The structures of sigma factors vary among bacterial strains, indicating that recognition rules may also vary. Therefore, we investigated the promoter structure of Bifidobacterium longum NCC2705 using a bioinformatics approach and wet analyses. The most frequent and optimal motifs were similar to other bacterial consensus motifs. The optimal spacer length between the two boxes was reported to be 17 bp. It is widely applied to a bioinformatics approach for other bacteria. Unexpectedly, conserved spacer lengths were 11 bp as well as 17 bp in the genus Bifidobacterium. Moreover, the sigma factor of the genus Bifidobacterium has a characteristic domain in the N terminus which may contribute to the additional functions. Hence, it would be valuable to reevaluate the promoter in other organisms.
KEYWORDS: bifidobacteria, promoters
ABSTRACT
Bacterial promoters consist of core sequence motifs termed –35 and –10 boxes. The consensus motifs are TTGACA and TATAAT, respectively, which were identified from leading investigations on Escherichia coli. However, the consensus sequences are not likely to fit genetically divergent bacteria. The sigma factor of the genus Bifidobacterium has a characteristic polar domain in the N terminus, suggesting the possibility of specific promoter recognition. We reevaluated the structure of Bifidobacterium longum NCC2705 promoters and compared them to other bacteria. Transcriptional start sites (TSSs) of the B. longum NCC2705 strain were identified using transcriptome sequencing (RNA-Seq) analysis to extract promoter regions. Conserved motifs of a bifidobacterial promoter were determined using regions upstream of TSSs and a hidden Markov model. As a result, consensus motifs of the –35 and –10 boxes were TTGTGC and TACAAT, respectively. To assess each base of both motifs, we constructed 37 plasmids based on pKO403-TPCTcon, including the hup promoter connected with a chloramphenicol acetyltransferase as a reporter gene. This reporter assay showed two optimal motifs of the –35 and –10 boxes, namely, TTGNNN and TANNNT, respectively. We further analyzed spacer lengths between the –35 and –10 boxes via a bioinformatics approach. The spacer lengths predominant in bacteria have been generally reported to be approximately 17 bp. In contrast, the predominant spacer lengths in the genus Bifidobacterium and related species were 11 bp, in addition to 17 bp. A reporter assay to assess the spacer lengths indicated that the 11-bp spacer length produced unusually high activity.
IMPORTANCE The structures of sigma factors vary among bacterial strains, indicating that recognition rules may also vary. Therefore, we investigated the promoter structure of Bifidobacterium longum NCC2705 using a bioinformatics approach and wet analyses. The most frequent and optimal motifs were similar to other bacterial consensus motifs. The optimal spacer length between the two boxes was reported to be 17 bp. It is widely applied to a bioinformatics approach for other bacteria. Unexpectedly, conserved spacer lengths were 11 bp as well as 17 bp in the genus Bifidobacterium. Moreover, the sigma factor of the genus Bifidobacterium has a characteristic domain in the N terminus which may contribute to the additional functions. Hence, it would be valuable to reevaluate the promoter in other organisms.
INTRODUCTION
The mechanisms of transcription and translation in bacteria have been elucidated over three decades (1–4). Transcription initiates when an RNA polymerase core enzyme and a sigma factor (σ factor) complex associate with double-stranded DNA at a promoter region. The primary σ factor of Escherichia coli (σ70, RpoD) recognizes the –35 box (TTGACA) and the –10 box (TATAAT), doublet 6-mer DNA motifs, which are named after their positions relative to the transcription start site (TSS). An approximately 17-bp spacer sequence exists between these two boxes (1, 2, 5).
For translation, AGGAGG is the widely accepted ribosomal binding sequence (RBS), or the Shine-Dalgarno sequence. The RBS is complementary to the 3′ end of 16S rRNA and binds to form the initiation complex of translation, as demonstrated using a limited number of model organisms, including E. coli and Bacillus subtilis. Today, a vast number of bacterial genome sequences have been analyzed using next-generation sequencing technology. Bioinformatics researchers have tried to use the consensus sequence of E. coli to predict promoter regions in other bacteria. For RBS identification, prediction using E. coli sequences appears effective, indicating the potential for an RBS detection algorithm based on the query sequences from the E. coli consensus genome.
In our previous study, we reported a critical alteration of the RBS structure in the whole-genome sequence of Bifidobacterium longum using bioinformatics analysis and a wet lab-based reporter assay (6). Unexpectedly, the most frequently appearing 6-mer consensus identified as a possible RBS in B. longum is not AGGAGG but rather AAGGAG and appears five or six bases upstream from the start codon (6). The AAGGAG version of the RBS shows the highest translation activity in our reporter assay. These data suggest that the rules for translation vary among genetically divergent bacteria and that the RBS sequence requires reevaluation for each species.
