‐associated lymphoid tissue (NALT) (Figure 7D) at a high frequency (ca. 5–10%), dominant signals were observed in various sinuses (ca. 10–20%) (Figure 7F). In serial sections of the lung from the mice which were administered with LE4/pCMV‐Luc+ complexes, the staining of exogenous luciferase was frequently observed in various types of cells (>20%), such as pneumocytes and monocytes (Figure 7H). No staining was detected in sequential tissue sections treated with a normal goat serum, demonstrating that the signal was specific to luciferase (Figures 7A, 7C, 7E, and 7G). Similarly, no signal was detected from mice transfected with complexes containing the empty plasmid, pcDNA 3.1, demonstrating the specificity to luciferase (data not shown).
Figure 7.
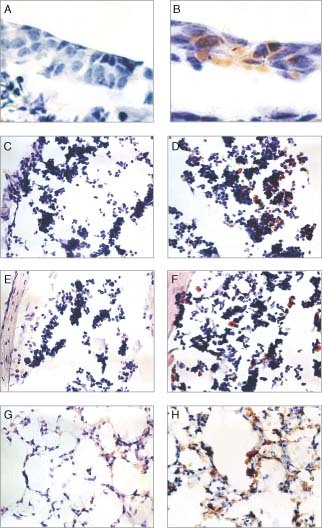
Immunohistochemical detection of luciferase in the nasal cavity and lung of BALB/c mice. Forty micrograms of pCMV‐Luc+ were complexed with LE3 (nasal cavity) and LE4 (lung), respectively, and in turn administered via intranasal instillation. The luciferase expression in the nasal cavity or lung was detected by immunoperoxidase staining. The sections shown in A, C, E, and G were treated with normal goat serum. All sections were counterstained with hematoxylin, and D was further stained with eosin. Magnification; A and B; ×1200, C–H; ×400
Discussion
In vivo transfection activity was evaluated for cationic emulsions with different lipid formulations after an intranasal instillation in mice. To our knowledge, this is one of the first successful demonstrations of a cationic o/w emulsion as an effective airway gene carrier in vivo. Notably, DOTAP became potent in delivering DNA to airway cells in vitro and in vivo after changing the formulation from liposome to emulsion.
The enhanced in vivo transfection activity with the emulsion in the nasal cavity and lung could be explained by a number of possibilities. One of these may be the difference of DNA degradation in lipid carrier/DNA complexes by mucosal secretions. The DNA in the complex must survive and deliver intact DNA to the target cells in the nasal mucosa. In mucosal secretions, there are many anionic materials that can compete with and substitute DNA in the complex. The released DNA is vulnerable to nuclease attack 15. Such a substitution has been assumed to be a major destabilization mechanism for the lipid/DNA complex 12, 15, 16. In addition to mucosal secretions, a recent report revealed that lung surfactants markedly inhibited cationic liposome‐mediated gene transfer 17, 18. The stability of the DNA/emulsion complexes was investigated by observing the competitive exchange between heparin or Newfectan and DNA in the complexes in the presence of DNase. For the counterpart liposome/DNA complex, the plasmid began to dissociate as the equivalency of heparin increased above unity. A similar pattern was observed with the commercialized liposome, Lipofectamine (data not shown). In distinct contrast, the emulsion did not release its plasmid in the presence of destabilizers. Thus, the complex between DNA and the emulsion is extremely stable and is resistant to the competitive exchange by an anionic proteoglycan. As a consequence, DNA complexed with emulsion could be protected from a DNase attack. From these observations, we conclude that the stable emulsion/DNA complex provided a necessary protection against inactivation by mucosal secretions and a higher accessibility of the active DNA to airway epithelia than the liposome/DNA complex.
