Type II toxin-antitoxin (TA) systems are small genetic elements composed of a toxic protein and its cognate antitoxin protein, the latter counteracting the toxicity of the former. While TA systems were initially discovered on plasmids, functioning as addiction modules through a phenomenon called postsegregational killing, they were later shown to be massively present in bacterial chromosomes, often in association with mobile genetic elements. Extensive research has been conducted in recent decades to better understand the physiological roles of these chromosomally encoded modules and to characterize the conditions leading to their activation.
KEYWORDS: persistence, proteolysis, stress responses, toxin-antitoxin, transcriptional regulation
ABSTRACT
Type II toxin-antitoxin (TA) systems are small genetic elements composed of a toxic protein and its cognate antitoxin protein, the latter counteracting the toxicity of the former. While TA systems were initially discovered on plasmids, functioning as addiction modules through a phenomenon called postsegregational killing, they were later shown to be massively present in bacterial chromosomes, often in association with mobile genetic elements. Extensive research has been conducted in recent decades to better understand the physiological roles of these chromosomally encoded modules and to characterize the conditions leading to their activation. The diversity of their proposed roles, ranging from genomic stabilization and abortive phage infection to stress modulation and antibiotic persistence, in conjunction with the poor understanding of TA system regulation, resulted in the generation of simplistic models, often refuted by contradictory results. This review provides an epistemological and critical retrospective on TA modules and highlights fundamental questions concerning their roles and regulations that still remain unanswered.
INTRODUCTION
In 1983, while assigning functions to the open reading frames (ORFs) of the F plasmid, the Couturier group mapped a conditional lethal amber mutation to a novel ORF close to the replication origin of this plasmid and encoding a polypeptide named H (1). This mutation led to the induction of resident prophages in a recA-dependent manner (1). Interestingly, suppressor mutations could be mapped to the neighboring gene encoding a polypeptide named G (1). The authors concluded that the G protein is a potent inducer of the SOS response, whose action is negatively controlled by the H polypeptide (1). A simultaneously released publication from Ogura and Hiraga showed that these two genes were able to inhibit cell division when plasmid replication was hampered and the copy number decreased (2). These authors concluded that this locus promotes stable plasmid maintenance by coupling plasmid replication to host cell division (2). They named these two genes ccdA (H-encoding gene) and ccdB (G-encoding gene) for coupled cell division (2). In 1985, Jaffé, in collaboration with the group of Hiraga, showed that the ccd locus greatly reduced the viability of cells that failed to inherit a plasmid copy during division and proposed the “nonviable segregant model” (3, 4) (Fig. 1). The ccd locus was then defined as control of cell death (5). These genes constitute the first identified toxin-antitoxin (TA) pair, although this term was first used much later (6). Subsequent studies from the Couturier and Horiuchi groups concomitantly showed that the CcdB protein poisons DNA gyrase much like quinolone antibiotics, leading to the generation of double-strand breaks and induction of the SOS response (7–10). This provided the link with earlier observations showing that the ccd locus induces resident prophages and produces long nonviable plasmid-free filaments (1, 3). CcdA was shown to inhibit this DNA-damaging activity by directly interacting with CcdB (11, 12). CcdA was also shown to be unstable due to constitutive degradation by the Lon ATP-dependent protease, refining the earlier model proposed by Jaffé et al. (5, 13). Cells devoid of the plasmid would stop synthesizing the Ccd proteins. CcdA would then be degraded and not replenished, leading to the liberation of CcdB and killing of plasmid-free segregants (Fig. 1A) (3, 13). Analogous systems located on different plasmids and phages were described concurrently, i.e., hok-sok (parB) and kis-kid (parD) on plasmid R1, pem on plasmid R100 (which proved to be identical to parD), stbDE on plasmid R485, parDE on plasmid RK2, and phd-doc on bacteriophage P1 (14–19). The mechanism by which TA systems kill plasmid-free cells is known as “postsegregational killing” (PSK) (Fig. 1A), and TAs themselves were referenced to as addiction modules (14, 20). Over the years, additional TAs were identified on plasmids but also on chromosomes (21–24). They were divided into different classes depending on the nature and mode of action of the antitoxin, the toxin always being a protein (for reviews, see references 25 and 26). This minireview will focus on type II TA systems in which both components are proteins. This class of TAs appears to be the most abundant in bacterial genomes, being heavily represented in mobile genetic elements such as plasmids and phages but also in bacterial chromosomes (21–24). Since TA systems were described as stabilizers of mobile DNA, those encoded on chromosomes piqued the curiosity of the microbiology community and the study of plasmid TAs became neglected to the profit of chromosomally encoded ones (27).
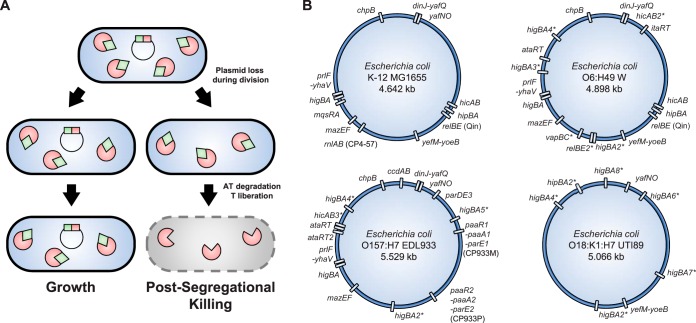
FIG 1.
Type II TA systems, postsegregational killing and distribution. (A) Nonviable segregant or postsegregational killing model. TA genes, as well as proteins, are represented in red (toxins) and green (antitoxins). Rectangles denote TA genes encoded on a plasmid, and round shapes denote TA proteins produced from these genes. A TA-encoding plasmid can be lost during division in a way that one of the daughter cells does not inherit a plasmid copy. In these cells, TA proteins cannot be replenished due to the absence of TA genes. Since the antitoxin is degraded while its cognate toxin is stable, the free toxin concentration will increase, exert its activity, and, in time, induce cell death, therefore killing plasmid-free segregants. (B) Distribution of type II TA systems in various E. coli reference strains generated by TAfinder (23). Asterisks indicate systems that were not validated experimentally. Parentheses include name of the prophage a TA is encoded on when applicable. The strains are MG1655 (NCBI U00096.3), a common lab strain from phylogroup A; W (CP002967.1), a soil isolate from phylogroup B1; EDL933 (AE005174.2), an enterohemorrhagic pathogen from phylogroup E; and UTI89 (CP000243.1), a uropathogen from phylogroup B2. No TA systems are conserved within these four distantly related E. coli strains.
TA systems are abundant and part of the prokaryotic accessory genome.
With the advent of the genomic era, TA systems were being discovered in nearly every bacterial chromosome and, moreover, often in multiple copies. A first report was published in 2005 by the Gerdes lab in which a total of 126 bacterial and archaeal chromosomes were analyzed (21). At this time, only 5 and 38 type II TA systems were identified in Escherichia coli K-12 and Mycobacterium tuberculosis H37Rv, respectively (21). Several studies followed up on this initial report, mining hundreds of genomes and leading to the detection of thousands of TA systems in bacterial and archaeal genomes, as well as to the identification of novel families (22, 24). As the number of annotated TA systems has grown exponentially, it is now recognized that prokaryotic genomes are literally invaded by these systems. For example, 12 and 76 type II TA systems are currently detected in E. coli K-12 and M. tuberculosis H37Rv, respectively (23) (Fig. 1B).
One of these genomic studies also observed that obligate intracellular bacteria tend to harbor none or fewer TA pairs compared to related free-living species (21). Pandey and Gerdes argue that this observation supports a role for TA systems in stress responses, the rationale being the following: as obligate intracellular species thrive in stable niches and do not need to cope with changing environments, the selective pressure on TA systems is reduced, and these modules are eliminated through evolution (21). The authors also argued that, in addition to TA systems, obligate intracellular bacteria have also lost the genes encoding enzymes regulating the metabolism of (p)ppGpp, an alarmone involved in the stringent response, which is activated when cells are deprived of nutrients like amino acids (21). This observation thus supported an implication of TA systems in stress responses, as well as a functional link between these systems and (p)ppGpp (21). Intriguingly, the small genomes of intracellular bacteria consist of a stable chromosome in which the proportion of accessory genes is drastically reduced compared to free-living organisms. They are mostly devoid of mobile genetic elements such as plasmids, phages, transposons or integrative and conjugative elements, which are assumed to be lost during the reductive genomic processes that characterize the evolution of these intracellular species (28). We thus argue that this observation indicates that TAs are intimately linked to mobile DNA elements as proposed in (22, 24). For example, the rnlAB and relBE modules are located on the CP4-57 and Qin cryptic prophages of E. coli K-12, respectively (29, 30) (Fig. 1B). Enterohemorrhagic E. coli O157:H7 encodes two homologous three-component paaR-paaA-parE systems in cryptic prophages CP933M and CP933P (Fig. 1B) (31). Comparison of the TA distribution among the E. coli chromosomes shows that TA content is highly variable from one isolate to another, with few to no systems being universally conserved in a given species (32, 33) (Fig. 1B). These observations suggest that chromosomal TAs are acquired by horizontal gene transfer and establish independently or as part of genomic islands (32, 33). Other studies aiming at building an inventory of the TA repertoire encoded on various E. coli chromosomes showed that their locations are conserved and that some of these systems are degenerating (32, 34). This decay—reflected by deletion or accumulation of nonsense mutations—would indicate that TA systems are progressively lost during evolution. The reasons for decay might be multiple and will be discussed in light of their proposed functions below.