For transcription, bioinformatics researchers have also attempted to predict promoter region sequences using the –35 and –10 box consensus sequences from E. coli. However, it has been proposed that each bacterial genus may have various local rules for promoter features because σ factors have evolutionarily diverse structures. In the genus Bifidobacterium, the σ factor has a characteristic polar domain in the N terminus and, therefore, may recognize a specific promoter structure.
Researchers have investigated specific highly expressed promoters in the Bifidobacterium genus (7–9). Comprehensive promoter analysis of the Bifidobacterium breve UCC2003 genome using both transcriptome sequencing (RNA-Seq) and tiling arrays identified the TSSs of over 400 genes and generated a predicted promoter consensus motif of TTGACA-(17-bp spacer)-TATAAT (10).
In this study, we analyzed the core promoter structures in B. longum NCC2705, including the –35 and –10 boxes and the relevant spacer lengths, to attempt to understand the transcription and the gene expression regulatory systems. To do this, we utilized RNA-Seq data and performed bioinformatics analyses using a hidden Markov model. For further analysis, we generated over 40 variants of the hup gene promoter driving expression of the chloramphenicol acetyltransferase (CAT) gene for reporter assay experiments.
RESULTS
Global analysis of the transcription start site.
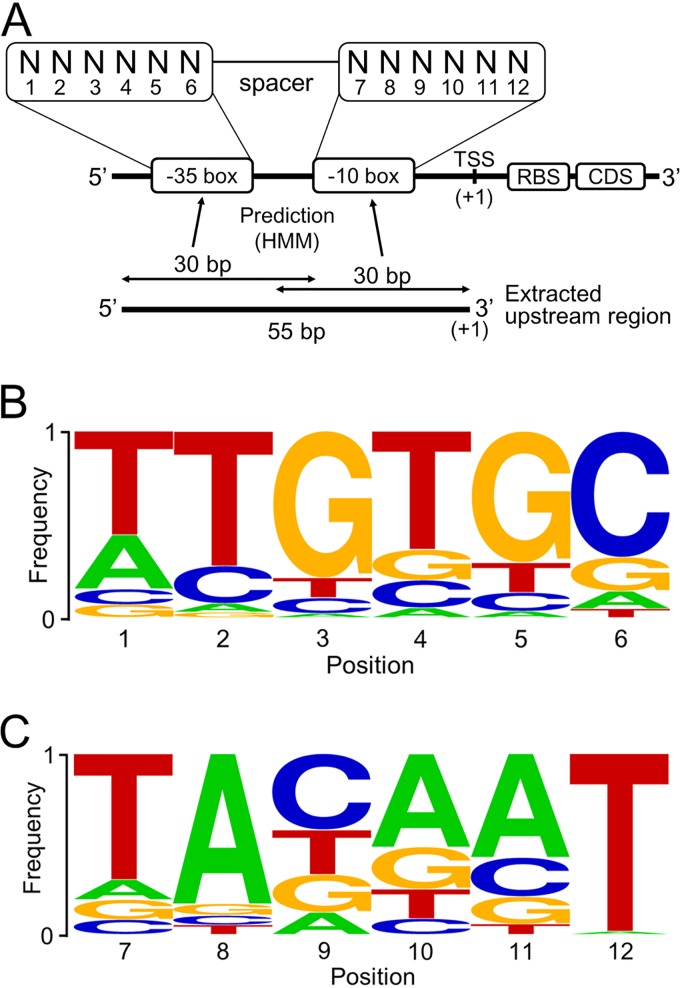
We first surveyed promoter motifs from the genomic sequence data of B. longum NCC2705 by extracting regions extending from the start codon to 150 bp upstream of 1,727 hypothetical genes. The base frequency of these regions did not show the expected promoter motifs but did show an AG-rich motif around the 3′ end, which was thought to be the RBS sequence. AAGGAG was the most frequent 6-base sequence, which is paired with the anti-Shine-Dalgarno sequence in the 16S rRNA and has been reported previously. Promoter motifs were unable to be identified solely using only the genomic sequence. We next attempted to identify promoter motifs using RNA-Seq analysis (GEO accession number GSE143410). Of the 1,727 genes, a total of 269 genes that showed relatively high levels of expression (>700 reads per kilobase per million [RPKM]) were selected to probe for conserved transcription-relevant sequences. We then performed manual curation to assign 130 TSSs of these transcripts to near their respective start codons, where the number of reads increased drastically (Fig. 1A; see Fig. S1 in the supplemental material). About two-thirds of the TSSs were positioned within 100 bp from the start codon.
FIG 1.
The consensus motifs in the –35 and –10 boxes using the hidden Markov model. (A) The putative structure of core promoter motifs. Each box contains a 6-base motif. The positions at –35 and –10 are numbered 1 to 6 and 7 to 12, respectively. The frequencies of residues at each position in the –35 and –10 boxes were predicted using 30-bp sequences from –26 to –55 and from –1 to –30, respectively. (B) Base frequencies in the –35 box. (C) Base frequencies in the –10 box.