It has not been reported clearly that the stability of complexes is relevant to transfection activity under in vivo conditions. Sternberg et al. have investigated the morphology and transfection activity both in vitro and in vivo 29. In that report, they were able to obtain stable complexes by substituting cholesterol for DOPE as a helper lipid, by adding PEG2000PE as steric stabilizer after the complex formation, and by pre‐condensing DNA with spermidine. These stability‐increased complexes showed a high expression of transgene in the lung after intravenous administration. We also have investigated the relationship between structural stability and in vivo transfection activity of complexes with stability‐modulated emulsions by changing the oil component in the emulsion 24. In that study, the most stable emulsion with squalene as the oil phase gave the most potent transfection activity after intravenous administration. These observations demonstrated that in vivo transfection activity was primarily associated with the stability of the complexes 30.
We further investigated the morphology of the emulsion/DNA complex using transmission electron microscopy (TEM). Trilaurin was selected as an oil phase for this particular experiment since it is solid during the sample preparation process due to its high melting temperature (ca. 46.5 °C). In this experiment, the emulsion and DNA appeared to be fully combined at C/D = 4 and formed a chromatin‐like structure. It has been reported that a liposome loses its structural integrity during the formation of liposome/DNA complexes in which DNA is sandwiched between lamellar lipid sheets or forms a hexagonal phase 31, 32. Unlike liposomal carriers, however, the cationic emulsion even retains its structural integrity during the formation of emulsion/DNA complexes. Therefore, we believe that the difference in structure of emulsion/DNA complexes might be one of possible explanations as to why they are superior to liposome/DNA complexes in airway gene transfer.
Intranasal immunization is associated with DNA delivery to mucosal surfaces throughout the respiratory and gastrointestinal tract 3. Interestingly, many transfected monocyte‐like cells were observed in NALT during immunohistochemical analysis. More importantly, in addition to NALT, a significant level of expression was observed in cervical lymph nodes from the mice administrated with the emulsion/DNA complexes (data not shown), indicating the cationic emulsion could be an in vivo delivery system for genetic vaccination against a pathogenic infection that may invade mucosal surfaces. It may be possible that the emulsion/DNA complexes can have inflammatory effects in vivo 33, 34, 35. The observed inflammation may be related to the presence of unmethylated CpG dinucleotide sequences in bacterially derived DNA 36, 37. No signs of severe toxicity, however, were noticed, but flu‐like symptoms including fever and myalgia was observed over a period of 24 h when the dose was below 40 µg per mouse.
In summary, we have demonstrated convincingly that the cationic lipid emulsion can be used as an effective mucosal gene carrier in vivo. The transfection efficiency of the emulsion carrier is ca. 200 times better than commonly used commercial liposome carriers. The increased transfection efficiency using the emulsion carrier may be ascribed to the increased probability of delivering intact DNA to the airway since the complex is strong and insensitive to mucosal destabilizers. Considering that there are numerous cationic lipids such as GL‐67, which have been reported to be more efficient in mucosal gene delivery than DOTAP, it would be an interesting exploration to formulate and evaluate them as a cationic emulsion for mucosal gene delivery 38.
Acknowledgements
This work was supported by a National Research Laboratory project from the Ministry of Science and Technology of Korea.