PROPOSED FUNCTIONS: THE MANY FACES OF TA SYSTEMS
The detection of TA systems on chromosomes naturally led to the idea that these systems could have functions beyond plasmid stabilization. Here, we briefly describe the proposed functions of TA systems in E. coli, unless otherwise specified, and the recent tumultuous developments in that respect (for more general reviews on TA systems, see references 25 and 26).
TA systems as stress response modules.
Early work on chromosomal TA systems showed that the activities of the RelE and MazF toxins in E. coli K-12 were triggered by amino acid starvation, leading to reduced translation during nutritional stress in a way that is independent of the (p)ppGpp alarmone (35, 36). This model was supported by the observation that amino acid starvation triggers mRNA cleavage in a mazEF- and relBE-dependent way (35, 36). Since RelE and MazE-induced toxicities are bacteriostatic and can be relieved by the ssrA tmRNA, bacteria could recover upon relief of the nutritional stress (36, 37). The authors proposed that in contrast to the stringent response, which inhibits translation indirectly, activation of TA systems would directly reduce translation and assist the stringent response in keeping energy consumption low under starvation conditions (35). In another series of studies, various stress conditions, such as nutritional stress, antibiotic exposure, oxidative stress, thymineless death, high temperature, and extracellular death factor (a linear signaling pentapeptide), were shown to mediate programmed cell death (PCD) solely through activation of the mazEF system in a (p)ppGpp-dependent way (38–43), in total disagreement with the data concerning mazEF involvement in nutritional stress management. Another series of studies claimed that the MazF RNase induces specific cleavages that can remodel cell physiology (44, 45). Transcriptome sequencing (RNA-seq) showed that MazF cleaves many messengers at or closely upstream of their translation initiation codon, thus generating leaderless mRNAs (44). This pool of leaderless transcripts was shown to be translated by a subpopulation of specialized ribosomes which were deprived of their anti-Shine-Dalgarno sequence by cleavage of the 23S rRNA by MazF (44). This was later shown to profoundly remodel the proteome of E. coli and to reflect a response induced by MazF when this system is activated under stress conditions (45). However, two independent studies failed to detect either leaderless transcripts or specialized ribosomes by RNA-seq (46, 47). Indeed, MazF was shown to cleave most messengers at multiple sites in their coding sequences as well as rRNA precursors, thus inhibiting ribosome biogenesis (46, 47). Reanalysis of previously published proteomic data using more powerful statistical analyses also failed to detect proteins upregulated in a MazF-dependent way during stress (48). Therefore, MazF was proposed to generally inhibit translation by cleaving transcripts indiscriminately and by cleaving rRNA precursors (46–49). The theory that TAs provide competitiveness under stress conditions was also put into question since deletion of five TA systems, including relBE and mazEF, did not cause any fitness deficiency under various stress conditions, such as nalidixic acid or rifampin exposure, amino acid starvation, nutritional downshift, low pH, or long-term stationary phase (50). Later on, the mqsRA system in E. coli was found to regulate a variety of biological processes such as biofilm formation and response to oxidative stress, notably by the capacity of the antitoxin MqsA to regulate expression of master regulators such as csgD or rpoS (51, 52). However, none of these phenotypes or regulations could be reproduced in an independent study (53). All of these observations call into question the implication of TA modules in various stress responses, whether through PCD, translation inhibition, or pleiotropic regulation.
Another pivotal role for TA systems consisted in the generation of persister cells, a subpopulation of supposedly dormant cells that is able to survive antimicrobial treatments. This hypothesis stemmed from the fact that missense mutations in the hipBA system induce a high proportion of persister cells and that other TA systems (dinJ-yafQ, mazEF, mqsRA, relBE, and yefM-yoeB) were highly expressed in populations enriched in persisters (54–56). A key study supporting this model showed that successive deletions of ten mRNase TA modules in E. coli K-12, which are among the most studied chromosomal TAs, lead to a proportional decrease in survival upon antimicrobial treatment (57). Complementary single-cell analyses based on fluorescent reporters concluded that antibiotic persister cells constitute a subpopulation in which the stringent response and TA systems are activated (58). This model was further refined by a study placing the hipBA module upstream and as a master regulator of the signaling cascade leading to persistence, proposing that stochastic activation of hipBA would lead to glutamate starvation since HipA inhibits glutamyl-tRNA charging (59). This would, in turn, activate the 10 TA-encoded mRNase toxins in a (p)ppGpp, polyphosphate and Lon-dependent manner, shutting down translation and leading to dormancy and antibiotic tolerance (58, 59). This model was, however, later refuted by multiple studies due to major experimental flaws (60–62). While these findings resolved the controversy surrounding the role of TA systems in antibiotic persistence in E. coli, a parallel story emerged in Salmonella enterica. Helaine et al. showed that, upon phagocytosis of S. enterica by macrophages, transient acidification and nutrient starvation would trigger activation of all 14 type II TAs, resulting in the generation of antibiotic persisters (63). An independent study was unable to observe reduced antibiotic persistence of TA-deleted S. enterica grown in medium mimicking conditions encountered in phagosomes such as low magnesium, amino-acid starvation or acid shock (64). A mutant deleted for three TA systems (ecnB, shpAB, and phd-doc) was also found to have no effect on virulence and persistence in a murine model (65). Further research will be required to clarify whether TA systems are involved in persistence into the host and whether they are involved in virulence, as is often claimed, although with very little mechanistic evidence (63, 66–68) or with experimental flaws, i.e., by ectopically overexpressing a toxin to study its effect on persistence (69–72; for a review, see reference 73).
As a conclusion, the lack of solid evidence to support the involvement of TA systems in the regulation of bacterial physiology, in spite of the attention accorded to these modules in the last 10 years, suggests that chromosomally encoded TA systems might provide other functions to their hosts.
TA systems as selfish genes driving competition between replicons.
Alternative TA system functions, in accordance with their mobile and addictive nature, have been proposed. Similar to what is observed for plasmids, TAs were shown to stabilize large genomic elements such as superintegron arrays in chromosome II of Vibrio cholerae in the absence of selection, potentially providing a fitness advantage when coping with changing environments (74). Similarly, some TAs promote maintenance of genetic mobile elements such as cryptic prophages and conjugative transposons: for instance, mosAT stabilizes the SXT conjugative transposon of V. cholerae (75) and paaR2-paaA2-parE2 is thought to stabilize the CP933P prophage of E. coli O157:H7 (31).
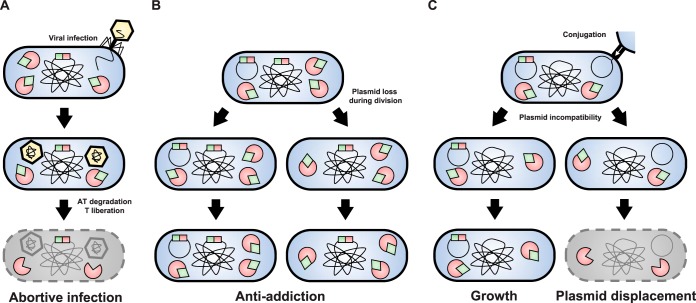
Research published by Fineran and colleagues showed that type III and IV TA modules function as “abortive infection” systems, inducing cell death in cells infected by phages to prevent infections from spreading (Fig. 2A) (76–78). In E. coli, similar mechanisms could involve type II TAs as a deletion of mazEF leads to increased P1 infection loads (79). Interestingly, the T4 phage harbors the dmd gene, which encodes a promiscuous antitoxin for two type II toxins: RnlA from the CP4-57 cryptic prophage of E. coli K-12 and LsoA from the pOSAK1_02 plasmid carried by E. coli O157:H7 (80). The rnlAB and lsoAB systems strongly reduce the growth of a dmd mutant of T4, suggesting that these two systems protect their hosts against phage infection (80). The ability of T4 to circumvent these two systems through dmd also suggests that phages and their hosts are involved in a continuous and ever-evolving arms race in which TA systems might play a significant role.
FIG 2.
Roles of TA systems regarding mobile genetic elements. TA genes, as well as proteins, are represented in red (toxins) and green (antitoxins). (A) Protection against phages. Some TA systems have been shown to contribute to viral defense through a process known as “abortive infection.” Viral infection would lead to a molar excess of toxin over its cognate antitoxin by yet-unknown mechanisms, leading to the killing of infected cells, preventing phage replication and propagation. (B) Antiaddiction. Chromosomal homologs of plasmid-encoded TA systems can cross-neutralize their toxic activities. Therefore, failure to inherit a TA-encoding plasmid will not lead to postsegregational killing if a homologous TA system is encoded on the chromosome. (C) Plasmid displacement. Cells that acquire more than one plasmid from the same incompatibility group through conjugation will partition these plasmids in different daughter cells. If one of such plasmids encodes a TA system, cells that fail to inherit this plasmid will still contain TA proteins in its cytoplasm and will be killed by postsegregational killing.
