Abstract
The adhesion G protein-coupled receptors (aGPCRs) are an evolutionarily ancient family of receptors that play key roles in many different physiological processes. These receptors are notable for their exceptionally long ectodomains, which span several hundred to several thousand amino acids and contain various adhesion-related domains as well as a GPCR Autoproteolysis Inducing (GAIN) domain. The GAIN domain is conserved throughout almost the entire family and undergoes autoproteolysis to cleave the receptors into two noncovalently-associated protomers. Recent studies have revealed that the signaling activity of aGPCRs is largely determined by changes in the interactions between these protomers. Here, we review recent advances in understanding aGPCR activation mechanisms and discuss the physiological roles and pharmacological properties of aGPCRs with an eye toward the potential utility of these receptors as drug targets.
Keywords: therapeutic, agonist, antagonist, ligand, antibody, pharmaceutical
Introduction
G protein-coupled receptors (GPCRs) are highly amenable to modulation by pharmaceuticals and it is estimated that one-third of currently-approved drugs target GPCRs (1). However, these therapeutics are focused on only a small fraction of the GPCR superfamily and no drugs to date have been approved to target any members of the second-largest GPCR family, the adhesion GPCRs (aGPCRs), which includes 33 members in humans. Thus, the aGPCR family possesses enormous potential for drug discovery, especially because aGPCRs have been linked to a wide variety of different diseases and regulate many important physiological processes throughout the body. In this review, we will discuss the relatively brief history of aGPCR research and describe the many recent advances that have shed light on aGPCR ligands, activation mechanisms and downstream signaling pathways. We will also discuss the roles of aGPCRs in various aspects of physiology and how these receptors might be targeted by novel therapeutics to treat human diseases.
Adhesion GPCR Fundamentals
Traditionally, aGPCRs have been named for idiosyncratic reasons, dependent more on the circumstances of their initial characterization than on the fact that they belonged to the aGPCR family. Recently, though, all aGPCRs were renamed with the ADGR prefix, followed by a letter indicating the receptor subfamily and a number for each receptor within that group (2). This new nomenclature has been adopted by the International Union of Basic and Clinical Pharmacology (IUPHAR) and will be used as the primary identifier for receptors discussed here. Table 1 lists the 33 human aGPCRs with their new and previous identifiers.
Table 1.
A summary of adhesion GPCR nomenclature and key signaling pathways
| Sub-group | New nomenclature | Previous identifier(s) | Signaling pathways | NTF interactors |
|---|---|---|---|---|
| III | ADGRA1 | GPR123 | ||
| ADGRA2 | GPR124, TEM5 | β-catenin (47), Cdc42 (99) | Heparin (71), αvβ3 Integrin (71), WNT7 (47) | |
| ADGRA3 | GPR125 | |||
| VII | ADGRB1 | BAI1 | Rac1 (45; 46), Gα12/13, RhoA (12; 19) | PtdSer (45), LPS (62), αvβ5 Integrin (63) |
| ADGRB2 | BAI2 | Gα16, NFAT-Luc (21) | ||
| ADGRB3 | BAI3 | C1ql1 (65), C1ql3 (64) | ||
| IV | ADGRC1 | CELSR1 | Rho kinase (51; 52) | |
| ADGRC2 | CELSR2 | Ca2+ (48) | C2 NTF (48) | |
| ADGRC3 | CELSR3 | Ca2+ (48) | C3 NTF (48) | |
| V | ADGRD1 | GPR133 | Gαs (161), cAMP (42; 161) | |
| ADGRD2 | GPR144 | |||
| II | ADGRE1 | EMR1 | ||
| ADGRE2 | EMR2 | Gα15 (42) | Chondroitin sulfate (68) | |
| ADGRE3 | EMR3 | |||
| ADGRE4 | EMR4 | |||
| ADGRE5 | CD97 | Gα12/13, RhoA (20) | CD55 (66; 67; 135), Chondroitin sulfate (68), Thy-1 (CD90) (69), α5β1 Integrin | |
| VI | ADGRF1 | GPR110 | Gαq (27), Gαs cAMP (34; 77) | Synaptamide (77) |
| ADGRF2 | GPR111 | |||
| ADGRF3 | GPR113 | |||
| ADGRF4 | GPR115 | Gα15 (42) | ||
| ADGRF5 | GPR116, Ig-Hepta | Gαq-RhoA/Rac1 (128) Gαq-IP1 (23) | Surfactant protein D (79) | |
| VIII | ADGRG1 | GPR56 | Gα12/13, RhoA (12; 18; 27; 37; 75), Gαq/11 (38) | Tissue transglutaminase (72; 73), Collagen III (74; 75) |
| ADGRG2 | GPR64, HE6 | cAMP (23; 24), MAPK, RhoA (14) | ||
| ADGRG3 | GPR97 | Gαo (42), RhoA and Cdc42 (43) | ||
| ADGRG4 | GPR112 | SRE-Luc (44) | ||
| ADGRG5 | GPR114 | Gαs (42) | ||
| ADGRG6 | GPR126 | cAMP (25; 26; 32; 39), | Collagen IV (25), Laminin-211 (32), Prion protein (76) | |
| ADGRG7 | GPR128 | |||
| I | ADGRL1 | LPHN1, CIRL, CL1 | Gαo (35; 36), Gαq (36) | α-latrotoxin (35; 54; 55), neurexins (56), teneurins (58), FLRT1 and 3 (57) |
| ADGRL2 | LPHN2, CIRL2, CL2 | teneurins (58), FLRT3 (57) | ||
| ADGRL3 | LPHN3, CIRL3, CL3 | teneurins (58), FLRT1 and 3(57), FLRT2 and Unc5 (60) | ||
| ADGRL4 | ELTD1 | |||
| IX | ADGRV1 | VLGR1, GPR98, MASS1 | Gαi (28), Gαs and Gαq (53) |
The first characterization of aGPCRs was in the area of immunology. A homolog of the mouse macrophage marker F4/80 was determined to be a seven-transmembrane-spanning (7-TM) protein and named EMR1 (for “epidermal growth factor-like molecule containing mucin-like hormone receptor 1”) (3). Shortly thereafter, independent work by a different group identified the leukocyte activation marker CD97 as a 7-TM receptor (4). These two receptors would later be grouped in the same subfamily and re-named ADGRE1 and ADGRE5, respectively (2). Soon, the discovery of additional 7-TM proteins with long extracellular domains and significant similarity in their transmembrane cores resulted in a grouping of the receptors to a family called LNB-TM7 for “long N-terminal domain 7-TM receptors with similarity to family B receptors” (5). In 2004, the family was renamed as the “adhesion G protein-coupled receptors” (6), the name that is still in widespread use today.
Adhesion GPCRs exhibit the classic seven-transmembrane architecture that is common to all members of the GPCR superfamily, featuring an extracellular N-terminus (NT) and a cytoplasmic C-terminus (CT). The NT regions of aGPCRs are quite large relative to most other GPCRs and possess a variety of conserved domains that can mediate adhesive interactions. The one NT domain that is common to almost all aGPCRs is the GPCR Autoproteolysis-Inducing (GAIN) domain (7), which contains a GPCR Proteolysis Site (GPS) motif (8). This domain is capable of autoproteolysis, resulting in two cleaved receptor protomers that have the ability to remain non-covalently associated with each other. By convention, the cleaved N-terminus following GAIN domain proteolysis is referred to as the N-terminal fragment (NTF), and the rest of the receptor, including the short N-terminal stalk, seven-transmembrane region and cytoplasmic C-terminus, is referred as the C-terminal fragment (CTF).
Adhesion GPCR Structure
Adhesion receptors were named due to the presence extracellular adhesion domains in the earliest-described members of the family (5; 9). However, the long amino termini of these proteins display a rich variety of protein domains with many properties beyond adhesion (Fig. 1). For example, 13 of the 33 human aGPCRs have a hormone-binding domain reminiscent of the Secretin family of GPCRs (10). It is currently unclear whether these domains might mediate the binding of key ligands to aGPCRs. Of all the N-terminal domains found in aGPCRs, the GAIN domain is the archetypal structural feature of the aGPCR family and is found in each member except for ADGRA1/GPR123 (2). The approximately 300-amino-acid domain is found close to the start of the first transmembrane domain. Arac and colleagues crystallized this domain from ADGRB3/BAI3 and ADGRL1 and called it the GPCR Autoproteolysis Inducing (GAIN) domain (7). Even though some aGPCRs undergo GAIN domain-mediated autoproteolysis in the endoplasmic reticulum (11), the N- and C-terminal protomers are known to traffic together to the cell membrane, bound to each other in a non-covalent manner (8). However, receptor cleavage is not necessary for aGPCRs to be expressed on the cell surface (12–14). The aforementioned crystal structure of the GAIN domain provided structural insight into how the NTF and CTF protomers remain associated, with the post-cleavage stalk tightly bound inside a hydrophobic groove in the cleaved GAIN domain (7).
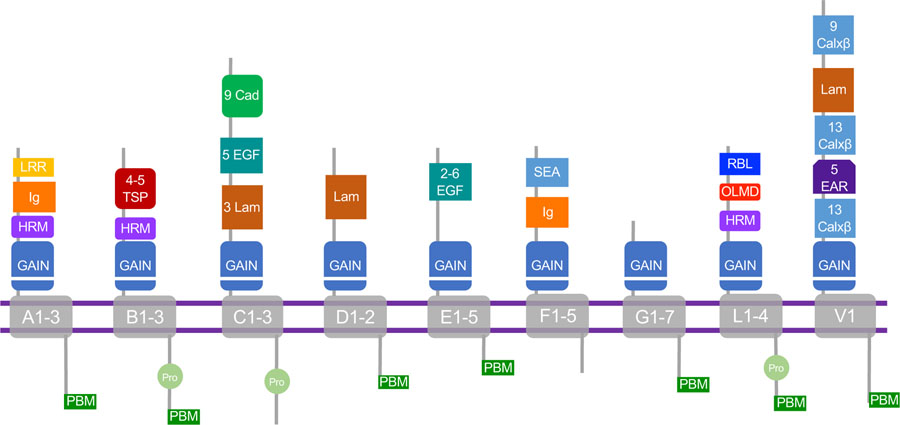
Figure 1. Adhesion GPCR sub-family structures.
The 9 adhesion GPCR sub-families are listed alphabetically with key protein domains depicted for each group. Some sub-family members have a slightly different structure from what is indicated in the figure: ADGRA1-No GAIN, HRM, IG, LRR; ADGRA2-No Ig; ADGRB3 has NT CUB but no Pro; ADGRB1–5xTSP1; ADGRC1 and C3 no Pro; ADGRF2–4 no SEA; ADGRG1–7: G1-PLL, G6-CUB, Laminin, PBM only in G1; ADGRL1–4: Pro only in L1, L4-Only EGF and GAIN. Abbreviations: GAIN: GPCR autoproteolysis-inducing domain; LRR: Leucine-rich repeat; Ig: Immunoglobulin-like; HRM: Hormone receptor motif; TSP: Type-1 thrombospondin repeat; Cad: Cadherin repeat; EGF: Epidermal Growth Factor-like (includes Calcium-binding EGF-like domains); Lam: Laminin; Pro: polyproline sequence; PBM: PDZ binding motif; SEA: Sperm protein/Enterokinase/Agrin domain; RBL: rhammose-binding lectin; OLMD; olfactomedin-like; EAR: Epilepsy-associated repeat.