References
- 1. Griesenbach U, Ferrari S, Geddes D, et al. Gene therapy progress and prospects: Cystic fibrosis. Gene Ther 2002; 9: 1344–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ferrari S, Geddes D, Alton E. Barriers to and new approaches for gene therapy and gene delivery in cystic fibrosis Adv Drug Deliv Rev 2002; 54: 1373–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hobson P, Barnfield C, Barnes A, et al. Mucosal immunization with DNA vaccines. Methods 2003; 31: 217–224. [DOI] [PubMed] [Google Scholar]
- 4. Klavinskis L, Barnfield C, Gao L, et al. Intranasal immunization with plasmid DNA‐lipid complexes elicits mucosal immunity in the female genital and rectal tracts. J Immunol 1999; 162: 254–262. [PubMed] [Google Scholar]
- 5. Robinson H, Montefiori D, Johnson R, et al. Neutralizing antibody‐independent containment of immunodeficiency virus challenges by DNA priming and recombinant pox virus booster immunizations. Nat Med 1999; 5: 526–534. [DOI] [PubMed] [Google Scholar]
- 6. Musey L, Hu Y, Eckert L, et al. HIV‐1 induces cytotoxic T lymphocytes in the cervix of infected women. J Exp Med 1997; 185: 293–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kim T, Lee J, Hung C, et al. Generation and characterization of DNA vaccines targeting the nucleocapsid protein of severe acute respiratory syndrome coronavirus. J Virol 2004; 78: 4638–4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kim T, Hung C, Ling M, et al. Enhancing DNA vaccine potency by co‐administration of DNA encoding anti‐apoptotic proteins. J Clin Invest 2003; 112: 109–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kim T, Hung C, Boyd D, et al. Enhancing DNA vaccine potency by combining a strategy to prolong dendritic cell life with intracellular targeting strategies. J Immunol 2003; 171: 2970–2976. [DOI] [PubMed] [Google Scholar]
- 10. Kim T, Kim J, Juang J, et al. Vaccination with DNA vaccine encoding HSV‐1 VP22 linked to antigen generates sustained antigen‐specific memory CD8+ T cell and long‐term protective immunity. Hum Gene Ther 2004; 15: 167–177. [DOI] [PubMed] [Google Scholar]
- 11. Woo P, Wong L, Zheng B, et al. Unique immunogenicity of hepatitis B virus DNA vaccine presented by live‐attenuated Salmonella typhimurium . Vaccine 2001; 19: 2945–2954. [DOI] [PubMed] [Google Scholar]
- 12. McCray P, Wang G, Kline J, et al. Alveolar macrophages inhibit retrovirus‐mediated gene transfer to airway epithelia. Hum Gene Ther 1997; 8: 1087–1093. [DOI] [PubMed] [Google Scholar]
- 13. Chillon M, Lee J, Fasbender A, et al. Adenovirus complexed with polyethylene glycol and cationic lipid is shielded from neutralizing antibodies in vitro . Gene Ther 1998; 5: 995–1002. [DOI] [PubMed] [Google Scholar]
- 14. Halbert C, Standaert T, Wilson C, et al. Successful readministration of adeno‐associated virus vectors to the mouse lung requires transient immunosuppression during the initial exposure. J Virol 1998; 72: 9795–9805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kim T, Chung H, Kwon I, et al. In vivo gene transfer to the mouse nasal cavity mucosa using a stable cationic lipid emulsion. Mol Cells 2000; 10: 142–147. [DOI] [PubMed] [Google Scholar]
- 16. Wiethoff C, Smith J, Koe G, et al. The potential role of proteoglycans in cationic lipid‐mediated gene delivery. Studies of the interaction of cationic lipid‐DNA complexes with model glycosaminoglycans. J Biol Chem 2001; 276: 32 806–32 813. [DOI] [PubMed] [Google Scholar]
- 17. Ernst N, Ulrichskotter S, Schmalix W, et al. Interaction of liposomal and polycationic transfection complexes with pulmonary surfactant. J Gene Med 1999; 1: 331–340. [DOI] [PubMed] [Google Scholar]
- 18. Duncan J, Whitsett J, Horowitz A. Pulmonary surfactant inhibits cationic liposome‐mediated gene delivery to respiratory epithelial cells in vitro . Hum Gene Ther 1997; 8: 431–438. [DOI] [PubMed] [Google Scholar]
- 19. Xu Y, Szoka F, Mechanism of DNA release from cationic liposome/DNA complexes used in cell transfection. Biochemistry 1996; 35: 5616–5623. [DOI] [PubMed] [Google Scholar]