Similarly, chromosomal TAs were proposed to protect cells from addiction by neutralizing plasmid-encoded homologous toxins (Fig. 2B). Specifically, the chromosomally encoded CcdA antitoxin of Erwinia chrysanthemi was shown to protect cells against the toxic effect of its F plasmid-encoded CcdB toxin homolog upon plasmid loss (81). Similarly, the ataRT system in the E. coli O157:H7 chromosome was shown to counteract toxicity of a plasmid-encoded homologous system, indicating that this system might play an antiaddictive role as well (82). Consequently, integration of TA systems from plasmids into chromosomes could be selected as a way to free a host from the burden of addiction (33, 83). The ability to neutralize addictive TAs could lead, in turn, to the selection of plasmid-encoded TAs that can circumvent neutralization by their chromosomal homologs, thus driving an arms race between the host and its accessory replicons and accounting for the diversity and multiplicity of TA systems, similarly to that proposed for restriction-modification (RM) systems (83, 84). As a result, chromosomal TA systems that would lose the capacity to counteract newly selected plasmid TA system will fall under neutral selection, albeit at a lower rate than other genes due to their addictive properties, explaining the decay previously observed for some of these systems (32, 34).
An interesting model suggested that TA systems could mediate competition between incompatible plasmids for the same host through PSK (Fig. 2C). TA systems allow a conjugative plasmid (TA+ plasmid) to outcompete a conjugative plasmid belonging to the same incompatibility group (identical replicon) but devoid of the TA system (TA− plasmid). Cells that do not inherit a TA-encoding plasmid during this “conflict” will be subjected to PSK provided they do not carry a copy of this TA (Fig. 2C) (85). Therefore, TA-encoding plasmids can mediate the displacement of plasmids devoid of TA, ensuring both their vertical and horizontal inheritance through the same addiction mechanism (85). Interestingly, similar displacement mechanisms were observed for plasmids encoding RM systems. Like RM systems, TA modules are refractory to gene efflux at the expense of their hosts and were dubbed “selfish genes” (84–88). In fact, plasmid stabilization, displacement and antiaddiction might just be the consequence of the selfish properties of TA systems. Selfishness might allow chromosomal TA systems to subsist in their host without providing any function. Time and genetic drift may lead to the appearance of inactive toxin mutants, explaining why some of these systems are under negative selection and slowly decaying (32, 34).
ACTIVATION OF TA SYSTEMS: FROM REGULATION TO PHENOTYPES
Transcriptional regulation of TA systems.
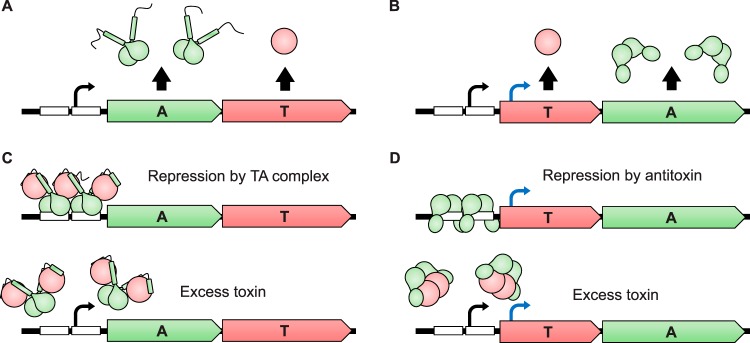
Although multiple functions were proposed for TA systems as described above, TA expression and regulation have been given little attention despite their proposed importance in bacterial physiology. It is important to note that among the 12 type II TA systems characterized in E. coli, eight adopt the “canonical” genetic organization in which the antitoxin gene precedes the toxin gene (Fig. 3A), while the four others (rnlAB, mqsRA, higBA, and hicAB) are in “reverse” organization with the toxin gene being upstream of the antitoxin gene (89) (Fig. 3B). In both configurations, TA systems are transcribed as bicistronic operons by an autoregulated promoter (89) (Fig. 3A and B). In the case of “reverse” systems, additional promoters have been shown to play a role in antitoxin expression (Fig. 3B). For instance, the rnlA and mqsA antitoxin genes are transcribed from additional constitutive promoters located in the toxin coding sequence (53, 90). Another layer of complexity is found in the hicAB system: the hicAB autoregulated promoter produces a transcript truncated at the position of the hicA ribosome-binding site, thus only supporting synthesis of the HicB antitoxin (91). Another constitutive promoter upstream of the former transcribes a messenger supporting translation of the whole system (91).
FIG 3.
Transcriptional regulation of type II TA systems. TA genes, as well as proteins, are represented in red (toxins) and green (antitoxins). (A) Transcription of canonically organized TA systems. The whole operon (antitoxin-toxin) is transcribed by a single autoregulated promoter. A lower translational efficiency of toxins ensures a molar excess of antitoxin. (B) Transcription of reverse-organized TA systems. The whole operon is generally transcribed by a single autoregulated promoter (black arrow). A molar excess of antitoxin is ensured through its exclusive transcription by other promoters (blue arrow). (C) Conditional cooperativity. Unsaturated toxin-antitoxin complexes tightly bind their operators (white boxes) to repress transcription. A molar excess of toxin leads to the formation of saturated complexes that do not bind their operators, leading to derepression of the promoter, transcription of the operon, and de novo antitoxin synthesis. (D) Repression of reverse-organized systems. Excess antitoxin binds operators (white boxes) to repress transcription of the whole operon while the antitoxin gene is still transcribed. A molar excess of toxin displaces the antitoxin from its operators, leading to derepression of the autoregulated promoter (black arrow) and transcription of the whole operon.
TA system expression is tightly autoregulated. Antitoxins generally comprise a DNA-binding domain that binds inverted repeats located in the operon promoter to repress transcription of the system, with low affinity in most described cases (92–95). This affinity can be enhanced by forming a complex with its cognate toxin (92–95). Toxin binding also allows cooperative recruitment of additional antitoxins to remaining operators, strengthening the repression (96) (Fig. 3C). Exceeding a certain toxin/antitoxin ratio will induce the formation of saturated complexes that cannot bind their operators (96, 97) (Fig. 3C). Since antitoxins bind cooperatively to their operators but only under specific conditions, i.e., in a range of toxin/antitoxin ratios, this mechanism was dubbed “conditional cooperativity” (97). This mode of regulation was proposed to form a tight negative-feedback loop that buffers the system against fluctuations in transcriptional activity (98). Indeed, since antitoxins appear to be more efficiently translated than their cognate toxins (see below), derepression of a TA operon by an excess of toxin should favor antitoxin neosynthesis, thus allowing homeostatic maintenance of a low toxin/antitoxin ratio (98). In some cases, i.e., mqsRA and hicAB—systems in which the toxin gene precedes that of the antitoxin—toxins were shown to displace their cognate antitoxins from operator sequences upon binding and not act as corepressor as mentioned above (91, 99) (Fig. 3D). Antitoxins of reverse-organized systems from other species were also shown to lose affinity for their operators upon toxin binding, i.e., GraA from Pseudomonas putida and HigA from Proteus vulgaris (100, 101). Therefore, while conditional cooperativity is supposedly the prevalent mode of TA autoregulation, other modes of regulation might exist, especially for systems in “reverse” configuration as described above.
Various TA systems were also shown to be regulated by factors in trans. The yafNO module was shown to be regulated by the SOS response (102). This is due to this system being inserted immediately downstream of the dinB gene, which encodes DNA polymerase IV, a translesion synthesis DNA polymerase implicated in adaptive mutation (102). The dinB gene is itself repressed by LexA and constitutes an operon with yafNO, thus explaining why this system is under SOS control (102). However, deletion of yafNO has no effect on the induction of the SOS system, in adaptive mutation or survival to DNA-damaging fluoroquinolone antibiotics (61, 102). IHF and Fis, two proteins that participate in the topology of the nucleoid, bind upstream of the hipBA and mazEF promoters, respectively, and upregulate their transcriptional activities (103, 104). Crp-Sxy, a supposed regulator of competence, was also shown to stimulate transcription of the hicAB transcript (91). However, the role of these transcriptional regulators in the functionality of the TA systems they regulate is unknown.
Posttranscriptional regulation of TA systems.
Ribosome profiling analysis performed with E. coli K-12 grown in synthetic medium suggests that all TA systems are expressed (89, 105). However, the translation efficiency of toxins appears to be lower relative to antitoxins. For example, the RelB antitoxin is translated six times more efficiently than its cognate toxin (105). This should allow a molar excess of antitoxin to ensure that the entire pool of cellular toxin is neutralized. Translational coupling and transcript cleavage at the level of the toxin sequence were shown to maintain a molar excess of antitoxin in the parD system (106). Analysis of RNA-seq data detected truncated transcripts of the dinJ-yafQ and yafNO systems in the toxin reading frame, suggesting that these two systems maintain higher antitoxin levels through mechanisms similar to parD (89).
Posttranslational regulation of TA systems.
Previous studies on plasmid-encoded TA systems have determined that antitoxins were proteolytically unstable. For example, CcdA is actively and constitutively degraded by the Lon protease (107). Other antitoxins, like Kis, from the parD system or Phd were shown to be degraded in a similar way by the ClpAP and ClpXP proteases, respectively (108, 109). Therefore, constitutive proteolysis of plasmid-encoded antitoxins appears as a rule since it supposedly allows the liberation of toxin molecules in plasmid-free segregants.