The 7-TM core of aGPCRs most closely resembles that of the Family B, Secretin-like receptors (15). Many rhodopsin family (Family A) GPCRs have a DRY motif in the third intracellular loop that is important in G protein coupling, but adhesion receptors do not adhere to this model (16). However, most (21 of 33) aGPCRs have an E-X-X-X-X-Y motif in the third intracellular loop where the position of the Y corresponds to that of the Y in the DRY motif of rhodopsin-like receptors (15). In the extracellular loops, aGPCRs somewhat resemble the metabotropic glutamate receptors with short first and third extracellular loops and conserved cysteine-tryptophan residues in the second extracellular loop (15). However, the functional contributions of these structural features are still largely untested for aGPCRs.
Adhesion GPCR activation mechanisms
For many years, no endogenous ligands for aGPCRs were known and relatively few specific tools, such as receptor antibodies, were available. Thus, initial progress in understanding the potential signaling activity of aGPCRs was slow. One early idea was that the long extracellular N termini might contain ligand binding sites, analogous to Family C GPCRs such as the metabotropic glutamate receptors (17). This hypothesis was tested for ADGRG1 and ADGRB1 via removal of the receptors’ NTF regions at the point of predicted GAIN domain cleavage, leaving just the CTF protomer intact. Surprisingly, instead of resulting in inactive receptors, the truncated forms of both G1 (18) and B1 (19) were found to be much more active than the full-length receptors. Parallel studies on truncated versions of ADGRE5 (20) and ADGRB2 (21) also resulted in large increases in receptor signaling activity. These findings led to the proposal of a disinhibition model of signaling (Fig. 2C) whereby the NTF regions of aGPCRs can inhibit the intrinsic signaling potential of the CTF (7-TM) region by locking the receptor in an inactive conformation (22). In addition to the receptors mentioned above, ADGRG2 (14; 23; 24), ADGRG6 (25), ADGRD1 (26), ADGRF1 (27) and ADGRV1 (28) have all subsequently been found to be highly active when the NTF is removed via truncation.
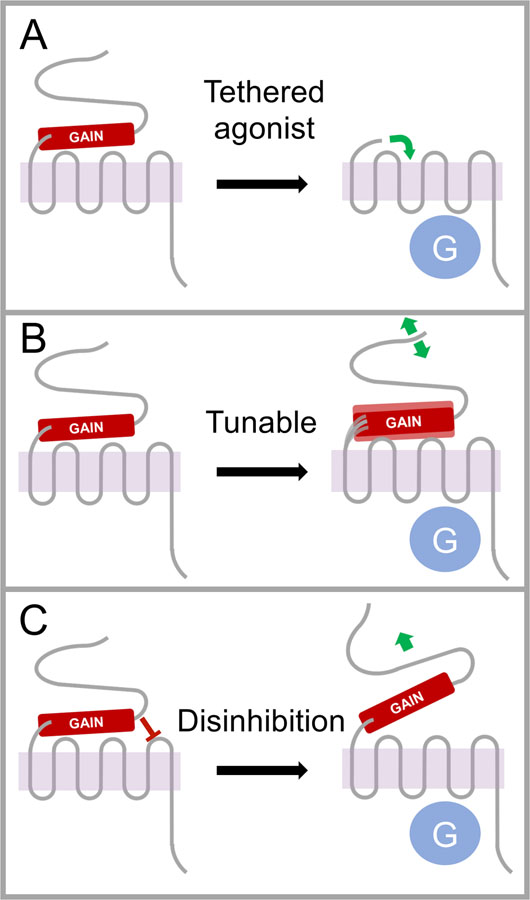
Figure 2. Models of the activation of adhesion GPCR signaling.
Generally speaking, adhesion GPCR activation is governed by interactions between the N-terminal fragment (NTF) and C-terminal fragment (CTF) protomers. A) The tethered agonist (e.g. stalk, stachel) can be unmasked when the NTF is completely removed. B) The stalk may also act as a lever, with its position being modulated by NTF movements to tune receptor activity. C) The NTF can also in some cases suppress signaling by the CTF protomer in ways that do not depend on the stalk, with disinhibition being achieved when the NTF is pulled away from the CTF by ligand interactions or shed completely from the protein complex. It should be noted that these mechanisms are not mutually exclusive, and in fact all three mechanisms may occur for any given aGPCR, although the relative importance of each mechanism may vary from receptor to receptor.
As early as 2002, the possibility was considered that aGPCR cleavage could play a role in receptor activation analogous to the role played by cleavage in activation of the protease-activated receptors (PARs) (8). However, it was not until 2014 that evidence was provided that exogenous peptides derived from the post-cleavage stalk sequence could activate ADGRG6 and ADGRD1 (26). Soon thereafter, it was independently reported that ADGRG1 and ADGRF1 could also be activated by peptides derived from the post-cleavage stalk region (27). This mechanism has been termed the tethered agonist model of aGPCR signaling, wherein the post-cleavage stalk acts as a tethered agonist to push the receptor into an active conformation (Fig 2A).
The generality of the tethered agonist model is an issue that is still being addressed in ongoing studies. A key hurdle for experiments in this area is that the stalk or “stachel” (German for “stinger”) peptides derived from the post-cleavage stalk are highly hydrophobic for almost all aGPCRs and therefore difficult to work with in aqueous solutions. Moreover, in some cases, only certain lengths of peptide can agonize the receptor, whereas peptides even a single residue longer or shorter can exhibit no agonistic action or even strongly inhibit activity by apparently acting as inverse agonists (27). A number of aGPCRs can clearly be activated by post-cleavage stalk/stachel peptides (29), but evidence has emerged that liberation of the post-cleavage stalk is not required for activation of all aGPCRs. For example, a truncated version of ADGRB1 that lacks the post-cleavage stalk exhibits no signaling deficits relative to a truncated version of ADGRB1 that mimics the GAIN-cleaved version of the receptor and possesses an intact stalk (12).
Another idea that has also gained experimental support is that the stalk can act more like a lever within the cleaved but associated NTF-CTF complex to tune signaling activity based on its position. This idea has been termed the tunable model (27) (Fig 2B). In this model, the position of the stalk may be dictated by NTF interactions with extracellular adhesive ligands and by mechanosensory stimuli. Indeed, various lines of evidence suggest that aGPCRs might generally act as metabotropic mechanosensors (30). For example, ADGRG5/GPR114 was found to be activated by mechanical stimulation in vitro in a manner that is dependent on a glutamine residue within the stachel sequence, which might be necessary for placing the stachel in the appropriate position (31). In addition, the signaling activity of ADGRG6 in response to its ligand laminin-211 is dependent on stimulation dynamics where under static conditions laminin-211 inhibits activity but becomes agonistic with increasing frequency of vibration or shaking (32). Along these same lines, the Drosophila homolog of the ADGRL receptors was shown to be a functional mechanoreceptor for multiple sensory modalities in the fly chordotonal organ (33). The localization of ADGRV1 to auditory hair cell stereocilia suggests that this receptor might also have a mechanosensory function, but this concept has not been demonstrated to date (30). The unusual ectodomain architecture of aGPCRs as well as the receptors’ known intercellular interactions (see below) invites the hypothesis that aGPCRs could be adept at conveying information related to external forces across the cell membrane (30).
The emerging consensus in the aGPCR field is that aGPCR signaling is activated by changes in the interactions between the NTF and CTF protomers. However, the specific changes in the NTF/CTF association necessary for activation may vary between different members of the family (Fig. 2), and ongoing work is addressing deeper mechanistic questions regarding how such activation occurs on a receptor-by-receptor basis. For aGPCRs that are activated via a tethered agonist-dependent mechanism, for example, there is interest in the specificity of tethered agonist binding to aGPCRs. One consequence of the high degree of sequence homology across the aGPCR family around the GAIN cleavage site is that the agonistic portion of stachel peptides from one receptor often closely or exactly matches that of another receptor. Indeed, this situation has recently been reported: activating peptides derived from ADGRF1/GPR110 can activate F1 and also the closely-related receptor ADGRF5/GPR116 as well as ADGRG2/GPR64, which resides in a separate sub-group of aGPCRs (34). Interestingly, it was also observed in some cases that only certain pathways could be activated when using peptides from one receptor to activate another; for example, peptides derived from ADGRF4/GPR115 and ADGRF5 were reported to stimulate IP1 accumulation but not cAMP in cells expressing ADGRF1 (34).
Adhesion GPCR signaling pathways
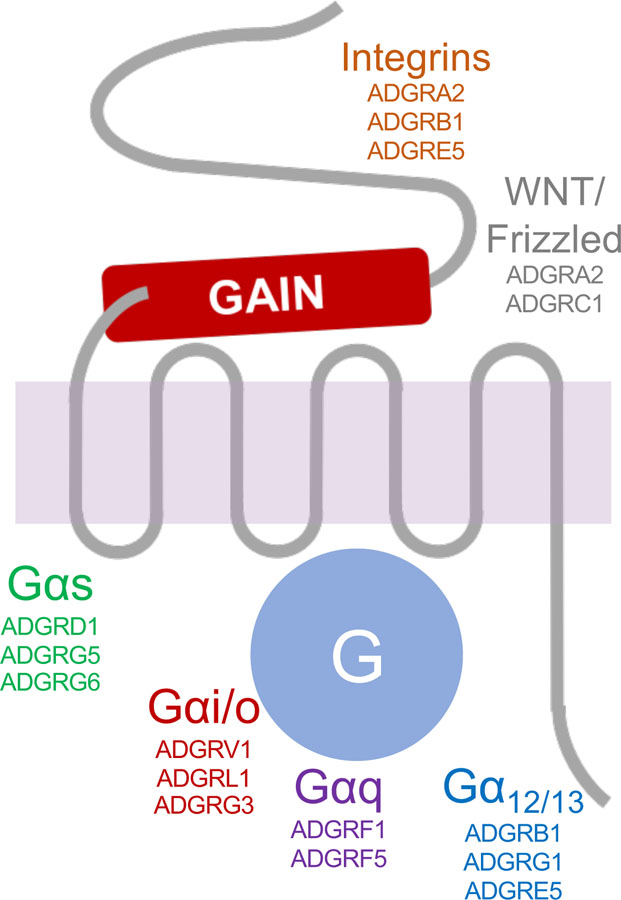
Understanding the signaling pathways downstream of aGPCRs is critical for drug discovery as well as for achieving a fundamental understanding of receptor function. Early, seminal signaling studies on ADGRL1 (35; 36) demonstrated clear G protein coupling and now many aGPCRs have been found to couple to G proteins. The currently-known G protein pathways activated by each aGPCR, both of the heterotrimeric and small GTPase variety, are described here and summarized in Table 1 and Figure 3.
Figure 3. Adhesion GPCR signaling pathways.
Adhesion GPCRs have been found to initiate a wide variety of downstream signaling cascades. This schematic diagram summarizes signaling pathways for which there is presently evidence of activation by more than one aGPCR.