Global analysis of core promoter motifs.
To identify two core promoter motifs, the –35 and –10 boxes, upstream regions of the predicted TSSs were analyzed using the hidden Markov model (HMM) (Fig. 1A). This yielded the predicted consensus motifs of the –35 and –10 boxes, which were TTGTGC and TACAAT, respectively (Fig. 1B and C). There were significant differences in the frequencies of the consensus bases. T12 had the highest frequency of 0.88, while C9 had the lowest frequency of 0.45. Frequencies of other bases varied from 0.52 to 0.73. Such differences may represent variations in the importance of different bases in promoter recognition.
As the global determination of TSSs and promoter motifs has been reported in scores of previous studies (10–33), we compiled the data from 26 bacteria belonging to several phyla, including Actinobacteria, Firmicutes, Cyanobacteria, Proteobacteria, and Chlamydiae (Table 1). Overall, TTGNNN in the –35 box and TANNNT in the –10 box were well conserved regardless of the strains, which fits our results shown in Fig. 1B and C.
TABLE 1.
Summary of consensus motifs of the –10 and –35 boxes in 26 bacteriaa
| Phylum | Bacterial strain | Sequence of: |
Reference | |
|---|---|---|---|---|
| –35 box | –10 box | |||
| Actinobacteria | Bifidobacterium longum NCC2705 | TTGTGC | TAYAAT | This study |
| Bifidobacterium breve UCC2003 | TTGACA | TATAAT | 10 | |
| Corynebacterium glutamicum ATCC 13032 | ttgnca | TAnnnT | 11 | |
| Mycolicibacterium smegmatis MC2 155 | Not mentioned | TAnnnT | 12 | |
| Mycobacterium tuberculosis H37Rv | Not mentioned | WAnnnT | 13 | |
| Streptomyces coelicolor A3(2) | nTGACC | TAnnnT | 14 | |
| Firmicutes | Bacillus methanolicus MGA3 | ttgana | TAtaaT | 15 |
| Enterococcus faecalis V583 | TTGACAA | GnTATAAT | 16 | |
| Streptococcus suis P1/7 | Not mentioned | tgnTAtAaT | 17 | |
| Lactococcus lactis subsp. cremoris MG1363 | ttga | tgnTAtAAT | 18 | |
| Cyanobacteria | Nostoc sp. strain PCC 7120 | Not mentioned | Not mentioned | 19 |
| Synechocystis sp. strain PCC 6803 | TTGnnn | Not mentioned | 20 | |
| Proteobacteria | Geobacter sulfurreducens PCA | Not mentioned | Not mentioned | 21 |
| Campylobacter jejuni NCTC 11168 | Not mentioned | TAwAaT | 22 | |
| Helicobacter pylori 26695 | Not mentioned | TAtaaT | 23 | |
| Bradyrhizobium japonicum USDA110 | TTG | AT-rich | 24 | |
| Agrobacterium fabrum C58 | cTTG | TATnnT | 25 | |
| Sinorhizobium meliloti 1021 | cTTGac | ctATat | 26 | |
| Salmonella enterica Typhimurium strain SL1344 | TTgc | TAnnnT | 27 | |
| Klebsiella pneumoniae MGH 78578 | cTTgaca | tgnTAnnnT | 28 | |
| Escherichia coli K-12 MG1655 | cTTgaca | tgnTAnnnT | 28 | |
| Vibrio harveyi FDAARGOS_107 | TTGM | TAnnnT | 29 | |
| Photobacterium profundum SS9 | Not mentioned | Not mentioned | 30 | |
| Xanthomonas campestris pv. vesicatoria 85-10 | Not mentioned | tAnnnT | 31 | |
| Neisseria gonorrhoeae MS11 | Not mentioned | TAHAAT | 32 | |
| Chlamydiae | Chlamydia pneumoniae CWL029 | TTGA | TAnnnT | 33 |
Many previous studies mentioned consensus motifs of the –10 and –35 boxes based on a determination of TSSs in scores of bacteria. In the case of previous reports which did not mention these motifs, “not mentioned” is listed. Uppercase letters indicate higher conservation than lowercase letters, as described in the associated reference.
Activity levels of putative promoter motifs.
We analyzed relationships between the –35 and –10 box sequences and promoter activities using the staphylococcal chloramphenicol acetyltransferase (CAT) gene as a reporter gene. We constructed 37 mutant plasmids, each with 1-bp replacement between the two boxes (Table 2).
TABLE 2.