As mentioned above, relBE and mazEF were first thought to be involved in nutritional stress responses (35, 36). Amino acid starvation induced by serine hydroxamate [SHX; which inhibits serine incorporation into proteins and triggers (p)ppGpp synthesis] was shown to activate the degradation of the RelB and MazE antitoxins independently of (p)ppGpp (35, 36). The authors of those studies proposed that amino acid starvation would trigger proteolysis of these two antitoxins. However, since SHX by itself is a translation inhibitor (35, 110), it is likely that the observed degradation of RelB and MazE under such treatment is simply a secondary effect of antitoxin instability and neosynthesis inhibition. Indeed, similar degradation kinetics were measured for RelB in the presence of chloramphenicol, a potent translation-inhibiting antibiotic which does not promote (p)ppGpp synthesis (35). A few years later, in the context of bacterial persistence, the same authors came up with a model proposing that (p)ppGpp (in contradiction to what is mentioned just above) induces antitoxin degradation through activation of the Lon protease by polyphosphate accumulation (58). This regulation scheme has been, however, contradicted by many studies. Antitoxin degradation under SHX treatment was shown to be (p)ppGpp independent on multiple occasions (35, 36, 62). In addition, it was shown than Lon is inhibited by polyphosphate in vitro and that (p)ppGpp does not affect polyphosphate synthesis (111, 112).
To further study antitoxin degradation, the overproduction of Lon was used as a tool to destabilize antitoxins, with the aim of releasing toxins and studying subsequent phenotypes. Under these overexpression conditions, growth and translation were inhibited and mRNA cleavage was detected as well (113). Deleting the five TA systems known at the time (yefM-yoeB, relBE, mazEF, chpB, and dinJ-yafQ) only partially rescued growth and translation inhibition but completely eliminated mRNA cleavage (113). Interestingly, rescue was solely provided by deletion of the yefM-yoeB system, while deletion of the four other TAs had no effect (113). This suggests that, under these conditions, only yefM-yoeB is activated through Lon-dependent proteolysis of the YefM antitoxin. However, while massive overproduction of Lon can bring some insight into its effects on the yefM-yoeB system, this does not constitute a physiologically relevant condition. A subsequent study from Janssen et al. demonstrated that yefM-yoeB-dependent mRNA cleavage is induced in cells grown at 42°C in a lon-dependent manner and in a background preventing ribosome rescue from YoeB (ssrA mutant) as well as stalled mRNA degradation (rnb mutant) (114). However, the authors found that yefM-yoeB-induced cleavage at 42°C was not correlated with growth inhibition and that yefM-yoeB did not give any fitness advantage or disadvantage when cells were grown at this temperature (114). Finally, another study showed that the MqsA antitoxin is rapidly degraded by Lon under oxidative stress (115), which was not reproduced by a subsequent study (53). To conclude, antitoxin proteolysis under stress conditions, leading to TA system activation, is far from being a general rule, and further research is needed to better understand the conditions leading to TA systems induction and phenotypes, probably on a case-by-case basis.
WHERE WE STAND, WHERE WE GO: QUESTIONS FOR THE FUTURE BASED ON LESSONS FROM THE PAST
First reports on chromosomally encoded TA systems date back to the late 1990s—about 10 years after their discovery on plasmids—when a homolog of R100 pem system (mazEF) and a new TA (relBE) were identified in the chromosome of E. coli K-12 laboratory strains (29, 38, 116). Mutants of the relB gene were actually first isolated in the late 1970s and conferred the so-called “delayed-relaxed” phenotype in which stable RNA synthesis continues for 10 min after amino acid starvation, accompanied by a reduction of (p)ppGpp concentration and synthesis of a translation inhibitor, most likely RelE (117). These mutants were phenotypically and genetically different from the classical “relaxed mutants” mapped in the relA gene, a finding which led to the suggestion that different loci could be involved in the stringent response (117). It was later shown that one of the relB alleles—relB101—encodes a destabilized RelB antitoxin that is more susceptible to Lon-mediated proteolysis than the wild-type RelB and that the “delayed-relaxed phenotype” is dependent on the RelE toxin (118). The mazE gene was first identified as an ORF of unknown function (ma-ze meaning “what is it” in Hebrew) located downstream of the relA gene (119). It was further shown that the mazEF locus is comprised in an operon together with relA (the latter having its own promoter) (38, 119).
These early observations naturally led to hypotheses linking TA systems to the stringent response that were compelling to investigate. Subsequent works proposed that TAs were linked to nutritional stress responses but with contradicting regulation and outcomes. In the case of mazEF, (p)ppGpp is thought to repress transcription of the operon, leading to the decay in mazEF mRNAs and degradation of MazE (38). Cells in which mazEF is “activated” would undergo programmed cell death due to MazF-dependent translation inhibition; cells in which mazEF is not activated would become necrophagic, surviving by feeding on their dead kin (41). In the case of relBE (also shown for mazEF, which directly contradicts the data mentioned just above), (p)ppGpp was proposed to increase transcription of the relBE and mazEF operons due to degradation of RelB and MazE (35, 36). In this model, cells in which relBE is “activated” would be metabolically dormant due to RelE-dependent translation inhibition and protected against stress; cells in which relBE is not activated would not survive (35). For both systems, the roles further expanded with time with, for example, MazEF becoming involved in an impressive number of stress responses—evolving into a “universal” stress-managing module capable of sensing many types of environmental changes and adjusting cell physiology accordingly, by generating specialized ribosomes and leaderless transcripts (38–45). The idea of a “stress regulon” controlled by TAs was further expanded with a TA system in M. tuberculosis that would hypothetically rewire the proteome toward the expression of stress proteins (120). Ultimately, relBE, as well as mazEF and other TA systems, became pivotal effectors of antibiotic persistence in a unified model in which all these systems would act synergistically and redundantly (58).
With time and in a common effort from independent groups, these models were questioned or disproved by different studies, shedding light on conceptual issues—notably, the absolute necessity that TA systems have to carry out important functions in cellular physiology since they are so widespread in bacterial chromosomes—as well as major experimental flaws like strain genotype issues, fluorescent reporter issues, and questionable data analysis (46, 47, 53, 60, 61). We thus propose to go back to the basics, ask crucial questions that still remain unanswered, and provide suggestions which could, hopefully, improve our understanding of TA systems.
When and how TA systems are activated?
Many studies claim that TA systems are activated under specific conditions by showing transcriptional upregulation of these systems under such conditions (55, 56, 63, 67, 70, 121, 122). As mentioned above, translation of antitoxins is more efficient than their cognate toxin. It is thus likely that transcriptional derepression and upregulation of TA systems allows the synthesis of supplemental antitoxin. Therefore, observing transcriptional upregulation of a TA system does not imply that free toxin is present in molar excess and is able to exert its activity. Another interesting observation is that toxins interact tightly with their cognate antitoxins, with dissociation constants in the nanomolar range (99, 123). Therefore, one could wonder whether toxin dissociation from its antitoxin is spontaneous and supported by antitoxin degradation or, rather, an active process involving factors in trans like chaperones.
What are the consequences of TA activation?
Should TA systems be activated, what would be the free toxin concentrations required to inhibit growth or induce cell death? As mentioned earlier, toxin activities were reported in physiologically relevant conditions but failed to induce any observable phenotypes (35, 36, 50, 114). Therefore, are there conditions in which free toxins are present at sufficient concentrations to have a discernible effect on cell physiology? Do TA genes necessarily need to be lost to see the negative effects of toxin liberation? If there are conditions in which the free toxin concentration is high enough to have an effect on cell physiology, would these toxins transiently inhibit growth or irreversibly affect cells? Can the effects of toxin activities be reversed? Can toxins be neutralized and cell growth resumed? These questions have been scarcely addressed and studied mainly by finding suppressors of toxin activity. For example, the ssrA tmRNA was shown to strongly curtail toxicity of translation-dependent mRNase toxins such as RelE or MazF, opening the possibility that the effects of these toxins can be reversed by trans translation (36, 37). In another study, Cheverton et al. showed that the toxicity of TacT, a tRNA-acetyltransferase toxin from Salmonella enterica, could be suppressed by the overproduction of peptidyl-tRNA hydrolase (Pth), suggesting that TacT-dependent growth inhibition and dormancy can be rescued (124). Interestingly, overproduction of Lon greatly reduced viability of E. coli, in a way that is dependent on the yefM-yoeB system, as mentioned above (113). Viability was not rescued upon plating on inducer-free medium. Therefore, the activation of yefM-yoeB under these conditions is partially responsible for this loss of viability, suggesting that activation of this system irreversibly kills cells (113). Since most toxins broadly inhibit translation and probably block de novo synthesis of their cognate antitoxins, how would cells be able to produce antitoxins after the toxins have reached growth-inhibiting concentrations? To date, the study of the reversibility of TA activation is challenging simply due to the lack of physiologically relevant conditions that activate TA systems and produce discernible phenotypes.
Why are TA systems so ubiquitous, highly dynamic, and mobile?
Since TA systems are part of the accessory genome, studying their origin and evolution may provide a way to understand their functions. As Fiedoruk and colleagues attempted to characterize the distribution and dynamics of TA systems in various E. coli strains, it was striking to see how heterogeneously these systems are distributed, with close to none of the 84 characterized strains having the same set of TA modules (32) (Fig. 1B). Thus, one can wonder how genes so erratically distributed and so impervious to fixation can be pivotal in essential processes such as stress responses. Thus, due to their heterogeneous distribution, it is likely that functions of TA systems are intimately linked to their mobile nature. Others have tried to assess functions of TA systems based on their locations, with the above-mentioned examples of mazEF and yafNO being part of larger operons (relA-mazEFG and dinB-yafNOP, respectively) but having no functional link with these other cistrons (50, 102, 119). It is, however, worth noting that in the many strains where these systems are absent, e.g., UTI89 (Fig. 1B), relA-mazG and dinB-yafP form undisturbed bicistronic operons (21, 32). It is thus likely that these operons are perfectly functional without TA pairs and that insertion of these systems in their intercistronic region occurred independently of their biological functions. In fact, TA insertion loci are quite plastic and can accommodate various mobile genetic elements. For example, a parDE system can be found in the relA-mazG intergenic region in Salmonella spp., while REP sequences can be found in the folA-apaH intergenic region that contains the ccd system in E. coli O157:H7, suggesting that TA insertion does not happen at random locations (21, 34). However, the mechanisms by which TA systems move between hosts and integrate into genomes remain unelucidated to this day.