ADGRG1 has been an intensively-studied aGPCR due to its involvement in human disease (discussed below) and a number of studies have investigated this receptor’s G protein coupling. An antibody directed at the G1 NTF was shown to activate Gα12/13-mediated RhoA signaling downstream of G1 as evidenced by inhibition with the RGS domain of p115RhoGEF, C3 exoenzyme, and dominant negative RhoA (37). Later work established that a version of G1 truncated at the predicted site of GAIN domain cleavage strongly activates the RhoA pathway (18), can be co-immunoprecipitated with Gα13 (12), and can directly stimulate Gα13 in reconstitution assays (27). Others have found that G1 can associate with Gαq (38), although G1-mediated activation of Gαq has not been observed (27).
Another ADGRG sub-family member, ADGRG6/GPR126 has been linked to both Gαs and Gαi (25; 26; 39). It is possible that G6 may couple differentially to these distinct G proteins in a manner that is dependent on the mechanism of receptor of activation and/or cellular context. Other aGPCRs have also exhibited the capacity to couple to multiple G proteins; for example, studies on ADGRG2 have provided evidence of coupling to Gαs, Gαi, Gα12/13 and Gαq (14; 23). Many traditional GPCRs can couple to multiple proteins, with coupling often strongly regulated by factors such as receptor phosphorylation (40) or association with scaffold proteins (41). In future studies it will be interesting to explore whether such regulation also occurs for aGPCRs.
Signaling pathways downstream of several other ADGRG sub-group receptors have also been documented. ADGRG2/GPR64 was reported to activate both SRE and NFκB luciferase reporters depending on NTF cleavage events (14) and was also found to stimulate cAMP when treated with stachel peptides (23). ADGRG3/GPR97 was found to couple to Gαo (42) and has also been associated with activation of the small GTPases Cdc42 and RhoA (43). Little is known about the function or activity of ADGRG4/GPR112 but the truncated CTF of this receptor was reported to activate SRE-luciferase (44). ADGRG5/GPR114 was found to stimulate cAMP, which was abolished by siRNA directed at Gαs (42), and a similar result was reported for ADGRD1/GPR133 (42), indicating that both of these receptors likely couple to Gαs.
Two ADGRB subfamily members have been demonstrated to couple to G proteins. ADGRB1 was found to activate the Rho pathway in a manner that was sensitive to the RGS domain of p115RhoGEF, thereby providing evidence for coupling to Gα12/13 (19). Furthermore, truncated constitutively-active versions of B1 can be co-immunoprecipitated in complex with Gα13 (12). Despite a high degree of similarity with the 7-TM structure of B1, ADGRB2/BAI2 has a unique signaling profile and its activation of an NFAT luciferase reporter was potentiated by the addition of Gα16, a promiscuous G protein (21). This finding demonstrates the receptor’s capacity for G protein coupling but not its specificity. The cognate G protein(s) for ADGRB3 have not yet been identified. In addition to the aforementioned coupling to heterotrimeric G proteins, ADGRB1 has also been shown to couple to guanine nucleotide exchange factors for Rac, including ELMO (45) and Tiam1 (46), to stimulate Rac in manner that does not depend on heterotrimeric G protein activation.
G protein coupling has yet to be shown for the ADGRA sub-family. Nonetheless, ADGRA2/GPR124 can be co-activated (with Frizzled) by WNT7A to stimulate β-catenin signaling and, unlike G protein signaling via many other aGPCRs, this activation was shown to be dependent on the presence of an intact N terminus (47). The members of the ADGRC sub-family can engage in homophilic interactions in trans (48) and are also involved in WNT-Frizzled signaling (49; 50), though it is not clear whether they function as co-receptors for WNT proteins like ADGRA2 above. ADGRC1 can signal to the Rho pathway (51; 52), although it is not known whether this occurs downstream of heterotrimeric G proteins or is dependent on some other mechanism such as direct recruitment of RhoGEFs. ADGRC2 and 3 (CELSR2 and 3) have been found to activate Ca2+ signaling in a phospholipase C and ER-calcium store dependent manner, suggestive of Gαq coupling, although it is unclear at present whether G proteins actually mediate this signaling activity (48). The largest aGPCR, ADGRV1, has been found to couple to Gαi (28) and also to signal to protein kinases A and C by way of Gαs and Gαq (53). There are still many aGPCRs for which G protein coupling has not been established (Table 1). This lack of basic understanding for many adhesion GPCRs reveals the enormous potential for basic discovery that still remains for these receptors.
Adhesion GPCR Ligands
While most aGPCRs are still considered to be orphan receptors, a number of extracellular interacting proteins have been identified, and some of these interacting partners may represent authentic endogenous agonists for the receptors (Table 1). Early on, ADGRL1 was found to be the calcium-independent receptor for the black widow spider venom neurotoxin α-latrotoxin (35; 54; 55). Subsequently, the ADGRLs have also been associated with endogenous ligands. Pre-synaptic neurexins (56), post-synaptic FLRT proteins (57), and teneurins (58; 59) have all been found to bind with high affinity to ADGRL1 and ADGRL3 extracellular regions and are thought to promote intercellular and perhaps transsynaptic adhesion. Furthermore, ADGRL3 was recently found to engage in a ternary complex with the transmembrane cell guidance protein Unc5 and FLRT2 (60). Binding of Lasso/teneurin-2 was found to stimulate Ca2+ signaling in hippocampal neurons expressing ADGRL1 (59).
Similarly, a number of interacting partners have been identified for the ADGRB/BAI sub-family of aGPCRs, but their effects on signaling still remain elusive. The N-terminal type-1 thrombospondin repeats of ADGRB1 bind to phosphatidylserine, thereby allowing the receptor to recognize and mediate the internalization of apoptotic cells (45). Moreover, as mentioned above, this function has been linked to intracellular signaling via the RacGEF ELMO/DOCK180. The role of B1 in macrophages has been further extended to include the recognition of Gram-negative bacteria, also via surface lipopolysaccharide (LPS) interaction with the thrombospondin repeats (61; 62). Like the ADGRL1–3 receptors, which share similar ectodomain structures, all three ADGRB proteins have at least 4 N-terminal type-1 thrombospondin repeats but it is unclear if ADGRB2 and B3 also bind phosphatidylserine and LPS. A unique feature of the ADGRB1 N terminus is an integrin-binding RGD motif, which has been shown to interact with αvβ5 integrin (63). Ectodomain interactions for ADGRB2 have not yet been reported. In the brain, ADGRB3 is a target of the C1ql proteins, which are secreted proteins bearing the complement pathway-like C1q globular domain. B3 was first reported to bind to C1ql3 (64) and subsequently was shown to bind to C1ql1 in the cerebellum in a manner that regulates synaptogenesis (65).
Another complement cascade protein, CD55/decay accelerating factor was found to be a ligand for ADGRE5/CD97 (66), though with relatively low affinity (67). The EGF-like domains of ADGRE2 and the longest form of ADGRE5 are nearly identical and both bind the glycosaminoglycan chondroitin sulfate (68). However, despite this high degree of ectodomain similarity, ADGRE2 only weakly interacts with CD55 (67) suggesting that even aGPCRs with very similar ectodomains may nonetheless have highly specific interactomes (68). E5 has also been shown to bind to Thy-1/CD90 (69) and α5β1 integrin (70) (see below). Similarly, ADGRA2 also has a cryptic RGD motif that can interact with αvβ3 integrin when it is unveiled by matrix metalloprotease cleavage (71).
The members of ADGRG sub-family of receptors have been found to bind to multiple extracellular proteins. G1 was shown to inhibit melanoma growth and metastasis by interacting with a component of the extracellular matrix (ECM), tissue transglutaminase (TG2), which binds to the extracellular N-terminus of G1 (72). Further work on this interaction revealed a unique paradigm where G1 prevents excess TG2-mediated ECM crosslinking by internalizing TG2 and degrading it, thereby retarding melanoma growth and progression (73). Additionally, G1 has been found to bind to collagen III in a manner that stimulates Gα12/13-mediated signaling to the Rho pathway (74; 75).
The N terminus of ADGRG6 has been found to bind to collagen IV (25) and laminin-211 (32), both of which may be key developmental signals that can stimulate receptor activation and increase cAMP levels. Interestingly, G6 expressed in Schwann cells was also found to be a target of the prion protein (76). The flexible tail of the prion protein contains a domain that is very similar to the G6-interacting motif of collagen IV, and indeed the prion protein was also found to stimulate cAMP signaling through interaction with G6 (76).
To date, ADGRF1/GPR110 and ADGRF5/GPR116 are the only receptors in the ADGRF sub-group for which extracellular interacting molecules have been identified. Full-length ADGRF1 was reported to be activated by synaptamide, a metabolite of the omega-3 fatty acid docosahexaenoic acid in brain tissue (77). Mice lacking ADGRF5 and surfactant protein D phenocopy each other in lung tissue and it has been found that surfactant protein D can interact the with ectodomain of F5 (78). However, as for many of the interactions described in this section, it is not yet clear what effect this interaction may have on G protein signaling or other signaling downstream of F5 (79).
Adhesion GPCRs in Nervous System Function and Disease
One criterion for assessing the therapeutic potential of a given drug target is association of the target with genetic diseases in humans (80). By this criterion, aGPCRs stack up well as potential therapeutic targets, as a number of aGPCRs have been associated with inherited human disorders. The most intensively-studied example in this regard has been ADGRG1, mutations of which cause a brain developmental disorder known as bilateral frontoparietal polymicrogyria (BFPP) (81). G1 is highly expressed in neural stem cells (NSCs) and plays a key role in inducing NSCs to stop migrating once they have found their proper position in the brain (37). Disease-associated mutations of G1 typically interfere with receptor folding, trafficking and/or signaling, and a number of distinct disease-causing mutations have been described (82; 83). Patients with BFPP also exhibit myelination deficits (81; 84), and indeed recent work in mice has revealed that G1 is expressed in oligodendrocytes during development and loss of G1 function results in deficient myelination (85; 86).
Another prominent disease-associated aGPCR is ADGRV1. Mutations to this receptor cause Usher syndrome type 2C, which is characterized by deafness and blindness (87). V1 is expressed at high levels in the stereocilia in the cochlea as well as the ciliary membrane of photoreceptors (88; 89). This receptor appears to be important for aspects of ciliary function. Many disease-causing mutations have been identified on the receptor’s massive (>5,000 amino acid) N-terminus (90). Some of these mutations introduce stop codons, meaning that the expressed receptor would be devoid of the seven-transmembrane region that is necessary for signaling (90). However, at least one disease-associated mutation is found on the receptor’s cytoplasmic C-terminus and has been shown to modulate V1 coupling to G proteins (28).
Mouse, zebrafish, and human studies have established that ADGRC1 is required for neural tube closure (91; 92). Similarly, the other two CELSR/ADGRC receptors are critical in neuronal migration and axon guidance (93), and ADGRC3/CELSR3 is important for excitatory synapse formation (94). Other aGPCRs that are important in distinct aspects of nervous system development include ADGRG6, which is required for peripheral nervous system myelination (39; 95; 96), and ADGRA2/GPR124, which regulates CNS angiogenesis. Knockout of A2 has been shown to result in embryonic death due to disruption of angiogenesis in the CNS and resultant hemorrhaging (97–99). A2 is widely expressed in the vasculature (99), where it can promote WNT signaling to regulate angiogenesis (47; 100; 101). Conditional knockout of A2 in adult mice compromised blood-brain barrier integrity and resulted in hemorrhage in models of stroke and glioblastoma (102).