- 20. Yi S, Yune T, Kim T, et al. A cationic lipid emulsion/DNA complex as a physically stable and serum‐resistant gene delivery system. Pharm Res 2000; 17: 313–319. [DOI] [PubMed] [Google Scholar]
- 21. Kim T, Chung H, Kwon I, et al. Optimization of lipid composition in cationic emulsion as in vitro and in vivo transfection agents. Pharm Res 2001; 18: 51–60. [DOI] [PubMed] [Google Scholar]
- 22. Chung H, Kim T, Kwon I, et al. Oil components modulate physical characteristics and function of the natural oil emulsions as drug or gene delivery system J Control Release 2001; 71: 339–350. [DOI] [PubMed] [Google Scholar]
- 23. Kim T, Kim Y, Chung H, et al. The role of non‐ionic surfactants on cationic lipid mediated gene transfer. J Control Release 2002; 82: 455–465. [DOI] [PubMed] [Google Scholar]
- 24. Kim Y, Kim T, Chung H, et al. Enhancement of cationic emulsion‐mediated gene transfer by stabilization of emulsion. Int J Pharm 2003; 252: 241–252.12550800 [Google Scholar]
- 25. Zelphati O, Uyechi L, Barron L, et al. Effect of serum components on the physico‐chemical properties of cationic lipid/oligonucleotide complexes and on their interactions with cells. Biochim Biophys Acta 1998; 1390: 119–133. [DOI] [PubMed] [Google Scholar]
- 26. Bradford M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein‐dye binding. Anal Biochem 1976; 72: 248–254. [DOI] [PubMed] [Google Scholar]
- 27. Stuart B. Deposition and clearance of inhaled particles. Environ Health Perspect 1984; 55: 369–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Janssens H, Krijgsman A, Verbraak T, et al. Determining factors of aerosol deposition for four pMDI‐spacer combinations in an infant upper airway model. J Aerosol Med 2004; 17: 51–61. [DOI] [PubMed] [Google Scholar]
- 29. Sternberg B, Sorgi F, Huang L. New structures in complex formation between DNA and cationic liposomes visualized by freeze‐fracture electron microscopy. FEBS Lett 1994; 356: 361–366. [DOI] [PubMed] [Google Scholar]
- 30. Lee E, Marshall J, Siegel C, et al. Detailed analysis of structures and formulations of cationic lipids for efficient gene transfer to the lung. Hum Gene Ther 1996; 7: 1701–1717. [DOI] [PubMed] [Google Scholar]
- 31. Koltover I, Salditt T, Radler J, et al. An inverted hexagonal phase of cationic liposome‐DNA complexes related to DNA release and delivery. Science 1998; 281: 78–81. [DOI] [PubMed] [Google Scholar]
- 32. Radler J, Koltover I, Salditt T, et al. Structure of DNA‐cationic liposome complexes: DNA intercalation in multilamellar membranes in distinct interhelical packing regimes. Science 1997; 275: 810–814. [DOI] [PubMed] [Google Scholar]
- 33. Scheule P, George J, Bagley R, et al. Basis of pulmonary toxicity associated with cationic lipid‐mediated gene transfer to the mammalian lung. Hum Gene Ther 1997; 8: 689–707. [DOI] [PubMed] [Google Scholar]
- 34. Chadwick S, Kingston H, Stern M, et al. Safety of a single aerosol administration of escalating doses of the cationic lipid GL‐67/DOPE/DMPE‐PEG5000 formulation to the lungs of normal volunteers. Gene Ther 1997; 4: 937–942. [DOI] [PubMed] [Google Scholar]
- 35. Yew N, Wang K, Przybylska M, et al. Contribution of plasmid DNA to inflammation in the lung after administration of cationic lipid : DNA complexes. Hum Gene Ther 1999; 10: 223–234. [DOI] [PubMed] [Google Scholar]
- 36. Schwartz D, Quinn T, Thorne P, et al. CpG motifs in bacterial DNA cause inflammation in the lower respiratory tract. J Clin Invest 1997; 100: 68–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yew N, Zhao H, Wu I, et al. Reduced inflammatory responses to plasmid DNA vectors by elimination and inhibition of immunostimulatory CpG motifs. Mol Ther 2000; 1: 255–262. [DOI] [PubMed] [Google Scholar]
- 38. Chadwick S, Kingston H, Stern M, et al. Safety of a single aerosol administration of escalating doses of the cationic lipid GL‐67/DOPE/DMPE‐PEG5000 formulation to the lungs of normal volunteers. Gene Ther 1997; 4: 937–942. [DOI] [PubMed] [Google Scholar]