What are the functions of TA systems?
We would also like to address a few more fundamental concepts in the study of TA systems. First, it is widely acknowledged that point mutations in TA systems can be selected during various genetic screens, when isolating mutants highly tolerant to antibiotics or deficient for the stringent response, for example (54, 117, 125). While this surely shows that TA systems are highly plastic and that phenotypes can be selected in a laboratory context with stringent conditions, it does not demonstrate a function for the wild-type variants of these systems. Another concerning approach is the use of toxin overproduction to study the functions of TA systems. While it may be amenable to find the target of this toxin, it is a questionable approach to study its implication in biological processes. Many studies ectopically overexpressed toxins and observed a phenotype, i.e., increased antimicrobial tolerance, thus claiming that TA systems are implicated in these processes; yet deleting these systems would not induce the opposite phenotype (69–72). However, artificially providing a molar excess of toxin disregards the fact that antitoxins are always produced in excess and that TA systems have to be activated to produce the phenotype observed under overproduction conditions. Therefore, instead of studying phenotype using ectopic toxin overexpression, one should determine whether toxin activity (e.g., RNA cleavage) and TA-dependent phenotypes (using properly designed deletion mutants of the whole module) can be detected in conditions of interest. Moreover, with the development of single-cell methods, observing TA activation with proper fluorescent reporters within individual cells should provide valuable insights about regulations at different levels.
To conclude on a positive note, many questionable studies and other controversies have set the TA field back to the questions it looked to address ten years ago (“TA systems, why so many, what for?”) (126). However, this step back is an opportunity to make two steps forward and to make new discoveries on TA systems with a clean slate and without the preconceived ideas that plagued the field for many years. Provided that TA systems are studied with a critical eye, we believe that the future holds great promises for these small but ubiquitous modules.
ACKNOWLEDGMENTS
We thank Mick Chandler and Clothilde Rousseau for critical reading of the manuscript.
Work in the Van Melderen lab is supported by the Fonds National de la Recherche Scientifique (FNRS; T.0147.15F PDR and J.0061.16F CDR), the ARC actions 2018-2023, the Fonds Jean Brachet, and the Wallonia Region (Algotech, convention 1510598).
REFERENCES
- 1.Karoui H, Bex F, Drèze P, Couturier M. 1983. Ham22, a mini-F mutation which is lethal to host cell and promotes recA-dependent induction of lambdoid prophage. EMBO J 2:1863–1868. doi: 10.1002/j.1460-2075.1983.tb01672.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ogura T, Hiraga S. 1983. Mini-F plasmid genes that couple host cell division to plasmid proliferation. Proc Natl Acad Sci U S A 80:4784–4788. doi: 10.1073/pnas.80.15.4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jaffé A, Ogura T, Hiraga S. 1985. Effects of the ccd function of the F plasmid on bacterial growth. J Bacteriol 163:841–849. doi: 10.1128/JB.163.3.841-849.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hiraga S, Jaffé A, Ogura T, Mori H, Takahashi H. 1986. F plasmid ccd mechanism in Escherichia coli. J Bacteriol 166:100–104. doi: 10.1128/jb.166.1.100-104.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van Melderen L, Bernard P, Couturier M. 1994. Lon-dependent proteolysis of CcdA is the key control for activation of CcdB in plasmid-free segregant bacteria. Mol Microbiol 11:1151–1157. doi: 10.1111/j.1365-2958.1994.tb00391.x. [DOI] [PubMed] [Google Scholar]
- 6.Johnson EP, Strom AR, Helinski DR. 1996. Plasmid RK2 toxin protein ParE: purification and interaction with the ParD antitoxin protein. J Bacteriol 178:1420–1429. doi: 10.1128/jb.178.5.1420-1429.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernard P, Couturier M. 1992. Cell killing by the F plasmid CcdB protein involves poisoning of DNA-topoisomerase II complexes. J Mol Biol 226:735–745. doi: 10.1016/0022-2836(92)90629-x. [DOI] [PubMed] [Google Scholar]
- 8.Miki T, Park JA, Nagao K, Murayama N, Horiuchi T. 1992. Control of segregation of chromosomal DNA by sex factor F in Escherichia coli: mutants of DNA gyrase subunit A suppress letD (ccdB) product growth inhibition. J Mol Biol 225:39–52. doi: 10.1016/0022-2836(92)91024-J. [DOI] [PubMed] [Google Scholar]
- 9.Maki S, Takiguchi S, Miki T, Horiuchi T. 1992. Modulation of DNA supercoiling activity of Escherichia coli DNA gyrase by F plasmid proteins: antagonistic actions of LetA (CcdA) and LetD (CcdB) proteins. J Biol Chem 267:12244–12251. [PubMed] [Google Scholar]
- 10.Bernard P, Kézdy KE, Van Melderen L, Steyaert J, Wyns L, Pato ML, Higgins PN, Couturier M. 1993. The F plasmid CcdB protein induces efficient ATP-dependent DNA cleavage by gyrase. J Mol Biol 234:534–541. doi: 10.1006/jmbi.1993.1609. [DOI] [PubMed] [Google Scholar]
- 11.Bernard P, Couturier M. 1991. The 41 carboxy-terminal residues of the miniF plasmid CcdA protein are sufficient to antagonize the killer activity of the CcdB protein. Mol Gen Genet 226:297–304. doi: 10.1007/bf00273616. [DOI] [PubMed] [Google Scholar]
- 12.Bahassi EM, O’Dea MH, Allali N, Messens J, Gellert M, Couturier M. 1999. Interactions of CcdB with DNA gyrase: inactivation of GyrA, poisoning of the gyrase-DNA complex, and the antidote action of CcdA. J Biol Chem 274:10936–10944. doi: 10.1074/jbc.274.16.10936. [DOI] [PubMed] [Google Scholar]
- 13.Van Melderen L, Thi MHD, Lecchi P, Gottesman S, Couturier M, Maurizi MR. 1996. ATP-dependent degradation of CcdA by Lon protease: effects of secondary structure and heterologous subunit interactions. J Biol Chem 271:27730–27738. doi: 10.1074/jbc.271.44.27730. [DOI] [PubMed] [Google Scholar]
- 14.Gerdes K, Rasmussen PB, Molin S. 1986. Unique type of plasmid maintenance function: postsegregational killing of plasmid-free cells. Proc Natl Acad Sci U S A 83:3116–3120. doi: 10.1073/pnas.83.10.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bravo A, de Torrontegui G, Díaz R. 1987. Identification of components of a new stability system of plasmid R1, ParD, that is close to the origin of replication of this plasmid. Mol Gen Genet 210:101–110. doi: 10.1007/bf00337764. [DOI] [PubMed] [Google Scholar]
- 16.Tsuchimoto S, Ohtsubo H, Ohtsubo E. 1988. Two genes, pemK and pemI, responsible for stable maintenance of resistance plasmid R100. J Bacteriol 170:1461–1466. doi: 10.1128/jb.170.4.1461-1466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lehnherr H, Maguin E, Jafri S, Yarmolinsky MB. 1993. Plasmid addiction genes of bacteriophage P1: doc, which causes cell death on curing of prophage, and phd, which prevents host death when prophage is retained. J Mol Biol 233:414–428. doi: 10.1006/jmbi.1993.1521. [DOI] [PubMed] [Google Scholar]
- 18.Jiang Y, Pogliano J, Helinski DR, Konieczny I. 2002. ParE toxin encoded by the broad-host-range plasmid RK2 is an inhibitor of Escherichia coli gyrase. Mol Microbiol 44:971–979. doi: 10.1046/j.1365-2958.2002.02921.x. [DOI] [PubMed] [Google Scholar]
- 19.Hayes F. 1998. A family of stability determinants in pathogenic bacteria. J Bacteriol 180:6415–6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yarmolinski MB. 1995. Programmed cell death in bacterial populations. Science 267:836–837. doi: 10.1126/science.7846528. [DOI] [PubMed] [Google Scholar]
- 21.Pandey DP, Gerdes K. 2005. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res 33:966–976. doi: 10.1093/nar/gki201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Makarova KS, Wolf YI, Koonin EV. 2009. Comprehensive comparative-genomic analysis of type 2 toxin-antitoxin systems and related mobile stress response systems in prokaryotes. Biol Direct 4:19. doi: 10.1186/1745-6150-4-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shao Y, Harrison EM, Bi D, Tai C, He X, Ou H-Y, Rajakumar K, Deng Z. 2011. TADB: a web-based resource for type 2 toxin-antitoxin loci in bacteria and archaea. Nucleic Acids Res 39:D606–D611. doi: 10.1093/nar/gkq908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leplae R, Geeraerts D, Hallez R, Guglielmini J, Dreze P, Van Melderen L. 2011. Diversity of bacterial type II toxin-antitoxin systems: a comprehensive search and functional analysis of novel families. Nucleic Acids Res 39:5513–5525. doi: 10.1093/nar/gkr131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van Melderen L, Jurenas D, Garcia-Pino A. 2018. Messing up translation from the start: how AtaT inhibits translation initiation in Escherichia coli. RNA Biol 15:303–307. doi: 10.1080/15476286.2017.1391439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harms A, Brodersen DE, Mitarai N, Gerdes K. 2018. Toxins, targets, and triggers: an overview of toxin-antitoxin biology. Mol Cell 70:768–784. doi: 10.1016/j.molcel.2018.01.003. [DOI] [PubMed] [Google Scholar]