The efficiencies of the promoter activities at each positiona
| Base | Relative CAT activity |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| –35 box at position: |
–10 box at position: |
|||||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | |
| A | 0.42 IJ | 0.38 JK | 0.34 KL | 0.77 G | 0.58 H | 0.77 G | <0.1 O | 1 D | 0.46 I | 1 D | 1 D | <0.1 O |
| C | 0.39 J | 0.38 JK | 0.32 L | 2.03 B | 0.88 E | 1 D | <0.1 O | <0.1 O | 1 D | 0.17 M | 0.54 H | <0.1 O |
| G | <0.1 O | 0.46 I | 1 D | 0.32 L | 1 D | 0.40 J | <0.1 O | <0.1 O | 0.77 G | 0.82 F | 0.20 M | <0.1 O |
| T | 1 D | 1 D | 0.53 H | 1 D | 0.78F G | 1.42 C | 1 D | <0.1 O | 2.98 A | 0.89 E | 0.12 N | 1 D |
| Consensus | T | T | G | T | G | C | T | A | C | A | A | T |
| Optimal | T | T | G | C | g | t | T | A | T | a | A | T |
The control strain had plasmid pKO403-TPCTcon (Fig. S3). The others had 1-bp mutations in the promoter regions of their plasmids. The position numbers are related to Fig. 1A. The results shown are relative to the value for the control. “Optimal” shows the highest bases in each position. Uppercase letters indicate that the highest bases showed 1.5 times as much activity as the second-highest bases. Boldface indicates the strongest activity in each position. Different letters indicate statistically distinguishable groups (P < 0.01; Tukey’s multiple-comparison test).
Table 2 showed that there were some commonalities between the optimal and consensus motifs. TA7–8 and AT11–12 in the –10 box and TTG1–3 in the –35 box were present in both the optimal and the consensus motifs and showed 1.85- to 10-times-higher activity than replacements at those sites. There were also some differences between the optimal and consensus promoter motifs. T9 in the –10 box and C4 and T6 in the –35 box conferred stronger activities to the optimal motifs than the consensus motifs. In particular, the activities of T9 and C4 were roughly 3 and 2 times higher, respectively, than those of consensus motifs.
Moreover, G5, T6, and A10 conferred stronger promoter activity than the second-strongest replacements of C5, C6, and T10, respectively. Considering the replacements of T9 and C4, we conducted a CAT assay using TTGCGC-(17-bp spacer)-TATAAT. However, the activity was comparable (94%) to the activity of TTGTGC-(17-bp spacer)-TATAAT. We, therefore, concluded the most important motifs for the –35 and –10 boxes in B. longum were TTGNNN and TANNNT, respectively.
Comparison of spacer lengths among bacteria.
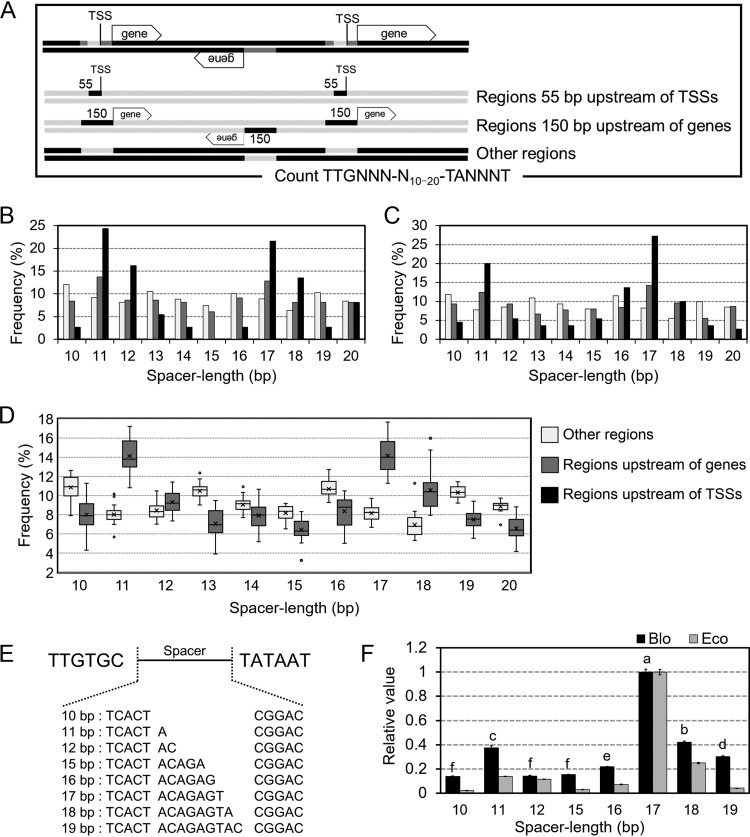
We investigated the spacer length between the –35 and –10 boxes among Bifidobacterium genus members (Fig. 2). Because the consensus promoter motif is rarely conserved, TTG1–3, TA7–8, and T12 were used in the query motifs 5′-TTGNNN-N10–20-TANNNT-3′, which were thought as a well-conserved motif among bacteria. This motif was searched for within regions of genes extending for 150 bp of the gene, the other regions, and extending 55 bp upstream of TSSs in B. longum NCC2705 and B. breve UCC2003, whose TSSs were determined in this study and a previous study, respectively (Fig. 2A to C) (10). Higher frequencies were found for spacer lengths of 17- and 11-bp regions upstream of genes and TSSs. Next, spacer lengths in regions upstream of genes and the other regions were analyzed among 18 Bifidobacterium genus members (34–50) (Fig. 2D; individual data are shown in Fig. S4 in the supplemental material). All strains showed 11- and 17-bp spacer lengths only in regions upstream of genes. To compare the promoter activities among various spacer lengths, the CAT assay was performed using 10- to 12-bp and 15- to 19-bp spacer lengths using B. longum NCC2705 and E. coli TOP10 as hosts (Fig. 2E and F). In B. longum, a 17-bp spacer length produced the highest activity, followed by the 18-bp and 11-bp spacer lengths with the second and third highest activity levels, respectively. In E. coli, although the 17- and 18-bp spacer lengths yielded higher activity, the 11-bp spacer length did not produce comparable activity to that in B. longum.