The latrophilins/ADGRL1–3 and BAIs/ADGRB1–3 appear to be critical for synapse formation and strengthening. ADGRL1–3 receptors are found in both pre- and post-synaptic compartments (103). Human studies have linked ADGRL3 to attention deficit hyperactivity disorder (ADHD) (104; 105) and animal studies have supported this finding. Studies in mice have revealed that targeted deletion of L3 results in hyperactivity and disrupted dopamine and serotonin transport (106), as well as significant changes in the relative strengths of connections between different layers of the neocortex (107). Moreover, deletion of L3 in zebrafish also results in hyperlocomotor behavior and other changes consistent with altered synaptic connections (108).
Parallel studies on ADGRB1 and B3 have revealed these receptors to be critical for dendritic maturation and stability. Knockdown of B1 in cultured neurons results in drastically altered dendritic spine morphology (46), and genetic deletion of B1 induced perturbations to the post-synaptic density (PSD) regions of excitatory synapses in vivo as well as impairments in synaptic plasticity and spatial memory (109). While less is known about the signaling properties of ADGRB3 relative to B1, it has been genetically linked to schizophrenia (110) and several studies have identified B3 as an important component of hippocampal and cerebellar synapses. In cultured neurons, C1ql3 was found to decrease spine density through actions at B3 (64). Additional experiments have shown that B3 is critical for dendritic development in vitro (111) and through interactions with C1ql proteins B3 is a necessary component of excitatory synapses on cerebellar Purkinje neurons (65). While ADGRB2 is also enriched in the nervous system (112; 113), and also contains these same RacGEF-binding motifs, it does not appear to be required for spine maturation and synaptic function. Mice lacking B2 were found to have no gross behavioral or anatomical defects but, surprisingly, displayed increased hippocampal neurogenesis and improved resilience compared to wild-type animals in mood disorder related behavioral tests (114).
Adhesion GPCRs in Cancer
Members of the adhesion GPCR family are among the most frequently mutated GPCRs in cancer (115; 116). Indeed, whole genome analyses revealed that ADGRV1, ADGRB3, and ADGRL3 genes are among the most significantly mutated genes in tumors (116; 117). In the late 1990s, ADGRE5 and ADGRG1 were found to be differentially expressed in various tumor cells: G1 was down-regulated in the most highly metastatic melanoma cell line in comparison to lines with less metastatic potential (118) and E5 was found to be undetectable in normal thyroid tissue but to be expressed in thyroid carcinomas with its expression level highest in aggressive tumors (119). ADGRG1 was later shown to constrain melanoma growth and metastasis (72) by inhibiting angiogenesis through VEGF suppression (120) and internalization of the tumor-promoting extracellular matrix enzyme tissue transglutaminase TG2 (mentioned above) (73).
Angiogenesis is known to be critical for tumor growth (121). In addition to ADGRG1, several other aGPCRs have been shown to regulate angiogenesis. For example, ADGRB1 inhibits angiogenesis through the release of N-terminal type-1 thrombospondin repeat fragments termed vasculostatins (122–124). The most N-terminal fragment is liberated by matrix metalloproteinase-14 cleavage to result in a 40 kDa protein (vasculostatin-40) whereas the entire N terminus (containing four additional thrombospondin repeat domains) can be released due to GAIN domain proteolysis. The ability of B1 to regulate angiogenesis may be especially important in brain tumors, as for example B1 is known to be lost in glioblastoma due to epigenetic silencing (125).
In contrast to ADGRB1, ADGRE5 is upregulated in glioblastoma (GBM) and increases the invasiveness of GBM cells (126). Moreover, rather than inhibit angiogenesis, the NT of ADGRE5 has been found to promote angiogenesis through chemotactic recruitment of endothelial cells, which is initiated by binding to integrins (70). In fact, ADGRE5 expression is induced in a wide range of cancer cell lines and correlates with metastatic aggressiveness (115). One possible mechanism linking ADGRE5 to invasiveness in prostate and thyroid cancer involves heterodimerization with the LPA receptor and signaling via Gα12/13 and the Rho pathway (20; 127). Similarly, ADGRF5 has been found to drive breast cancer metastasis through Rho pathway signaling but via Gαq/p63RhoGEF rather than Gα12/13 (128).
ADGRL4/ELTD1 has also been found to be a pro-angiogenic adhesion GPCR that is upregulated in GBM tumor and endothelial cells (129–131). Little is known about this receptor’s signaling activity, but ADGRL4 expression was observed to be regulated by multiple pro-angiogenic factors including the Notch ligand DLL4, VEGF, and bFGF (basic Fibroblast Growth Factor) (130). Importantly, siRNA knockdown of ADGRL4 attenuated vascular endothelial cell sprouting in vitro and inhibited tumor growth in vivo, indicating that this receptor may also be a therapeutic target for multiple human cancers (130).
Recently, ADGRD1/GPR133 was implicated in GBM as well (132). D1 was found to be expressed in hypoxic GBM cells, with the receptor’s expression levels inversely correlated with patient survival. In addition, it was found that knockdown of D1 in mouse brain limited tumor growth and improved survival. These data suggest that ADGRD1 inhibitors could provide a novel therapeutic avenue for treating GBM by limiting the ability of GBM cells to survive in a hypoxic environment (132).
Adhesion GPCRs in the Function and Diseases of the Immune, Cardiac, Pulmonary and Musculoskeletal Systems
Adhesion GPCR research began in the immunology field, and thus it is not surprising that many types of immune cells express aGPCRs. For example, expression in immune tissues is a common feature for all five members of the ADGRE family (EMR1–4, CD97), two ADGRG receptors (GPR56, GPR97), and ADGRB1 (133). ADGRE2/EMR2 is widely expressed in myeloid tissue including macrophages, monocytes, and mast cells. A recent study found that a missense mutation that switches a cysteine to tyrosine upstream of the cleavage site in ADGRE2 co-segregated in two large families with vibratory urticaria, a condition in which hives develops on the skin from typically innocuous stimuli (134). This study concluded that the loss of the cysteine residue destabilized the interactions between the extracellular NTF and the 7-TM domain, thereby lowering the threshold of mechanical stimulation that induces mast cell degranulation. As mentioned above, ADGRE5 can bind to CD55 on T-cells (135), and macrophage-expressed ADGRB1 can recognize phosphatidylserine (45) and lipopolysaccharides on bacteria (62). In general, while aGPCRs have been found to be involved in myriad immune functions, in most cases the relative contributions of receptor-mediated adhesion versus G protein-dependent signaling are not yet clear.
The importance of ADGRG6 for peripheral nervous system myelination was described above, but complete loss of G6 in mice also results in embryonic lethality due to cardiovascular failure (136). However, in zebrafish, reintroduction of the G6 ectodomain up to the GPS motif rescued the cardiac defect but not myelination, indicating that the NTF alone is sufficient for cardiac functions but that the CTF is required for myelination (137). Mice lacking ADGRL4 displayed exaggerated cardiac hypertrophy following pressure overload (138), which suggests that this receptor could be targeted in hypertrophic cardiomyopathy (139). Another ADGRL receptor, ADGRL2, is involved in the epithelial-mesenchymal transition in heart valve development (140).
Several aGPCRs are expressed in the lung (78) but only ADGRF5/GPR116 has been studied in detail. Loss of functional F5 dramatically disrupts lung surfactant homeostasis in multiple mouse models (141; 142). In mice lacking F5, pulmonary surfactant, which is required for efficient respiration, accumulates in the lung (78; 141). Although mice lacking surfactant protein D have a similar phenotype to F5-null mice and these proteins interact (co-immunoprecipitate), it is unclear if surfactant protein D is an endogenous ligand for ADGRF5 (79).
Several aGPCRs mediate important regulation of the musculoskeletal system. For example, ADGRB1, ADGRB3, and ADGRG1 have key functions in skeletal muscle. B1 and B3 are critical in the fusion of myoblasts to form myofibers (143; 144) whereas G1 signaling is a key mediator of muscle hypertrophy (145). Mutations to ADGRG6 have been shown to result in a severe form of arthrogryposis multiplex congenita (AMC), a disease characterized by lack of normal joint flexibility and muscle strength in newborns (146). The disease-causing mutations in G6 impair cleavage of the receptor’s GAIN domain, implicating GAIN autoproteolysis as essential for the receptor’s function in vivo. Certain variants of G6 have also been associated with adolescent-onset idiopathic scoliosis, which is the most common skeletal disease in children (147; 148).
Many aGPCRs are not ubiquitously expressed in every tissue in the body, but rather are selectively distributed in a limited number of tissues. Indeed, some aGPCRs appear to play highly-specific physiological roles. For example, expression of ADGRG2/GPR64 is restricted to the epididymis (this receptor is also known as “HE6’” for human epididymal protein 6) and male mice lacking the gene for this receptor are infertile (149). The discrete expression of many aGPCRs in a limited number of tissues is another property that makes them such attractive drug targets, as the identification of drug targets with limited distribution patterns allows for the development of drugs with tissue-specific actions.
Targeting Adhesion GPCRs with Drugs
Due to their extraordinary size and complexity, aGPCRs present a variety of opportunities for therapeutic targeting (Fig. 4). First, similar to rhodopsin-like Family A GPCRs, the 7-TM domains of aGPCRs can be modulated by small molecules. Two examples thus far demonstrate the feasibility of this approach; beclomethasone diproprionate was identified as a small molecule agonist of ADGRG3/GPR97 (42) and dihydromunduletone was found to be an antagonist of G1 (150). As more pathways downstream of aGPCRs are elucidated, it is likely that additional aGPCR family members will be identified in drug screening efforts as targets of small molecules.
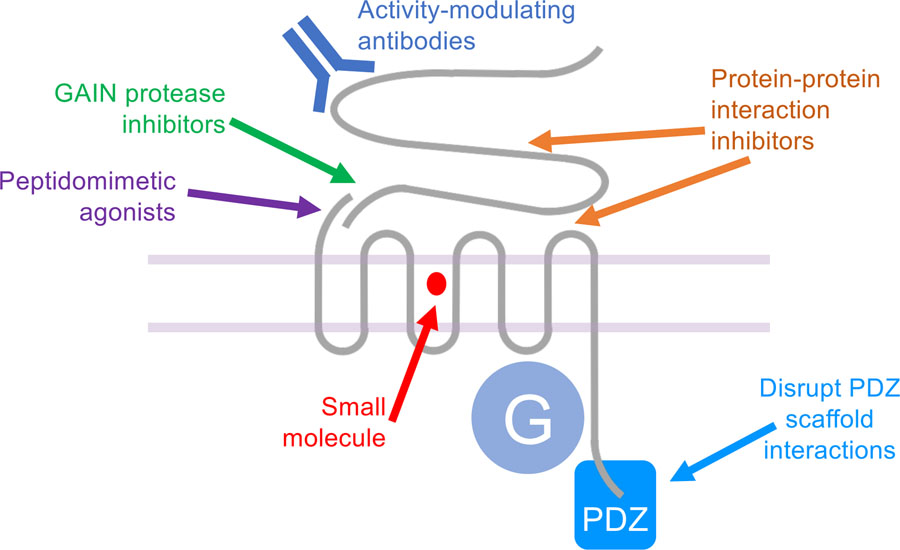
Figure 4. Targeting adhesion GPCRs with therapeutics.