- 27.Gerdes K, Christensen SK, Løbner-Olesen A. 2005. Prokaryotic toxin-antitoxin stress response loci. Nat Rev Microbiol 3:371–382. doi: 10.1038/nrmicro1147. [DOI] [PubMed] [Google Scholar]
- 28.Bordenstein SR, Reznikoff WS. 2005. Mobile DNA in obligate intracellular bacteria. Nat Rev Microbiol 3:688–699. doi: 10.1038/nrmicro1233. [DOI] [PubMed] [Google Scholar]
- 29.Gotfredsen M, Gerdes K. 1998. The Escherichia coli relBE genes belong to a new toxin-antitoxin gene family. Mol Microbiol 29:1065–1076. doi: 10.1046/j.1365-2958.1998.00993.x. [DOI] [PubMed] [Google Scholar]
- 30.Koga M, Otsuka Y, Lemire S, Yonesaki T. 2011. Escherichia coli rnlA and rnlB compose a novel toxin-antitoxin system. Genetics 187:123–130. doi: 10.1534/genetics.110.121798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hallez R, Geeraerts D, Sterckx Y, Mine N, Loris R, Van Melderen L. 2010. New toxins homologous to ParE belonging to three-component toxin-antitoxin systems in Escherichia coli O157:H7. Mol Microbiol 76:719–732. doi: 10.1111/j.1365-2958.2010.07129.x. [DOI] [PubMed] [Google Scholar]
- 32.Fiedoruk K, Daniluk T, Swiecicka I, Sciepuk M, Leszczynska K. 2015. Type II toxin-antitoxin systems are unevenly distributed among Escherichia coli phylogroups. Microbiology 161:158–167. doi: 10.1099/mic.0.082883-0. [DOI] [PubMed] [Google Scholar]
- 33.Ramisetty BC, Santhosh RS. 2016. Horizontal gene transfer of chromosomal type II toxin-antitoxin systems of Escherichia coli. FEMS Microbiol Lett 363:fnv238. doi: 10.1093/femsle/fnv238. [DOI] [PubMed] [Google Scholar]
- 34.Mine N, Guglielmini J, Wilbaux M, Van Melderen L. 2009. The decay of the chromosomally encoded ccdO157 toxin-antitoxin system in the Escherichia coli species. Genetics 181:1557–1566. doi: 10.1534/genetics.108.095190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Christensen SK, Mikkelsen M, Pedersen K, Gerdes K. 2001. RelE, a global inhibitor of translation, is activated during nutritional stress. Proc Natl Acad Sci U S A 98:14328–14333. doi: 10.1073/pnas.251327898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Christensen SK, Pedersen K, Hansen FG, Gerdes K. 2003. Toxin-antitoxin loci as stress response elements: ChpAK/MazF and ChpBK cleave translated RNAs and are counteracted by tmRNA. J Mol Biol 332:809–819. doi: 10.1016/s0022-2836(03)00922-7. [DOI] [PubMed] [Google Scholar]
- 37.Christensen SK, Gerdes K. 2003. RelE toxins from Bacteria and Archaea cleave mRNAs on translating ribosomes, which are rescued by tmRNA. Mol Microbiol 48:1389–1400. doi: 10.1046/j.1365-2958.2003.03512.x. [DOI] [PubMed] [Google Scholar]
- 38.Aizenman E, Engelberg-Kulka H, Glaser G. 1996. An Escherichia coli chromosomal “addiction module” regulated by guanosine [corrected] 3′,5′-bispyrophosphate: a model for programmed bacterial cell death. Proc Natl Acad Sci U S A 93:6059–6063. doi: 10.1073/pnas.93.12.6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Engelberg-Kulka H, Amitai S, Kolodkin-Gal I, Hazan R. 2006. Bacterial programmed cell death and multicellular behavior in bacteria. PLoS Genet 2:e135. doi: 10.1371/journal.pgen.0020135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hazan R, Sat B, Engelberg-Kulka H. 2004. Escherichia coli mazEF-mediated cell death is triggered by various stressful conditions. J Bacteriol 186:3663–3669. doi: 10.1128/JB.186.11.3663-3669.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kolodkin-Gal I, Engelberg-Kulka H. 2006. Induction of Escherichia coli chromosomal mazEF by stressful conditions causes an irreversible loss of viability. J Bacteriol 188:3420–3423. doi: 10.1128/JB.188.9.3420-3423.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kolodkin-Gal I, Hazan R, Gaathon A, Carmeli S, Engelberg-Kulka H. 2007. A linear pentapeptide is a quorum-sensing factor required for mazEF-mediated cell death in Escherichia coli. Science 318:652–655. doi: 10.1126/science.1147248. [DOI] [PubMed] [Google Scholar]
- 43.Sat B, Reches M, Engelberg-Kulka H. 2003. The Escherichia coli mazEF suicide module mediates thymineless death. J Bacteriol 185:1803–1807. doi: 10.1128/jb.185.6.1803-1807.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vesper O, Amitai S, Belitsky M, Byrgazov K, Kaberdina AC, Engelberg-Kulka H, Moll I. 2011. Selective translation of leaderless mRNAs by specialized ribosomes generated by MazF in Escherichia coli. Cell 147:147–157. doi: 10.1016/j.cell.2011.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nigam A, Ziv T, Oron-Gottesman A, Engelberg-Kulka H, Nigam A, Ziv T, Oron-Gottesman A, Engelberg-Kulka H. 2019. Stress-induced MazF-mediated proteins in Escherichia coli. mBio 10:e00340-19. doi: 10.1128/mBio.00340-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Culviner PH, Laub MT. 2018. Global analysis of the Escherichia coli toxin MazF reveals widespread cleavage of mRNA and the inhibition of rRNA maturation and ribosome biogenesis. Mol Cell 70:868–880. doi: 10.1016/j.molcel.2018.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mets T, Kasvandik S, Saarma M, Maiväli Ü, Tenson T, Kaldalu N. 2019. Fragmentation of Escherichia coli mRNA by MazF and MqsR. Biochimie 156:79–91. doi: 10.1016/j.biochi.2018.10.004. [DOI] [PubMed] [Google Scholar]
- 48.Kaldalu N, Maiväli Ü, Hauryliuk V, Tenson T, Kaldalu N, Maiväli Ü, Hauryliuk V, Tenson T. 2019. Reanalysis of proteomics results fails to detect MazF-mediated stress proteins. mBio 10:e00949-19. doi: 10.1128/mBio.00949-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wade JT, Laub MT, Wade JT, Laub MT. 2019. Concerns about “stress-induced MazF-mediated proteins in Escherichia coli. mBio 10:e00825-19. doi: 10.1128/mBio.00825-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsilibaris V, Maenhaut-Michel G, Mine N, Van Melderen L. 2007. What is the benefit to Escherichia coli of having multiple toxin-antitoxin systems in its genome? J Bacteriol 189:6101–6108. doi: 10.1128/JB.00527-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Soo VW, Wood TK. 2013. Antitoxin MqsA represses curli formation through the master biofilm regulator CsgD. Sci Rep 3:3186. doi: 10.1038/srep03186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang X, Wood TK. 2011. Toxin-antitoxin systems influence biofilm and persister cell formation and the general stress response. Appl Environ Microbiol 77:5577–5583. doi: 10.1128/AEM.05068-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fraikin N, Rousseau CJ, Goeders N, Van Melderen L, Fraikin N, Rousseau CJ, Goeders N, Van Melderen L. 2019. Reassessing the role of the type II MqsRA toxin-antitoxin system in stress response and biofilm formation: mqsA is transcriptionally uncoupled from mqsR. mBio 10:e02678-19. doi: 10.1128/mBio.02678-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moyed HS, Bertrand KP. 1983. hipA, a newly recognized gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J Bacteriol 155:768–775. doi: 10.1128/JB.155.2.768-775.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Keren I, Shah D, Spoering A, Kaldalu N, Lewis K. 2004. Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J Bacteriol 186:8172–8180. doi: 10.1128/JB.186.24.8172-8180.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shah D, Zhang Z, Khodursky A, Kaldalu N, Kurg K, Lewis K. 2006. Persisters: a distinct physiological state of Escherichia coli. BMC Microbiol 6:53. doi: 10.1186/1471-2180-6-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maisonneuve E, Shakespeare LJ, Jorgensen MG, Gerdes K. 2011. Bacterial persistence by RNA endonucleases. Proc Natl Acad Sci U S A 108:13206–13211. doi: 10.1073/pnas.1100186108. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 58.Maisonneuve E, Castro-Camargo M, Gerdes K. 2013. (p)ppGpp controls bacterial persistence by stochastic induction of toxin-antitoxin activity. Cell 154:1140–1150. doi: 10.1016/j.cell.2013.07.048. [DOI] [PubMed] [Google Scholar]