FIG 2.
Comparison of the frequencies and functions of B. longum NCC2705 and other Bifidobacterium members. (A) Flowchart of the spacer length analysis. Promoter motifs were identified in three sections of genomic sequences, namely, other regions, regions 150 bp upstream of genes, and regions 55 bp upstream of TSSs. (B) The frequency of spacer lengths in B. longum NCC2705. A total of 130 TSSs identified from RNA-Seq results were used to extract regions 55 bp upstream of TSSs. (C) Same as panel B but for B. breve UCC2003. TSSs of 418 genes were obtained from a previous study (10). (D) Frequency of spacer lengths in 18 members of the genus Bifidobacterium. White, gray, and black bars or boxes in panels B, C, and D represent frequencies in other regions, regions 150 bp upstream of genes, and regions 55 bp upstream of TSSs, respectively. (E) The sequences used for reporter assays to assess the function of spacer lengths in vivo. (F) The promoter activity using each spacer length. Plasmids with different spacer lengths were designed based on pKO403-TPCT9. The resulting plasmids were transformed into B. longum NCC2705 and E. coli cells. Results are shown as relative to the results obtained using 17-bp spacer lengths. Values are presented as mean values (±2 standard deviations [SD]). Different letters indicate statistically distinguishable groups (P < 0.01; Tukey’s multiple-comparison test).
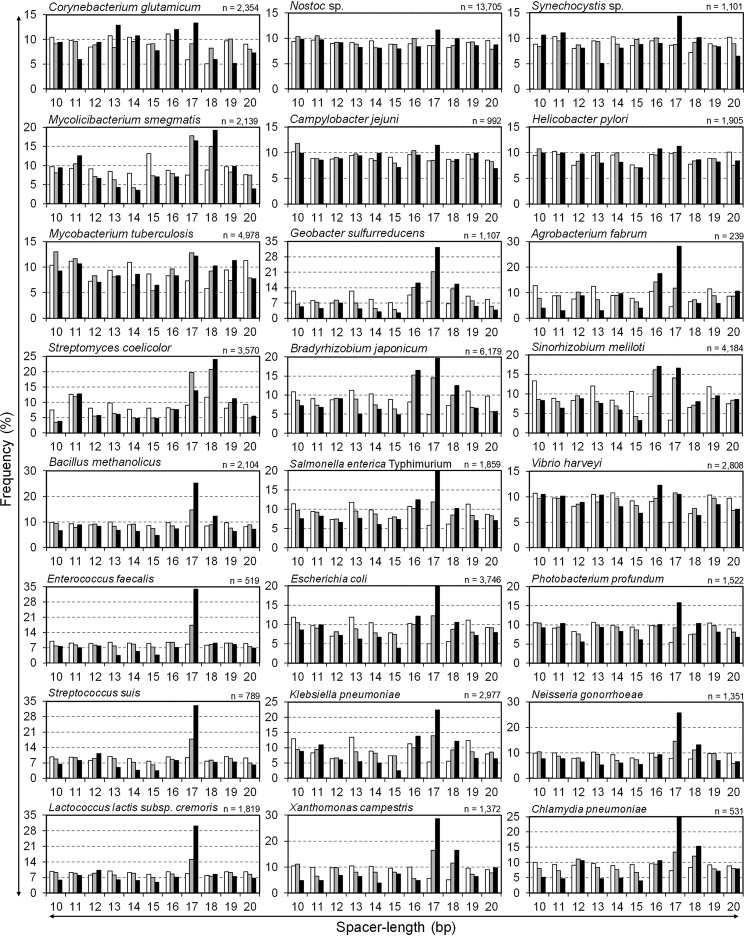
Next, spacer lengths were analyzed in over 70 bacterial species (data partially listed in Fig. 3 and Fig. S4). In particular, the spacer length of the upstream of TSSs was analyzed in 24 bacterial strains shown in Table 1, whose TSSs were previously determined with genome-wide analysis. No bacterial strain in Fig. 3 showed a remarkable peak of 11 bp, whereas peaks of 16 to 18 bp, especially 17 bp, were remarkable in most strains. In particular, strains in the phylum Firmicutes tended to show a single high peak of 17 bp. Among bacteria of the family Bifidobacteriaceae, to which the genus Bifidobacterium belongs, both 11- and 17-bp spacer lengths were often detected to various degrees (Fig. S4). In particular, spacer lengths of Parascardovia denticolens were most often 11 bp long. In contrast, among the phylum Actinobacteria, to which the Bifidobacterium genus belongs, spacer lengths of 17 and 18 bp were common in regions upstream of genes and TSSs, whereas the prevalence of 11-bp spacer lengths was not as clear as the strains in the family Bifidobacteriaceae (Fig. 2 and 3; Fig. S4). As an eccentric strain, Mycoplasma genitalium exhibits a high frequency of 13-bp spacer lengths (Fig. S4).