Adhesion GPCRs offer many opportunities for targeted drug development, including i) small molecule agonists, antagonists or allosteric modulators (red), ii) peptides or peptidomimetic agonists (purple) that act at the tethered agonist interaction site, iii) modulators of GAIN proteolysis (green), iv) antibodies that modulate receptor activity (dark blue), and v) regulators of protein-protein interactions both extracellularly (orange) and intracellularly, for example with small molecules that disrupt binding to PDZ scaffold proteins (light blue).
The long ectodomains of aGPCRs provide opportunities for interventions that could alter receptor activity. First, the sizeable surfaces of aGPCR ectodomains are potential sites for antibody interaction. As mentioned previously, a polyclonal antibody directed at the ectodomain of G1 can activate heterotrimeric G protein signaling by the receptor (37). Similarly, ADGRE2/EMR2-mediated signaling in neutrophils is enhanced by antibodies recognizing the ADGRE2 N-terminus (151; 152). Presumably this agonistic action of anti-aGPCR antibodies occurs by inducing a rearrangement in the NTF to relieve its inhibition on signaling by the 7-TM domain, by promoting ectodomain shedding, and/or by changing the orientation of the tethered agonist peptide. Beyond the concept of antibodies acting as agonists or antagonists, antibody-drug conjugates have been successful in specifically delivering cytotoxic compounds to target cells in cancer (153). In such cases, an anti-aGPCR antibody that does not alter receptor activity could be useful as a therapeutic agent.
Many of the ligands identified thus far for aGPCRs are large transmembrane or extracellular matrix proteins, for example neurexins (56) and teneurins (59) for ADGRL1, collagens and laminins for ADGRG1 (74) and ADGRG6 (25; 32). Therefore, molecules that disrupt these protein-protein interactions could potentially regulate receptor activity. In addition, the aforementioned disinhibition model of aGPCR activity posits that the NTF exerts an inhibitory constraint on the 7-TM region, possibly through NTF interactions with the extracellular loops. If these interactions could be disrupted, this could also lead to receptor activation. While protein-protein interactions with large interfaces can be challenging to target therapeutically, drugs that disrupt protein-protein interaction interfaces have entered clinical trials and a number of advances have been made in this area in recent years (154). Progress in obtaining crystal structures of aGPCR ectodomains will likely aid in these efforts (7; 155). Moreover, intracellular protein-protein interactions could also be targeted by molecules that can cross the plasma membrane. One possible target for such molecules would be the disruption of PDZ scaffold protein interactions at the C terminus of aGPCRs. Approximately half of human aGPCRs have C-terminal PDZ binding motifs and a number of aGPCRs interact with PDZ proteins (19; 46; 156; 157). PDZ interactions are some of the most feasible protein-protein interfaces to target with small molecules because only the final few amino acids of the receptor C terminus are involved and act like a ligand in the binding pocket of the PDZ protein (158; 159).
The GAIN domain also provides multiple avenues for the therapeutic targeting of aGPCRs. First, because this domain acts as a protease, it should be possible to identify compounds that both inhibit and enhance ectodomain proteolysis. Protease inhibitors have been in the clinic since the early 1980s and are widely utilized to treat disorders such as hypertension, cancer and HIV infection (160). Second, a number of aGPCRs can be activated by peptides derived from the post-cleavage receptor stalk (stachel) sequence, and peptidomimetic drugs could presumably be developed to agonize these receptors as well. Further design of these molecules might also provide greater receptor specificity than can be attained with peptides, which have been shown to have extensive crossover activity at multiple receptors (34).
Conclusions
Adhesion G protein-coupled receptors have enormous potential as drug targets. Their large ectodomains and complex structural dynamics create a multitude of drug targeting possibilities in addition to their canonical heptahelical transmembrane domains. A growing body of work over the last two decades has established that this family of receptors is important in a wide range of physiological functions, yet there are currently no drugs used in the clinic that target these proteins. Recent advances in understanding both the physiological functions of many aGPCRs and the molecular dynamics that control their activation have brought the therapeutic targeting of these receptors closer to reality.
References
- 1.Roth BL, Kroeze WK. 2015. Integrated Approaches for Genome-wide Interrogation of the Druggable Non-olfactory G Protein-coupled Receptor Superfamily. J Biol Chem 290:19471–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hamann J, Aust G, Arac D, Engel FB, Formstone C, et al. 2015. International Union of Basic and Clinical Pharmacology. XCIV. Adhesion G protein-coupled receptors. Pharmacol Rev 67:338–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baud V, Chissoe SL, Viegas-Pequignot E, Diriong S, N’Guyen VC, et al. 1995. EMR1, an unusual member in the family of hormone receptors with seven transmembrane segments. Genomics 26:334–44 [DOI] [PubMed] [Google Scholar]
- 4.Hamann J, Eichler W, Hamann D, Kerstens HM, Poddighe PJ, et al. 1995. Expression cloning and chromosomal mapping of the leukocyte activation antigen CD97, a new seven-span transmembrane molecule of the secretion receptor superfamily with an unusual extracellular domain. J Immunol 155:1942–50 [PubMed] [Google Scholar]
- 5.Stacey M, Lin HH, Gordon S, McKnight AJ. 2000. LNB-TM7, a group of seven-transmembrane proteins related to family-B G-protein-coupled receptors. Trends Biochem Sci 25:284–9 [DOI] [PubMed] [Google Scholar]
- 6.Bjarnadottir TK, Fredriksson R, Hoglund PJ, Gloriam DE, Lagerstrom MC, Schioth HB. 2004. The human and mouse repertoire of the adhesion family of G-protein-coupled receptors. Genomics 84:23–33 [DOI] [PubMed] [Google Scholar]
- 7.Arac D, Boucard AA, Bolliger MF, Nguyen J, Soltis SM, et al. 2012. A novel evolutionarily conserved domain of cell-adhesion GPCRs mediates autoproteolysis. EMBO J 31:1364–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krasnoperov V, Lu Y, Buryanovsky L, Neubert TA, Ichtchenko K, Petrenko AG. 2002. Post-translational proteolytic processing of the calcium-independent receptor of alpha-latrotoxin (CIRL), a natural chimera of the cell adhesion protein and the G protein-coupled receptor. Role of the G protein-coupled receptor proteolysis site (GPS) motif. J Biol Chem 277:46518–26 [DOI] [PubMed] [Google Scholar]
- 9.McKnight AJ, Gordon S. 1998. The EGF-TM7 family: unusual structures at the leukocyte surface. J Leukoc Biol 63:271–80 [DOI] [PubMed] [Google Scholar]
- 10.Krishnan A, Nijmeijer S, de Graaf C, Schioth HB. 2016. Classification, Nomenclature, and Structural Aspects of Adhesion GPCRs. Handb Exp Pharmacol 234:15–41 [DOI] [PubMed] [Google Scholar]
- 11.Lin HH, Chang GW, Davies JQ, Stacey M, Harris J, Gordon S. 2004. Autocatalytic cleavage of the EMR2 receptor occurs at a conserved G protein-coupled receptor proteolytic site motif. J Biol Chem 279:31823–32 [DOI] [PubMed] [Google Scholar]
- 12.Kishore A, Purcell RH, Nassiri-Toosi Z, Hall RA. 2016. Stalk-dependent and Stalk-independent Signaling by the Adhesion G Protein-coupled Receptors GPR56 (ADGRG1) and BAI1 (ADGRB1). J Biol Chem 291:3385–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Promel S, Waller-Evans H, Dixon J, Zahn D, Colledge WH, et al. 2012. Characterization and functional study of a cluster of four highly conserved orphan adhesion-GPCR in mouse. Dev Dyn 241:1591–602 [DOI] [PubMed] [Google Scholar]
- 14.Peeters MC, Fokkelman M, Boogaard B, Egerod KL, van de Water B, et al. 2015. The adhesion G protein-coupled receptor G2 (ADGRG2/GPR64) constitutively activates SRE and NFkappaB and is involved in cell adhesion and migration. Cell Signal 27:2579–88 [DOI] [PubMed] [Google Scholar]
- 15.de Graaf C, Nijmeijer S, Wolf S, Ernst OP. 2016. 7TM Domain Structure of Adhesion GPCRs. Handb Exp Pharmacol 234:43–66 [DOI] [PubMed] [Google Scholar]
- 16.Rovati GE, Capra V, Neubig RR. 2007. The highly conserved DRY motif of class A G protein-coupled receptors: beyond the ground state. Mol Pharmacol 71:959–64 [DOI] [PubMed] [Google Scholar]
- 17.Pin JP, Galvez T, Prezeau L. 2003. Evolution, structure, and activation mechanism of family 3/C G-protein-coupled receptors. Pharmacol Ther 98:325–54 [DOI] [PubMed] [Google Scholar]
- 18.Paavola KJ, Stephenson JR, Ritter SL, Alter SP, Hall RA. 2011. The N terminus of the adhesion G protein-coupled receptor GPR56 controls receptor signaling activity. J Biol Chem 286:28914–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stephenson JR, Paavola KJ, Schaefer SA, Kaur B, Van Meir EG, Hall RA. 2013. Brain-specific angiogenesis inhibitor-1 signaling, regulation, and enrichment in the postsynaptic density. J Biol Chem 288:22248–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ward Y, Lake R, Yin JJ, Heger CD, Raffeld M, et al. 2011. LPA receptor heterodimerizes with CD97 to amplify LPA-initiated RHO-dependent signaling and invasion in prostate cancer cells. Cancer Res 71:7301–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okajima D, Kudo G, Yokota H. 2010. Brain-specific angiogenesis inhibitor 2 (BAI2) may be activated by proteolytic processing. J Recept Signal Transduct Res 30:143–53 [DOI] [PubMed] [Google Scholar]
- 22.Paavola KJ, Hall RA. 2012. Adhesion G protein-coupled receptors: signaling, pharmacology, and mechanisms of activation. Mol Pharmacol 82:777–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Demberg LM, Rothemund S, Schoneberg T, Liebscher I. 2015. Identification of the tethered peptide agonist of the adhesion G protein-coupled receptor GPR64/ADGRG2. Biochem Biophys Res Commun 464:743–7 [DOI] [PubMed] [Google Scholar]
- 24.Balenga N, Azimzadeh P, Hogue JA, Staats PN, Shi Y, et al. 2016. Orphan Adhesion GPCR GPR64/ADGRG2 Is Overexpressed in Parathyroid Tumors and Attenuates Calcium-Sensing Receptor-Mediated Signaling. J Bone Miner Res [DOI] [PMC free article] [PubMed]