- 59.Germain E, Roghanian M, Gerdes K, Maisonneuve E. 2015. Stochastic induction of persister cells by HipA through (p)ppGpp-mediated activation of mRNA endonucleases. Proc Natl Acad Sci U S A 112:5171–5176. doi: 10.1073/pnas.1423536112. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 60.Harms A, Fino C, Sørensen MA, Semsey S, Gerdes K, Harms A, Fino C, Sørensen MA, Semsey S, Gerdes K. 2017. Prophages and growth dynamics confound experimental results with antibiotic-tolerant persister cells. mBio 8:e01964-17. doi: 10.1128/mBio.01964-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Goormaghtigh F, Fraikin N, Putrins M, Hallaert T, Hauryliuk V, Garcia-Pino A, Sjodin A, Kasvandik S, Udekwu K, Tenson T, Kaldalu N, Van Melderen L. 2018. Reassessing the role of type II toxin-antitoxin systems in formation of Escherichia coli type II persister cells. mBio 9:e00640-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ramisetty BC, Ghosh D, Roy Chowdhury M, Santhosh RS. 2016. What is the link between stringent response, endoribonuclease encoding type II toxin-antitoxin systems and persistence? Front Microbiol 7:1882. doi: 10.3389/fmicb.2016.01882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Helaine S, Cheverton AM, Watson KG, Faure LM, Matthews SA, Holden DW. 2014. Internalization of Salmonella by macrophages induces formation of nonreplicating persisters. Science 343:204–208. doi: 10.1126/science.1244705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pontes MH, Groisman EA. 2019. Slow growth determines nonheritable antibiotic resistance in Salmonella enterica. Sci Signal 12:eaax3938. doi: 10.1126/scisignal.aax3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Claudi B, Sprote P, Chirkova A, Personnic N, Zankl J, Schurmann N, Schmidt A, Bumann D. 2014. Phenotypic variation of Salmonella in host tissues delays eradication by antimicrobial chemotherapy. Cell 158:722–733. doi: 10.1016/j.cell.2014.06.045. [DOI] [PubMed] [Google Scholar]
- 66.Norton JP, Mulvey MA. 2012. Toxin-antitoxin systems are important for niche-specific colonization and stress resistance of uropathogenic Escherichia coli. PLoS Pathog 8:e1002954. doi: 10.1371/journal.ppat.1002954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lobato-Márquez D, Moreno-Córdoba I, Figueroa V, Díaz-Orejas R, García-del Portillo F. 2015. Distinct type I and type II toxin-antitoxin modules control Salmonella lifestyle inside eukaryotic cells. Sci Rep 5:9374. doi: 10.1038/srep09374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.De la Cruz MA, Zhao W, Farenc C, Gimenez G, Raoult D, Cambillau C, Gorvel J-P, Méresse S. 2013. A toxin-antitoxin module of Salmonella promotes virulence in mice. PLoS Pathog 9:e1003827. doi: 10.1371/journal.ppat.1003827. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 69.Muthuramalingam M, White JC, Murphy T, Ames JR, Bourne CR. 2019. The toxin from a ParDE toxin-antitoxin system found in Pseudomonas aeruginosa offers protection to cells challenged with anti-gyrase antibiotics. Mol Microbiol 111:441–454. doi: 10.1111/mmi.14165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dufour D, Mankovskaia A, Chan Y, Motavaze K, Gong S-G, Lévesque CM. 2018. A tripartite toxin-antitoxin module induced by quorum sensing is associated with the persistence phenotype in Streptococcus mutans. Mol Oral Microbiol 33:420–429. doi: 10.1111/omi.12245. [DOI] [PubMed] [Google Scholar]
- 71.Rycroft JA, Gollan B, Grabe GJ, Hall A, Cheverton AM, Larrouy-Maumus G, Hare SA, Helaine S. 2018. Activity of acetyltransferase toxins involved in Salmonella persister formation during macrophage infection. Nat Commun 9:1993. doi: 10.1038/s41467-018-04472-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mok WWK, Brynildsen MP. 2018. Timing of DNA damage responses impacts persistence to fluoroquinolones. Proc Natl Acad Sci U S A 115:E6301–E6309. doi: 10.1073/pnas.1804218115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fraikin N, Goormaghtigh F, Van Melderen L. 2019. Toxin-antitoxin systems and persistence, p 181–202. In Lewis K. (ed), Persister cells and infectious disease. Springer International Publishing, Cham, Switzerland. [Google Scholar]
- 74.Szekeres S, Dauti M, Wilde C, Mazel D, Rowe-Magnus DA. 2007. Chromosomal toxin-antitoxin loci can diminish large-scale genome reductions in the absence of selection. Mol Microbiol 63:1588–1605. doi: 10.1111/j.1365-2958.2007.05613.x. [DOI] [PubMed] [Google Scholar]
- 75.Wozniak RA, Waldor MK. 2009. A toxin-antitoxin system promotes the maintenance of an integrative conjugative element. PLoS Genet 5:e1000439. doi: 10.1371/journal.pgen.1000439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Blower TR, Fineran PC, Johnson MJ, Toth IK, Humphreys DP, Salmond GP. 2009. Mutagenesis and functional characterization of the RNA and protein components of the toxIN abortive infection and toxin-antitoxin locus of Erwinia. J Bacteriol 191:6029–6039. doi: 10.1128/JB.00720-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fineran PC, Blower TR, Foulds IJ, Humphreys DP, Lilley KS, Salmond GP. 2009. The phage abortive infection system, ToxIN, functions as a protein-RNA toxin-antitoxin pair. Proc Natl Acad Sci U S A 106:894–899. doi: 10.1073/pnas.0808832106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dy RL, Przybilski R, Semeijn K, Salmond GPC, Fineran PC. 2014. A widespread bacteriophage abortive infection system functions through a type IV toxin-antitoxin mechanism. Nucleic Acids Res 42:4590–4605. doi: 10.1093/nar/gkt1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hazan R, Engelberg-Kulka H. 2004. Escherichia coli mazEF-mediated cell death as a defense mechanism that inhibits the spread of phage P1. Mol Genet Genomics 272:227–234. doi: 10.1007/s00438-004-1048-y. [DOI] [PubMed] [Google Scholar]
- 80.Otsuka Y, Yonesaki T. 2012. Dmd of bacteriophage T4 functions as an antitoxin against Escherichia coli LsoA and RnlA toxins. Mol Microbiol 83:669–681. doi: 10.1111/j.1365-2958.2012.07975.x. [DOI] [PubMed] [Google Scholar]
- 81.Saavedra De Bast M, Mine N, Van Melderen L. 2008. Chromosomal toxin-antitoxin systems may act as anti-addiction modules. J Bacteriol 190:4603–4609. doi: 10.1128/JB.00357-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jurėnas D, Garcia-Pino A, Van Melderen L. 2017. Novel toxins from type II toxin-antitoxin systems with acetyltransferase activity. Plasmid 93:30–35. doi: 10.1016/j.plasmid.2017.08.005. [DOI] [PubMed] [Google Scholar]
- 83.Ramisetty BCM, Santhosh RS. 2017. Endoribonuclease type II toxin-antitoxin systems: functional or selfish? Microbiology 163:931–939. doi: 10.1099/mic.0.000487. [DOI] [PubMed] [Google Scholar]
- 84.Mruk I, Kobayashi I. 2014. To be or not to be: regulation of restriction-modification systems and other toxin-antitoxin systems. Nucleic Acids Res 42:70–86. doi: 10.1093/nar/gkt711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cooper TF, Heinemann JA. 2000. Postsegregational killing does not increase plasmid stability but acts to mediate the exclusion of competing plasmids. Proc Natl Acad Sci U S A 97:12643–12648. doi: 10.1073/pnas.220077897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Naito T, Kusano K, Kobayashi I. 1995. Selfish behavior of restriction-modification systems. Science 267:897–899. doi: 10.1126/science.7846533. [DOI] [PubMed] [Google Scholar]
- 87.Naito Y, Naito T, Kobayashi I. 1998. Selfish restriction modification genes: resistance of a resident R/M plasmid to displacement by an incompatible plasmid mediated by host killing. Biol Chem 379:429–436. doi: 10.1515/bchm.1998.379.4-5.429. [DOI] [PubMed] [Google Scholar]
- 88.Van Melderen L, Saavedra De Bast M. 2009. Bacterial toxin-antitoxin systems: more than selfish entities? PLoS Genet 5:e1000437. doi: 10.1371/journal.pgen.1000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Deter HS, Jensen RV, Mather WH, Butzin NC. 2017. Mechanisms for differential protein production in toxin-antitoxin systems. Toxins 9:211. doi: 10.3390/toxins9070211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Otsuka Y, Miki K, Koga M, Katayama N, Morimoto W, Takahashi Y, Yonesaki T. 2010. IscR regulates RNase LS activity by repressing rnlA transcription. Genetics 185:823–830. doi: 10.1534/genetics.110.114462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Turnbull KJ, Gerdes K. 2017. HicA toxin of Escherichia coli derepresses hicAB transcription to selectively produce HicB antitoxin. Mol Microbiol 104:781–792. doi: 10.1111/mmi.13662. [DOI] [PubMed] [Google Scholar]
- 92.Tam JE, Kline BC. 1989. The F plasmid ccd autorepressor is a complex of CcdA and CcdB proteins. Mol Gen Genet 219:26–32. doi: 10.1007/bf00261153. [DOI] [PubMed] [Google Scholar]