FIG 3.
Comparison of the frequencies of spacer lengths among bacteria. This figure is similar to Fig. 2B and C but for 24 bacterial strains shown in Table 1 whose TSSs were isolated experimentally. The number in the top right of each graph shows the number of TSSs used for the spacer length analysis. The figure includes 4 bacteria in the phylum Actinobacteria, Corynebacterium glutamicum ATCC 13032, Mycolicibacterium smegmatis MC2 155, Mycobacterium tuberculosis H37Rv, and Streptomyces coelicolor A3(2), 4 bacteria in the phylum Firmicutes, Bacillus methanolicus MGA3, Enterococcus faecalis V583, Streptococcus suis P1/7, and Lactococcus lactis subsp. cremoris MG1363, 2 bacteria in the phylum Cyanobacteria, Nostoc sp. strain PCC 7120 and Synechocystis sp. strain PCC 6803, 13 bacteria in the phylum Proteobacteria, Geobacter sulfurreducens PCA, Campylobacter jejuni subsp. jejuni NCTC 11168, Helicobacter pylori 26695, Bradyrhizobium japonicum USDA110, Agrobacterium fabrum C58, Sinorhizobium meliloti 1021, Salmonella enterica serovar Typhimurium strain SL1344, Klebsiella pneumoniae subsp. pneumoniae MGH 78578, E. coli K-12 MG1655, Vibrio harveyi FDAARGOS_107, Photobacterium profundum SS9, Xanthomonas campestris pv. vesicatoria 85-10, and Neisseria gonorrhoeae MS11, and 1 bacterium in the phylum Chlamydiae, Chlamydia pneumoniae CWL029.
DISCUSSION
We attempted to utilize a bioinformatics approach to predict promoter consensus sequences of B. longum. However, no methods or conditions that we utilized yielded meaningful results. We then attempted to assign TSSs using RNA-Seq analysis. We instead used a standard mRNA preparation and RNA-Seq method, and it was relatively easy to assign the TSSs using read depth profiles manually from the results obtained. We then used the resulting TSSs to determine consensus sequences of the core promoter. In general, mRNA was readily processed in the bacterial cells. As we utilized raw data from RNA-Seq to determine TSSs, our data are likely to have been affected by the process. Our method isolated TSSs only in the half number of genes, which show an expression level of >700 RPKM, which should be caused by the processing of mRNA. To isolate TSSs of such processed mRNA, mRNA must be prepared with intact 5′ termini. In a previous study, a 5′ triphosphate capturing RNA preparation was used for bacterial or mitochondrial RNA-Seq (51).
About two-thirds of the TSSs were located within 40 bp of the start codon. The remaining TSSs, located >40 bp from the start codon, may be associated with other transcriptional regulators, such as unknown short peptide-coding regions, small regulatory RNAs, or riboswitches and require further investigation (52, 53).