- 25.Paavola KJ, Sidik H, Zuchero JB, Eckart M, Talbot WS. 2014. Type IV collagen is an activating ligand for the adhesion G protein-coupled receptor GPR126. Sci Signal 7:ra76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liebscher I, Schon J, Petersen SC, Fischer L, Auerbach N, et al. 2014. A tethered agonist within the ectodomain activates the adhesion G protein-coupled receptors GPR126 and GPR133. Cell Rep 9:2018–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stoveken HM, Hajduczok AG, Xu L, Tall GG. 2015. Adhesion G protein-coupled receptors are activated by exposure of a cryptic tethered agonist. Proc Natl Acad Sci U S A 112:6194–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu QX, Dong JH, Du HB, Zhang DL, Ren HZ, et al. 2014. Constitutive Galphai coupling activity of very large G protein-coupled receptor 1 (VLGR1) and its regulation by PDZD7 protein. J Biol Chem 289:24215–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liebscher I, Schoneberg T. 2016. Tethered Agonism: A Common Activation Mechanism of Adhesion GPCRs. Handb Exp Pharmacol 234:111–25 [DOI] [PubMed] [Google Scholar]
- 30.Scholz N, Monk KR, Kittel RJ, Langenhan T. 2016. Adhesion GPCRs as a Putative Class of Metabotropic Mechanosensors. Handb Exp Pharmacol 234:221–47 [DOI] [PubMed] [Google Scholar]
- 31.Wilde C, Fischer L, Lede V, Kirchberger J, Rothemund S, et al. 2016. The constitutive activity of the adhesion GPCR GPR114/ADGRG5 is mediated by its tethered agonist. FASEB J 30:666–73 [DOI] [PubMed] [Google Scholar]
- 32.Petersen SC, Luo R, Liebscher I, Giera S, Jeong SJ, et al. 2015. The adhesion GPCR GPR126 has distinct, domain-dependent functions in Schwann cell development mediated by interaction with laminin-211. Neuron 85:755–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scholz N, Gehring J, Guan C, Ljaschenko D, Fischer R, et al. 2015. The adhesion GPCR latrophilin/CIRL shapes mechanosensation. Cell Rep 11:866–74 [DOI] [PubMed] [Google Scholar]
- 34.Demberg LM, Winkler J, Wilde C, Simon KU, Schon J, et al. 2017. Activation of adhesion G protein-coupled receptors: agonist specificity of Stachel sequence-derived peptides. J Biol Chem [DOI] [PMC free article] [PubMed]
- 35.Lelianova VG, Davletov BA, Sterling A, Rahman MA, Grishin EV, et al. 1997. Alpha-latrotoxin receptor, latrophilin, is a novel member of the secretin family of G protein-coupled receptors. J Biol Chem 272:21504–8 [DOI] [PubMed] [Google Scholar]
- 36.Rahman MA, Ashton AC, Meunier FA, Davletov BA, Dolly JO, Ushkaryov YA. 1999. Norepinephrine exocytosis stimulated by alpha-latrotoxin requires both external and stored Ca2+ and is mediated by latrophilin, G proteins and phospholipase C. Philos Trans R Soc Lond B Biol Sci 354:379–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iguchi T, Sakata K, Yoshizaki K, Tago K, Mizuno N, Itoh H. 2008. Orphan G protein-coupled receptor GPR56 regulates neural progenitor cell migration via a G alpha 12/13 and Rho pathway. J Biol Chem 283:14469–78 [DOI] [PubMed] [Google Scholar]
- 38.Little KD, Hemler ME, Stipp CS. 2004. Dynamic regulation of a GPCR-tetraspanin-G protein complex on intact cells: central role of CD81 in facilitating GPR56-Galpha q/11 association. Mol Biol Cell 15:2375–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mogha A, Benesh AE, Patra C, Engel FB, Schoneberg T, et al. 2013. Gpr126 functions in Schwann cells to control differentiation and myelination via G-protein activation. J Neurosci 33:17976–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Daaka Y, Luttrell LM, Lefkowitz RJ. 1997. Switching of the coupling of the beta2-adrenergic receptor to different G proteins by protein kinase A. Nature 390:88–91 [DOI] [PubMed] [Google Scholar]
- 41.Mahon MJ, Donowitz M, Yun CC, Segre GV. 2002. Na(+)/H(+ ) exchanger regulatory factor 2 directs parathyroid hormone 1 receptor signalling. Nature 417:858–61 [DOI] [PubMed] [Google Scholar]
- 42.Gupte J, Swaminath G, Danao J, Tian H, Li Y, Wu X. 2012. Signaling property study of adhesion G-protein-coupled receptors. FEBS Lett 586:1214–9 [DOI] [PubMed] [Google Scholar]
- 43.Valtcheva N, Primorac A, Jurisic G, Hollmen M, Detmar M. 2013. The orphan adhesion G protein-coupled receptor GPR97 regulates migration of lymphatic endothelial cells via the small GTPases RhoA and Cdc42. J Biol Chem 288:35736–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peeters MC, Mos I, Lenselink EB, Lucchesi M, AP IJ, Schwartz TW. 2016. Getting from A to B-exploring the activation motifs of the class B adhesion G protein-coupled receptor subfamily G member 4/GPR112. FASEB J 30:1836–48 [DOI] [PubMed] [Google Scholar]
- 45.Park D, Tosello-Trampont AC, Elliott MR, Lu M, Haney LB, et al. 2007. BAI1 is an engulfment receptor for apoptotic cells upstream of the ELMO/Dock180/Rac module. Nature 450:430–4 [DOI] [PubMed] [Google Scholar]
- 46.Duman JG, Tzeng CP, Tu YK, Munjal T, Schwechter B, et al. 2013. The adhesion-GPCR BAI1 regulates synaptogenesis by controlling the recruitment of the Par3/Tiam1 polarity complex to synaptic sites. J Neurosci 33:6964–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Posokhova E, Shukla A, Seaman S, Volate S, Hilton MB, et al. 2015. GPR124 functions as a WNT7-specific coactivator of canonical beta-catenin signaling. Cell Rep 10:123–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shima Y, Kawaguchi SY, Kosaka K, Nakayama M, Hoshino M, et al. 2007. Opposing roles in neurite growth control by two seven-pass transmembrane cadherins. Nat Neurosci 10:963–9 [DOI] [PubMed] [Google Scholar]
- 49.Morgan R, El-Kadi AM, Theokli C. 2003. Flamingo, a cadherin-type receptor involved in the Drosophila planar polarity pathway, can block signaling via the canonical wnt pathway in Xenopus laevis. Int J Dev Biol 47:245–52 [PubMed] [Google Scholar]
- 50.Usui T, Shima Y, Shimada Y, Hirano S, Burgess RW, et al. 1999. Flamingo, a seven-pass transmembrane cadherin, regulates planar cell polarity under the control of Frizzled. Cell 98:585–95 [DOI] [PubMed] [Google Scholar]
- 51.Nishimura T, Honda H, Takeichi M. 2012. Planar cell polarity links axes of spatial dynamics in neural-tube closure. Cell 149:1084–97 [DOI] [PubMed] [Google Scholar]
- 52.Yates LL, Schnatwinkel C, Murdoch JN, Bogani D, Formstone CJ, et al. 2010. The PCP genes Celsr1 and Vangl2 are required for normal lung branching morphogenesis. Hum Mol Genet 19:2251–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shin D, Lin ST, Fu YH, Ptacek LJ. 2013. Very large G protein-coupled receptor 1 regulates myelin-associated glycoprotein via Galphas/Galphaq-mediated protein kinases A/C. Proc Natl Acad Sci U S A 110:19101–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krasnoperov VG, Bittner MA, Beavis R, Kuang Y, Salnikow KV, et al. 1997. alpha-Latrotoxin stimulates exocytosis by the interaction with a neuronal G-protein-coupled receptor. Neuron 18:925–37 [DOI] [PubMed] [Google Scholar]
- 55.Sugita S, Ichtchenko K, Khvotchev M, Sudhof TC. 1998. alpha-Latrotoxin receptor CIRL/latrophilin 1 (CL1) defines an unusual family of ubiquitous G-protein-linked receptors. G-protein coupling not required for triggering exocytosis. J Biol Chem 273:32715–24 [DOI] [PubMed] [Google Scholar]
- 56.Boucard AA, Ko J, Sudhof TC. 2012. High affinity neurexin binding to cell adhesion G-protein-coupled receptor CIRL1/latrophilin-1 produces an intercellular adhesion complex. J Biol Chem 287:9399–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.O’Sullivan ML, de Wit J, Savas JN, Comoletti D, Otto-Hitt S, et al. 2012. FLRT proteins are endogenous latrophilin ligands and regulate excitatory synapse development. Neuron 73:903–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Boucard AA, Maxeiner S, Sudhof TC. 2014. Latrophilins function as heterophilic cell-adhesion molecules by binding to teneurins: regulation by alternative splicing. J Biol Chem 289:387–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Silva JP, Lelianova VG, Ermolyuk YS, Vysokov N, Hitchen PG, et al. 2011. Latrophilin 1 and its endogenous ligand Lasso/teneurin-2 form a high-affinity transsynaptic receptor pair with signaling capabilities. Proc Natl Acad Sci U S A 108:12113–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jackson VA, Mehmood S, Chavent M, Roversi P, Carrasquero M, et al. 2016. Super-complexes of adhesion GPCRs and neural guidance receptors. Nat Commun 7:11184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Billings EA, Lee CS, Owen KA, D’Souza RS, Ravichandran KS, Casanova JE. 2016. The adhesion GPCR BAI1 mediates macrophage ROS production and microbicidal activity against Gram-negative bacteria. Sci Signal 9:ra14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Das S, Owen KA, Ly KT, Park D, Black SG, et al. 2011. Brain angiogenesis inhibitor 1 (BAI1) is a pattern recognition receptor that mediates macrophage binding and engulfment of Gram-negative bacteria. Proc Natl Acad Sci U S A 108:2136–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Koh JT, Kook H, Kee HJ, Seo YW, Jeong BC, et al. 2004. Extracellular fragment of brain-specific angiogenesis inhibitor 1 suppresses endothelial cell proliferation by blocking alphavbeta5 integrin. Exp Cell Res 294:172–84 [DOI] [PubMed] [Google Scholar]
- 64.Bolliger MF, Martinelli DC, Sudhof TC. 2011. The cell-adhesion G protein-coupled receptor BAI3 is a high-affinity receptor for C1q-like proteins. Proc Natl Acad Sci U S A 108:2534–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sigoillot SM, Iyer K, Binda F, Gonzalez-Calvo I, Talleur M, et al. 2015. The Secreted Protein C1QL1 and Its Receptor BAI3 Control the Synaptic Connectivity of Excitatory Inputs Converging on Cerebellar Purkinje Cells. Cell Rep [DOI] [PubMed]
- 66.Hamann J, Vogel B, van Schijndel GM, van Lier RA. 1996. The seven-span transmembrane receptor CD97 has a cellular ligand (CD55, DAF). J Exp Med 184:1185–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lin HH, Stacey M, Saxby C, Knott V, Chaudhry Y, et al. 2001. Molecular analysis of the epidermal growth factor-like short consensus repeat domain-mediated protein-protein interactions: dissection of the CD97-CD55 complex. J Biol Chem 276:24160–9 [DOI] [PubMed] [Google Scholar]
- 68.Stacey M, Chang GW, Davies JQ, Kwakkenbos MJ, Sanderson RD, et al. 2003. The epidermal growth factor-like domains of the human EMR2 receptor mediate cell attachment through chondroitin sulfate glycosaminoglycans. Blood 102:2916–24 [DOI] [PubMed] [Google Scholar]