- 93.Magnuson R, Lehnherr H, Mukhopadhyay G, Yarmolinsky MB. 1996. Autoregulation of the plasmid addiction operon of bacteriophage P1. J Biol Chem 271:18705–18710. doi: 10.1074/jbc.271.31.18705. [DOI] [PubMed] [Google Scholar]
- 94.Ruiz-Echevarría MJ, Berzal-Herranz A, Gerdes K, Díaz-Orejas R. 1991. The kis and kid genes of the parD maintenance system of plasmid R1 form an operon that is autoregulated at the level of transcription by the coordinated action of the Kis and Kid proteins. Mol Microbiol 5:2685–2693. doi: 10.1111/j.1365-2958.1991.tb01977.x. [DOI] [PubMed] [Google Scholar]
- 95.Tsuchimoto S, Ohtsubo E. 1993. Autoregulation by cooperative binding of the PemI and PemK proteins to the promoter region of the pem operon. Mol Gen Genet 237:81–88. doi: 10.1007/bf00282787. [DOI] [PubMed] [Google Scholar]
- 96.Garcia-Pino A, Balasubramanian S, Wyns L, Gazit E, De Greve H, Magnuson RD, Charlier D, van Nuland NAJ, Loris R. 2010. Allostery and intrinsic disorder mediate transcription regulation by conditional cooperativity. Cell 142:101–111. doi: 10.1016/j.cell.2010.05.039. [DOI] [PubMed] [Google Scholar]
- 97.Overgaard M, Borch J, Jørgensen MG, Gerdes K. 2008. Messenger RNA interferase RelE controls relBE transcription by conditional cooperativity. Mol Microbiol 69:841–857. doi: 10.1111/j.1365-2958.2008.06313.x. [DOI] [PubMed] [Google Scholar]
- 98.Cataudella I, Trusina A, Sneppen K, Gerdes K, Mitarai N. 2012. Conditional cooperativity in toxin-antitoxin regulation prevents random toxin activation and promotes fast translational recovery. Nucleic Acids Res 40:6424–6434. doi: 10.1093/nar/gks297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Brown BL, Lord DM, Grigoriu S, Peti W, Page R. 2013. The Escherichia coli toxin MqsR destabilizes the transcriptional repression complex formed between the antitoxin MqsA and the mqsRA operon promoter. J Biol Chem 288:1286–1294. doi: 10.1074/jbc.M112.421008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Schureck MA, Meisner J, Hoffer ED, Wang D, Onuoha N, Ei Cho S, Lollar P III, Dunham CM. 2019. Structural basis of transcriptional regulation by the HigA antitoxin. Mol Microbiol 111:1449–1462. doi: 10.1111/mmi.14229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Talavera A, Tamman H, Ainelo A, Konijnenberg A, Hadži S, Sobott F, Garcia-Pino A, Hõrak R, Loris R. 2019. A dual role in regulation and toxicity for the disordered N terminus of the toxin GraT. Nat Commun 10:972. doi: 10.1038/s41467-019-08865-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Singletary LA, Gibson JL, Tanner EJ, McKenzie GJ, Lee PL, Gonzalez C, Rosenberg SM. 2009. An SOS-regulated type 2 toxin-antitoxin system. J Bacteriol 191:7456–7465. doi: 10.1128/JB.00963-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Black DS, Irwin B, Moyed HS. 1994. Autoregulation of hip, an operon that affects lethality due to inhibition of peptidoglycan or DNA synthesis. J Bacteriol 176:4081–4091. doi: 10.1128/jb.176.13.4081-4091.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Marianovsky I, Aizenman E, Engelberg-Kulka H, Glaser G. 2001. The regulation of the Escherichia coli mazEF promoter involves an unusual alternating palindrome. J Biol Chem 276:5975–5984. doi: 10.1074/jbc.M008832200. [DOI] [PubMed] [Google Scholar]
- 105.Li G-W, Burkhardt D, Gross C, Weissman JS. 2014. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell 157:624–635. doi: 10.1016/j.cell.2014.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ruiz-Echevarría MJ, de la Cueva G, Díaz-Orejas R. 1995. Translational coupling and limited degradation of a polycistronic messenger modulate differential gene expression in the parD stability system of plasmid R1. Mol Gen Genet 248:599–609. doi: 10.1007/bf02423456. [DOI] [PubMed] [Google Scholar]
- 107.Salmon MA, Van Melderen L, Bernard P, Couturier M. 1994. The antidote and autoregulatory functions of the F plasmid CcdA protein: a genetic and biochemical survey. Mol Gen Genet 244:530–538. doi: 10.1007/bf00583904. [DOI] [PubMed] [Google Scholar]
- 108.Diago-Navarro E, Hernández-Arriaga AM, Kubik S, Konieczny I, Díaz-Orejas R. 2013. Cleavage of the antitoxin of the parD toxin-antitoxin system is determined by the ClpAP protease and is modulated by the relative ratio of the toxin and the antitoxin. Plasmid 70:78–85. doi: 10.1016/j.plasmid.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 109.Lehnherr H, Yarmolinsky MB. 1995. Addiction protein Phd of plasmid prophage P1 is a substrate of the ClpXP serine protease of Escherichia coli. Proc Natl Acad Sci U S A 92:3274–3277. doi: 10.1073/pnas.92.8.3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tosa T, Pizer LI. 1971. Biochemical bases for the antimetabolite action of l-serine hydroxamate. J Bacteriol 106:972–982. doi: 10.1128/JB.106.3.972-982.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gray MJ, Gray MJ. 2019. Inorganic polyphosphate accumulation in Escherichia coli is regulated by DksA but not by (p)ppGpp. J Bacteriol 201:e00664-18. doi: 10.1128/JB.00664-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Osbourne DO, Soo VW, Konieczny I, Wood TK. 2014. Polyphosphate, cyclic AMP, guanosine tetraphosphate, and c-di-GMP reduce in vitro Lon activity. Bioengineered 5:264–268. doi: 10.4161/bioe.29261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Christensen SK, Maenhaut-Michel G, Mine N, Gottesman S, Gerdes K, Van Melderen L. 2004. Overproduction of the Lon protease triggers inhibition of translation in Escherichia coli: involvement of the yefM-yoeB toxin-antitoxin system. Mol Microbiol 51:1705–1717. doi: 10.1046/j.1365-2958.2003.03941.x. [DOI] [PubMed] [Google Scholar]
- 114.Janssen BD, Garza-Sánchez F, Hayes CS. 2015. YoeB toxin is activated during thermal stress. MicrobiologyOpen 4:682–697. doi: 10.1002/mbo3.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wang X, Kim Y, Hong SH, Ma Q, Brown BL, Pu M, Tarone AM, Benedik MJ, Peti W, Page R, Wood TK. 2011. Antitoxin MqsA helps mediate the bacterial general stress response. Nat Chem Biol 7:359–366. doi: 10.1038/nchembio.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Masuda Y, Miyakawa K, Nishimura Y, Ohtsubo E. 1993. chpA and chpB, Escherichia coli chromosomal homologs of the pem locus responsible for stable maintenance of plasmid R100. J Bacteriol 175:6850–6856. doi: 10.1128/jb.175.21.6850-6856.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Diderichsen B, Fiil NP, Lavallé R. 1977. Genetics of the relB locus in Escherichia coli. J Bacteriol 131:30–33. doi: 10.1128/JB.131.1.30-33.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Christensen SK, Gerdes K. 2004. Delayed-relaxed response explained by hyperactivation of RelE. Mol Microbiol 53:587–597. doi: 10.1111/j.1365-2958.2004.04127.x. [DOI] [PubMed] [Google Scholar]
- 119.Metzger S, Dror IB, Aizenman E, Schreiber G, Toone M, Friesen JD, Cashel M, Glaser G. 1988. The nucleotide sequence and characterization of the relA gene of Escherichia coli. J Biol Chem 263:15699–15704. [PubMed] [Google Scholar]
- 120.Barth VC, Zeng J-M, Vvedenskaya IO, Ouyang M, Husson RN, Woychik NA. 2019. Toxin-mediated ribosome stalling reprograms the Mycobacterium tuberculosis proteome. Nat Commun 10:3035. doi: 10.1038/s41467-019-10869-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ramage HR, Connolly LE, Cox JS. 2009. Comprehensive functional analysis of Mycobacterium tuberculosis toxin-antitoxin systems: implications for pathogenesis, stress responses, and evolution. PLoS Genet 5:e1000767. doi: 10.1371/journal.pgen.1000767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Keren I, Minami S, Rubin E, Lewis K. 2011. Characterization and transcriptome analysis of Mycobacterium tuberculosis persisters. mBio 2:e00100-11. doi: 10.1128/mBio.00100-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Overgaard M, Borch J, Gerdes K. 2009. RelB and RelE of Escherichia coli form a tight complex that represses transcription via the ribbon-helix-helix motif in RelB. J Mol Biol 394:183–196. doi: 10.1016/j.jmb.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Cheverton AM, Gollan B, Przydacz M, Wong CT, Mylona A, Hare SA, Helaine S. 2016. A Salmonella toxin promotes persister formation through acetylation of tRNA. Mol Cell 63:86–96. doi: 10.1016/j.molcel.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Levin-Reisman I, Ronin I, Gefen O, Braniss I, Shoresh N, Balaban NQ. 2017. Antibiotic tolerance facilitates the evolution of resistance. Science 355:826–830. doi: 10.1126/science.aaj2191. [DOI] [PubMed] [Google Scholar]
- 126.Van Melderen L. 2010. Toxin-antitoxin systems: why so many, what for? Curr Opin Microbiol 13:781–785. doi: 10.1016/j.mib.2010.10.006. [DOI] [PubMed] [Google Scholar]