Although the B. longum NCC2705 genome is relatively GC rich for bacteria (60%), TA7–8 and T12 had the highest base occurrence frequencies (0.67 to 0.88) (Fig. 1C) and had the most dramatic decreases in activity upon mutation (Table 2). The frequency of C9 was the lowest (∼0.45) and had increased activity upon mutation to T, which is the second-highest base (Fig. 1C; Table 2). It was suggested that there might be a positive correlation between the occurrence frequency of a particular base in the –10 box and its impact on promoter activity. Based on this idea, the most important motifs in the –10 box are TA7–8 and T12. In contrast, for the TTG1–3 and TGC4–6 motifs in the –35 box, the frequency of each base was 0.52 to 0.70 (Fig. 1B), which was a smaller range than that of the –10 box (0.45 to 0.88) (Fig. 1C). From the reporter assay, the consensus motif TTG1–3 functioned as the optimal motif (Table 2). Previous studies also indicated that TTG1–3, TA7–8, and T12 were the most highly conserved motifs in the promoters of highly expressed genes in B. breve UCC2003 (10), E. coli (1, 2), and other bacteria (Table 1) and are likely universal motifs. Overall, replacements of sequences in the –10 box had a more significant impact on promoter activities than replacements in the –35 box, indicating that the –10 box, especially TANNNT, is the minimum structure of the core promoter because the –10 box is the DNA motif bound by the σ70 factor that becomes the origin of transcription in E. coli and all promoters do not have a –35 box. Positions 4 to 6 in the –35 box (Fig. 1A) showed uncertain activities because the complete optimal motif from Table 2, TTGCGC-(17-bp spacer)-TATAAT, resembles the activity of TTGTGC-(17-bp spacer)-TATAAT. In general, the consensus motif of positions 4 to 6 was ACA, but the degree of conservation was lower than any other position (1, 2). Moreover, promoter activity is affected by several factors, such as the up-element interacting alpha subunit and the extended –10 box (TGNTATAAT), among others, as well as the –35 and the –10 boxes (54, 55). Positions 4 to 6 in the –35 box may not have an absolute motif contributing to promoter activity, and the optimal motif is likely to be affected by the sequences or structure around the box and variable according to each promoter.
The consensus motif of the –35 box differed little from that identified by a previous study of B. breve UCC2003. This finding could be the result of the differences in analyzing consensus motifs. A previous study in B. breve was based on the supposition that spacer length should be 14 to 20 bp. In contrast, as our analysis with HMM isolated the motifs separately in individual boxes, our –35 box consensus motif in Fig. 1B shows the best hit 6-bp motif from −26 to −55 of the upstream regions of TSSs. Moreover, we utilized 130 TSSs, which is a lower number than that in previous studies of B. breve and other bacteria. To determine consensus motifs precisely, more information regarding TSSs must be elucidated.
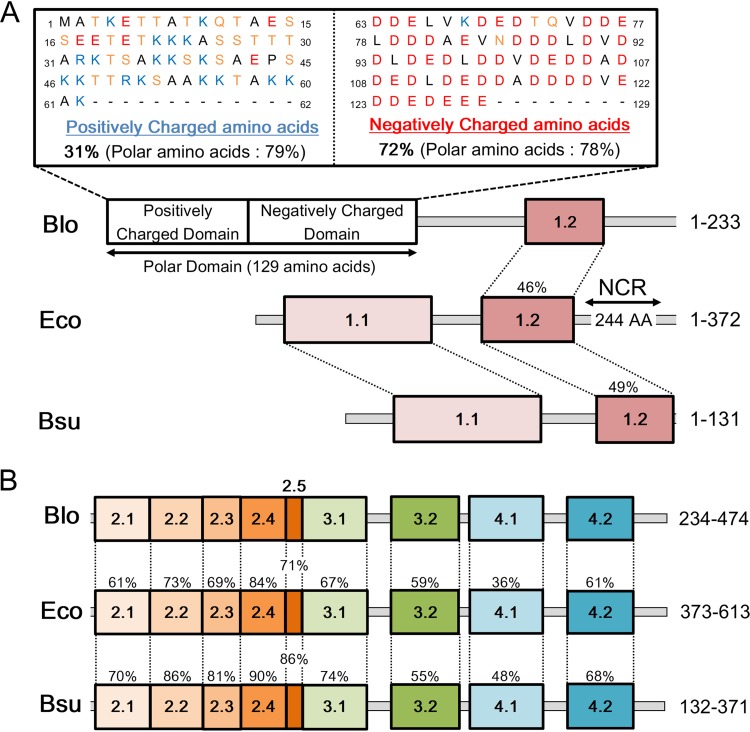
We further compared the structures of the primary σ factor from B. longum NCC2705, E. coli K-12 MG1655, and Bacillus subtilis 168 (Fig. 4) (56, 57). The primary σ factor in B. longum has a distinct polar domain at the N terminus consisting of a positively charged and a negatively charged domain (Fig. 4A). Such a domain does not exist in the σ factors of E. coli and B. subtilis but is conserved among other members of the genus Bifidobacterium and the phylum Actinobacteria (see Fig. S7 in the supplemental material). Domains 2 and 4, especially 2.4 and 4.2, are thought to be essential regions for binding to the –10 and –35 boxes of promoters, respectively (5, 58–60). Domain 2.4 of bifidobacterial σ70 shares 84% and 90% identity with E. coli and B. subtilis, respectively (Fig. 4B). In contrast, domain 4.2 of bifidobacterial σ70 shares only 61% and 68% homology with its E. coli and B. subtilis counterparts, respectively (Fig. 4B). Moreover, domains 2 to 4 of the primary σ factors show high homology among bacteria (Fig. 4B; see Fig. S6 in the supplemental material), which results in high conservation of the promoter motifs, as shown in Table 1.