- 69.Wandel E, Saalbach A, Sittig D, Gebhardt C, Aust G. 2012. Thy-1 (CD90) is an interacting partner for CD97 on activated endothelial cells. J Immunol 188:1442–50 [DOI] [PubMed] [Google Scholar]
- 70.Wang T, Ward Y, Tian L, Lake R, Guedez L, et al. 2005. CD97, an adhesion receptor on inflammatory cells, stimulates angiogenesis through binding integrin counterreceptors on endothelial cells. Blood 105:2836–44 [DOI] [PubMed] [Google Scholar]
- 71.Vallon M, Essler M. 2006. Proteolytically processed soluble tumor endothelial marker (TEM) 5 mediates endothelial cell survival during angiogenesis by linking integrin alpha(v)beta3 to glycosaminoglycans. J Biol Chem 281:34179–88 [DOI] [PubMed] [Google Scholar]
- 72.Xu L, Begum S, Hearn JD, Hynes RO. 2006. GPR56, an atypical G protein-coupled receptor, binds tissue transglutaminase, TG2, and inhibits melanoma tumor growth and metastasis. Proc Natl Acad Sci U S A 103:9023–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang L, Friedland S, Corson N, Xu L. 2014. GPR56 inhibits melanoma growth by internalizing and degrading its ligand TG2. Cancer Res 74:1022–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Luo R, Jeong SJ, Jin Z, Strokes N, Li S, Piao X. 2011. G protein-coupled receptor 56 and collagen III, a receptor-ligand pair, regulates cortical development and lamination. Proc Natl Acad Sci U S A 108:12925–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Luo R, Jeong SJ, Yang A, Wen M, Saslowsky DE, et al. 2014. Mechanism for adhesion G protein-coupled receptor GPR56-mediated RhoA activation induced by collagen III stimulation. PLoS One 9:e100043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kuffer A, Lakkaraju AK, Mogha A, Petersen SC, Airich K, et al. 2016. The prion protein is an agonistic ligand of the G protein-coupled receptor Adgrg6. Nature 536:464–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lee JW, Huang BX, Kwon H, Rashid MA, Kharebava G, et al. 2016. Orphan GPR110 (ADGRF1) targeted by N-docosahexaenoylethanolamine in development of neurons and cognitive function. Nat Commun 7:13123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ludwig MG, Seuwen K, Bridges JP. 2016. Adhesion GPCR Function in Pulmonary Development and Disease. Handb Exp Pharmacol 234:309–27 [DOI] [PubMed] [Google Scholar]
- 79.Fukuzawa T, Ishida J, Kato A, Ichinose T, Ariestanti DM, et al. 2013. Lung surfactant levels are regulated by Ig-Hepta/GPR116 by monitoring surfactant protein D. PLoS One 8:e69451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Overington JP, Al-Lazikani B, Hopkins AL. 2006. How many drug targets are there? Nat Rev Drug Discov 5:993–6 [DOI] [PubMed] [Google Scholar]
- 81.Piao X, Hill RS, Bodell A, Chang BS, Basel-Vanagaite L, et al. 2004. G protein-coupled receptor-dependent development of human frontal cortex. Science 303:2033–6 [DOI] [PubMed] [Google Scholar]
- 82.Chiang NY, Hsiao CC, Huang YS, Chen HY, Hsieh IJ, et al. 2011. Disease-associated GPR56 mutations cause bilateral frontoparietal polymicrogyria via multiple mechanisms. J Biol Chem 286:14215–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jin Z, Tietjen I, Bu L, Liu-Yesucevitz L, Gaur SK, et al. 2007. Disease-associated mutations affect GPR56 protein trafficking and cell surface expression. Hum Mol Genet 16:1972–85 [DOI] [PubMed] [Google Scholar]
- 84.Piao X, Chang BS, Bodell A, Woods K, Benzeev B, et al. 2005. Genotype-phenotype analysis of human frontoparietal polymicrogyria syndromes. Ann Neurol 58:680–7 [DOI] [PubMed] [Google Scholar]
- 85.Ackerman SD, Garcia C, Piao X, Gutmann DH, Monk KR. 2015. The adhesion GPCR Gpr56 regulates oligodendrocyte development via interactions with Galpha12/13 and RhoA. Nat Commun 6:6122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Giera S, Deng Y, Luo R, Ackerman SD, Mogha A, et al. 2015. The adhesion G protein-coupled receptor GPR56 is a cell-autonomous regulator of oligodendrocyte development. Nat Commun 6:6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.McMillan DR, White PC. 2010. Studies on the very large G protein-coupled receptor: from initial discovery to determining its role in sensorineural deafness in higher animals. Adv Exp Med Biol 706:76–86 [DOI] [PubMed] [Google Scholar]
- 88.McGee J, Goodyear RJ, McMillan DR, Stauffer EA, Holt JR, et al. 2006. The very large G-protein-coupled receptor VLGR1: a component of the ankle link complex required for the normal development of auditory hair bundles. J Neurosci 26:6543–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.van Wijk E, van der Zwaag B, Peters T, Zimmermann U, Te Brinke H, et al. 2006. The DFNB31 gene product whirlin connects to the Usher protein network in the cochlea and retina by direct association with USH2A and VLGR1. Hum Mol Genet 15:751–65 [DOI] [PubMed] [Google Scholar]
- 90.Weston MD, Luijendijk MW, Humphrey KD, Moller C, Kimberling WJ. 2004. Mutations in the VLGR1 gene implicate G-protein signaling in the pathogenesis of Usher syndrome type II. Am J Hum Genet 74:357–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Curtin JA, Quint E, Tsipouri V, Arkell RM, Cattanach B, et al. 2003. Mutation of Celsr1 disrupts planar polarity of inner ear hair cells and causes severe neural tube defects in the mouse. Curr Biol 13:1129–33 [DOI] [PubMed] [Google Scholar]
- 92.Robinson A, Escuin S, Doudney K, Vekemans M, Stevenson RE, et al. 2012. Mutations in the planar cell polarity genes CELSR1 and SCRIB are associated with the severe neural tube defect craniorachischisis. Hum Mutat 33:440–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tissir F, Goffinet AM. 2013. Shaping the nervous system: role of the core planar cell polarity genes. Nat Rev Neurosci 14:525–35 [DOI] [PubMed] [Google Scholar]
- 94.Thakar S, Wang L, Yu T, Ye M, Onishi K, et al. 2017. Evidence for opposing roles of Celsr3 and Vangl2 in glutamatergic synapse formation. Proc Natl Acad Sci U S A 114:E610–E8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Monk KR, Naylor SG, Glenn TD, Mercurio S, Perlin JR, et al. 2009. A G protein-coupled receptor is essential for Schwann cells to initiate myelination. Science 325:1402–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Monk KR, Oshima K, Jors S, Heller S, Talbot WS. 2011. Gpr126 is essential for peripheral nerve development and myelination in mammals. Development 138:2673–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Anderson KD, Pan L, Yang XM, Hughes VC, Walls JR, et al. 2011. Angiogenic sprouting into neural tissue requires Gpr124, an orphan G protein-coupled receptor. Proc Natl Acad Sci U S A 108:2807–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cullen M, Elzarrad MK, Seaman S, Zudaire E, Stevens J, et al. 2011. GPR124, an orphan G protein-coupled receptor, is required for CNS-specific vascularization and establishment of the blood-brain barrier. Proc Natl Acad Sci U S A 108:5759–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kuhnert F, Mancuso MR, Shamloo A, Wang HT, Choksi V, et al. 2010. Essential regulation of CNS angiogenesis by the orphan G protein-coupled receptor GPR124. Science 330:985–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Vanhollebeke B, Stone OA, Bostaille N, Cho C, Zhou Y, et al. 2015. Tip cell-specific requirement for an atypical Gpr124- and Reck-dependent Wnt/beta-catenin pathway during brain angiogenesis. Elife 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhou Y, Nathans J. 2014. Gpr124 controls CNS angiogenesis and blood-brain barrier integrity by promoting ligand-specific canonical wnt signaling. Dev Cell 31:248–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chang J, Mancuso MR, Maier C, Liang X, Yuki K, et al. 2017. Gpr124 is essential for blood-brain barrier integrity in central nervous system disease. Nat Med [DOI] [PMC free article] [PubMed]
- 103.Meza-Aguilar DG, Boucard AA. 2014. Latrophilins updated. Biomol Concepts 5:457–78 [DOI] [PubMed] [Google Scholar]
- 104.Arcos-Burgos M, Jain M, Acosta MT, Shively S, Stanescu H, et al. 2010. A common variant of the latrophilin 3 gene, LPHN3, confers susceptibility to ADHD and predicts effectiveness of stimulant medication. Mol Psychiatry 15:1053–66 [DOI] [PubMed] [Google Scholar]
- 105.Ribases M, Ramos-Quiroga JA, Sanchez-Mora C, Bosch R, Richarte V, et al. 2011. Contribution of LPHN3 to the genetic susceptibility to ADHD in adulthood: a replication study. Genes Brain Behav 10:149–57 [DOI] [PubMed] [Google Scholar]
- 106.Wallis D, Hill DS, Mendez IA, Abbott LC, Finnell RH, et al. 2012. Initial characterization of mice null for Lphn3, a gene implicated in ADHD and addiction. Brain Res 1463:85–92 [DOI] [PubMed] [Google Scholar]
- 107.O’Sullivan ML, Martini F, von Daake S, Comoletti D, Ghosh A. 2014. LPHN3, a presynaptic adhesion-GPCR implicated in ADHD, regulates the strength of neocortical layer 2/3 synaptic input to layer 5. Neural Dev 9:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lange M, Norton W, Coolen M, Chaminade M, Merker S, et al. 2012. The ADHD-linked gene Lphn3.1 controls locomotor activity and impulsivity in zebrafish. Mol Psychiatry 17:855. [DOI] [PubMed] [Google Scholar]
- 109.Zhu D, Li C, Swanson AM, Villalba RM, Guo J, et al. 2015. BAI1 regulates spatial learning and synaptic plasticity in the hippocampus. J Clin Invest 125:1497–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.DeRosse P, Lencz T, Burdick KE, Siris SG, Kane JM, Malhotra AK. 2008. The genetics of symptom-based phenotypes: toward a molecular classification of schizophrenia. Schizophr Bull 34:1047–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lanoue V, Usardi A, Sigoillot SM, Talleur M, Iyer K, et al. 2013. The adhesion-GPCR BAI3, a gene linked to psychiatric disorders, regulates dendrite morphogenesis in neurons. Mol Psychiatry 18:943–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kee HJ, Ahn KY, Choi KC, Won Song J, Heo T, et al. 2004. Expression of brain-specific angiogenesis inhibitor 3 (BAI3) in normal brain and implications for BAI3 in ischemia-induced brain angiogenesis and malignant glioma. FEBS Lett 569:307–16 [DOI] [PubMed] [Google Scholar]
- 113.Shiratsuchi T, Nishimori H, Ichise H, Nakamura Y, Tokino T. 1997. Cloning and characterization of BAI2 and BAI3, novel genes homologous to brain-specific angiogenesis inhibitor 1 (BAI1). Cytogenet Cell Genet 79:103–8 [DOI] [PubMed] [Google Scholar]
- 114.Okajima D, Kudo G, Yokota H. 2011. Antidepressant-like behavior in brain-specific angiogenesis inhibitor 2-deficient mice. J Physiol Sci 61:47–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Aust G, Zhu D, Van Meir EG, Xu L. 2016. Adhesion GPCRs in Tumorigenesis. Handb Exp Pharmacol 234:369–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.O’Hayre M, Vazquez-Prado J, Kufareva I, Stawiski EW, Handel TM, et al. 2013. The emerging mutational landscape of G proteins and G-protein-coupled receptors in cancer. Nat Rev Cancer 13:412–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kan Z, Jaiswal BS, Stinson J, Janakiraman V, Bhatt D, et al. 2010. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature 466:869–73 [DOI] [PubMed] [Google Scholar]
- 118.Zendman AJ, Cornelissen IM, Weidle UH, Ruiter DJ, van Muijen GN. 1999. TM7XN1, a novel human EGF-TM7-like cDNA, detected with mRNA differential display using human melanoma cell lines with different metastatic potential. FEBS Lett 446:292–8 [DOI] [PubMed] [Google Scholar]
- 119.Aust G, Eichler W, Laue S, Lehmann I, Heldin NE, et al. 1997. CD97: a dedifferentiation marker in human thyroid carcinomas. Cancer Res 57:1798–806 [PubMed] [Google Scholar]
- 120.Yang L, Chen G, Mohanty S, Scott G, Fazal F, et al. 2011. GPR56 Regulates VEGF production and angiogenesis during melanoma progression. Cancer Res 71:5558–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Weis SM, Cheresh DA. 2011. Tumor angiogenesis: molecular pathways and therapeutic targets. Nat Med 17:1359–70 [DOI] [PubMed] [Google Scholar]
- 122.Cork SM, Kaur B, Devi NS, Cooper L, Saltz JH, et al. 2012. A proprotein convertase/MMP-14 proteolytic cascade releases a novel 40 kDa vasculostatin from tumor suppressor BAI1. Oncogene 31:5144–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kaur B, Brat DJ, Devi NS, Van Meir EG. 2005. Vasculostatin, a proteolytic fragment of brain angiogenesis inhibitor 1, is an antiangiogenic and antitumorigenic factor. Oncogene 24:3632–42 [DOI] [PubMed] [Google Scholar]
- 124.Kaur B, Cork SM, Sandberg EM, Devi NS, Zhang Z, et al. 2009. Vasculostatin inhibits intracranial glioma growth and negatively regulates in vivo angiogenesis through a CD36-dependent mechanism. Cancer Res 69:1212–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zhu D, Hunter SB, Vertino PM, Van Meir EG. 2011. Overexpression of MBD2 in glioblastoma maintains epigenetic silencing and inhibits the antiangiogenic function of the tumor suppressor gene BAI1. Cancer Res 71:5859–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Safaee M, Clark AJ, Oh MC, Ivan ME, Bloch O, et al. 2013. Overexpression of CD97 confers an invasive phenotype in glioblastoma cells and is associated with decreased survival of glioblastoma patients. PLoS One 8:e62765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ward Y, Lake R, Martin PL, Killian K, Salerno P, et al. 2013. CD97 amplifies LPA receptor signaling and promotes thyroid cancer progression in a mouse model. Oncogene 32:2726–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Tang X, Jin R, Qu G, Wang X, Li Z, et al. 2013. GPR116, an adhesion G-protein-coupled receptor, promotes breast cancer metastasis via the Galphaq-p63RhoGEF-Rho GTPase pathway. Cancer Res 73:6206–18 [DOI] [PubMed] [Google Scholar]
- 129.Dieterich LC, Mellberg S, Langenkamp E, Zhang L, Zieba A, et al. 2012. Transcriptional profiling of human glioblastoma vessels indicates a key role of VEGF-A and TGFbeta2 in vascular abnormalization. J Pathol 228:378–90 [DOI] [PubMed] [Google Scholar]
- 130.Masiero M, Simoes FC, Han HD, Snell C, Peterkin T, et al. 2013. A core human primary tumor angiogenesis signature identifies the endothelial orphan receptor ELTD1 as a key regulator of angiogenesis. Cancer Cell 24:229–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Towner RA, Jensen RL, Colman H, Vaillant B, Smith N, et al. 2013. ELTD1, a potential new biomarker for gliomas. Neurosurgery 72:77–90; discussion 1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bayin NS, Frenster JD, Kane JR, Rubenstein J, Modrek AS, et al. 2016. GPR133 (ADGRD1), an adhesion G-protein-coupled receptor, is necessary for glioblastoma growth. Oncogenesis 5:e263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hamann J, Hsiao CC, Lee CS, Ravichandran KS, Lin HH. 2016. Adhesion GPCRs as Modulators of Immune Cell Function. Handb Exp Pharmacol 234:329–50 [DOI] [PubMed] [Google Scholar]
- 134.Boyden SE, Desai A, Cruse G, Young ML, Bolan HC, et al. 2016. Vibratory Urticaria Associated with a Missense Variant in ADGRE2. N Engl J Med 374:656–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Capasso M, Durrant LG, Stacey M, Gordon S, Ramage J, Spendlove I. 2006. Costimulation via CD55 on human CD4+ T cells mediated by CD97. J Immunol 177:1070–7 [DOI] [PubMed] [Google Scholar]
- 136.Waller-Evans H, Promel S, Langenhan T, Dixon J, Zahn D, et al. 2010. The orphan adhesion-GPCR GPR126 is required for embryonic development in the mouse. PLoS One 5:e14047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Patra C, van Amerongen MJ, Ghosh S, Ricciardi F, Sajjad A, et al. 2013. Organ-specific function of adhesion G protein-coupled receptor GPR126 is domain-dependent. Proc Natl Acad Sci U S A 110:16898–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Xiao J, Jiang H, Zhang R, Fan G, Zhang Y, et al. 2012. Augmented cardiac hypertrophy in response to pressure overload in mice lacking ELTD1. PLoS One 7:e35779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Musa G, Engel FB, Niaudet C. 2016. Heart Development, Angiogenesis, and Blood-Brain Barrier Function Is Modulated by Adhesion GPCRs. Handb Exp Pharmacol 234:351–68 [DOI] [PubMed] [Google Scholar]
- 140.Doyle SE, Scholz MJ, Greer KA, Hubbard AD, Darnell DK, et al. 2006. Latrophilin-2 is a novel component of the epithelial-mesenchymal transition within the atrioventricular canal of the embryonic chicken heart. Dev Dyn 235:3213–21 [DOI] [PubMed] [Google Scholar]
- 141.Bridges JP, Ludwig MG, Mueller M, Kinzel B, Sato A, et al. 2013. Orphan G protein-coupled receptor GPR116 regulates pulmonary surfactant pool size. Am J Respir Cell Mol Biol 49:348–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Yang MY, Hilton MB, Seaman S, Haines DC, Nagashima K, et al. 2013. Essential regulation of lung surfactant homeostasis by the orphan G protein-coupled receptor GPR116. Cell Rep 3:1457–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Hochreiter-Hufford AE, Lee CS, Kinchen JM, Sokolowski JD, Arandjelovic S, et al. 2013. Phosphatidylserine receptor BAI1 and apoptotic cells as new promoters of myoblast fusion. Nature 497:263–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hamoud N, Tran V, Croteau LP, Kania A, Cote JF. 2014. G-protein coupled receptor BAI3 promotes myoblast fusion in vertebrates. Proc Natl Acad Sci U S A 111:3745–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.White JP, Wrann CD, Rao RR, Nair SK, Jedrychowski MP, et al. 2014. G protein-coupled receptor 56 regulates mechanical overload-induced muscle hypertrophy. Proc Natl Acad Sci U S A 111:15756–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Ravenscroft G, Nolent F, Rajagopalan S, Meireles AM, Paavola KJ, et al. 2015. Mutations of GPR126 are responsible for severe arthrogryposis multiplex congenita. Am J Hum Genet 96:955–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kou I, Takahashi Y, Johnson TA, Takahashi A, Guo L, et al. 2013. Genetic variants in GPR126 are associated with adolescent idiopathic scoliosis. Nat Genet 45:676–9 [DOI] [PubMed] [Google Scholar]
- 148.Xu JF, Yang GH, Pan XH, Zhang SJ, Zhao C, et al. 2015. Association of GPR126 gene polymorphism with adolescent idiopathic scoliosis in Chinese populations. Genomics 105:101–7 [DOI] [PubMed] [Google Scholar]
- 149.Davies B, Baumann C, Kirchhoff C, Ivell R, Nubbemeyer R, et al. 2004. Targeted deletion of the epididymal receptor HE6 results in fluid dysregulation and male infertility. Mol Cell Biol 24:8642–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Stoveken HM, Bahr LL, Anders MW, Wojtovich AP, Smrcka AV, Tall GG. 2016. Dihydromunduletone Is a Small-Molecule Selective Adhesion G Protein-Coupled Receptor Antagonist. Mol Pharmacol 90:214–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Huang YS, Chiang NY, Hu CH, Hsiao CC, Cheng KF, et al. 2012. Activation of myeloid cell-specific adhesion class G protein-coupled receptor EMR2 via ligation-induced translocation and interaction of receptor subunits in lipid raft microdomains. Mol Cell Biol 32:1408–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Yona S, Lin HH, Dri P, Davies JQ, Hayhoe RP, et al. 2008. Ligation of the adhesion-GPCR EMR2 regulates human neutrophil function. FASEB J 22:741–51 [DOI] [PubMed] [Google Scholar]
- 153.Zolot RS, Basu S, Million RP. 2013. Antibody-drug conjugates. Nat Rev Drug Discov 12:259–60 [DOI] [PubMed] [Google Scholar]
- 154.Arkin MR, Tang Y, Wells JA. 2014. Small-molecule inhibitors of protein-protein interactions: progressing toward the reality. Chem Biol 21:1102–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Salzman GS, Ackerman SD, Ding C, Koide A, Leon K, et al. 2016. Structural Basis for Regulation of GPR56/ADGRG1 by Its Alternatively Spliced Extracellular Domains. Neuron 91:1292–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Tobaben S, Sudhof TC, Stahl B. 2000. The G protein-coupled receptor CL1 interacts directly with proteins of the Shank family. J Biol Chem 275:36204–10 [DOI] [PubMed] [Google Scholar]
- 157.Kreienkamp HJ, Zitzer H, Gundelfinger ED, Richter D, Bockers TM. 2000. The calcium-independent receptor for alpha-latrotoxin from human and rodent brains interacts with members of the ProSAP/SSTRIP/Shank family of multidomain proteins. J Biol Chem 275:32387–90 [DOI] [PubMed] [Google Scholar]
- 158.Blazer LL, Neubig RR. 2009. Small molecule protein-protein interaction inhibitors as CNS therapeutic agents: current progress and future hurdles. Neuropsychopharmacology 34:126–41 [DOI] [PubMed] [Google Scholar]
- 159.Wang NX, Lee HJ, Zheng JJ. 2008. Therapeutic use of PDZ protein-protein interaction antagonism. Drug News Perspect 21:137–41 [PMC free article] [PubMed] [Google Scholar]
- 160.Drag M, Salvesen GS. 2010. Emerging principles in protease-based drug discovery. Nat Rev Drug Discov 9:690–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Bohnekamp J, Schoneberg T. 2011. Cell adhesion receptor GPR133 couples to Gs protein. J Biol Chem 286:41912–6 [DOI] [PMC free article] [PubMed] [Google Scholar]