FIG 4.
Comparison of the σ factors of B. longum NCC2705 (Blo), E. coli K-12 MG1655 (Eco), and B. subtilis 168 (Bsu). (A) N-terminal region to domain 2.1. NCR, nonconserved region. (B) C-terminal region starting from domain 2.1. Percentages indicate the conservation of each domain. A typical polar domain, consisting of a positively charged domain and a negatively charged domain, existing only in Blo; positively charged amino acids are K, R, and H (blue); negatively charged amino acids are D and E (red); and noncharged polar amino acids are N, S, T, and Q (yellow). The numbers on the right side indicate the amino acid positions of the regions.
Our results (Fig. 2F and 3) indicate that 17 bp is the optimal spacer length of the –35 and –10 boxes in all bacteria, which agrees with a previous report (2). In contrast, both 11-bp and 17-bp spacer lengths were detected in the regions upstream of genes and TSSs in members of the genus Bifidobacterium (Fig. 2B to D). Figure 2F shows our results indicating that a 17-bp spacer length produces the highest activity, while an 11-bp spacer length produces unusually high activity that is posited to be associated with the frequency of this spacer length. A previous study reported the identification of over 400 TSSs in B. breve UCC2003 and identified an optimal spacer length of 17 bp based on a bioinformatics approach (10). However, we reevaluated spacer lengths using these TSSs and found higher frequencies of 11-bp spacers as well as 17-bp spacers (Fig. 2C).
In general, the effectiveness of transcription using –35 and –10 boxes depends on the identity of the primary σ factor. Studies so far have not indicated multiple optimal spacer lengths for a given primary σ factor. Bacteria belonging to the genus Bifidobacterium possess at least two types of the conserved σ factors, with one serving as the primary σ factor like the σ70 of E. coli, and the other acting as an alternative σ factor. At least, the primary σ factor (BL1428 in B. longum NCC2705) should recognize the 17-bp spacer length because of the commonality of domains 2 to 4 (Fig. 4). In contrast, the alternative σ factor (BL1357 in B. longum NCC2705) is significantly different from the primary σ factor in terms of amino acid sequences (the detailed alignment of domains 2 and 4 is shown in Fig. S5 in the supplemental material), indicating that the alternative σ factor might not be related to the recognition of the motif TTGNNN-spacer-TANNNT.
A feasible explanation for multiple spacer lengths in the genus Bifidobacterium is that the primary σ factor can recognize each spacer type. The structures of σ factors in the genus Bifidobacterium include a polar domain in the N-terminal region (Fig. 4A; Fig. S6). However, as this polar domain has no relationship to any other studied protein, it is difficult to predict the three-dimensional (3D) structure of the domain and analyze its interaction with DNA sequences or RNA polymerase. A recent study indicated that the nonconserved region (NCR) of the E. coli σ factor, which is not conserved in B. longum and B. subtilis (Fig. 1A), interacts with a region upstream of the –10 box (61). This finding suggests that unknown domains, such as polar domains, also serve specific functions like DNA recognition or small-molecule binding. Moreover, various types of additional N-terminal polar domains are found in bacteria belonging to the phylum Actinobacteria and others (Fig. S7). These domains lack amino acid sequence homology, and their function remains unknown.
In contrast, the 11-bp spacer length is conserved only among the family Bifidobacteriaceae, especially in P. denticolens (Fig. S4), whereas other bacteria in the phylum Actinobacteria lack 11-bp spacers but have 17- to 18-bp spacers (Fig. 3), indicating that the 11-bp spacer length is only conserved in Bifidobacterium and related species. For these reasons, the polar domain is likely not related to the recognition of the 11-bp spacer length. For the further elucidation of the 11-bp spacer length, we consider a different point of view, such as specific growth stages, in the presence of a specific cofactor or during certain environmental stresses.
Bacteria in the phylum Firmicutes have simple σ factors with no unknown domains like B. subtilis and tend to have a significant and single peak at 17 bp, indicating a single spacer length (Fig. 3 and 4). In contrast, M. genitalium uniquely shows three prominent spacer lengths of 4 bp, 13 bp, and 22 bp, and its primary σ factor lacks domains 1.1 to 1.2 but includes an additional unknown 249-amino-acid sequence (Fig. S4).
In this study, we used transcriptome analysis, bioinformatics approaches, and a reporter-based assay and showed that the bifidobacterial σ factor and promoter structure include an unknown polar domain and two core sequence boxes, respectively, and the preferred spacer length is likely different from that of E. coli. These results highlight the possibility of diverse promoter structures among bacterial species. Moreover, we also showed that raw RNA-Seq data could be used to assign TSSs and to determine the consensus sequences of core promoter motifs. The recognition rules of σ factors with additional N-terminal domains should be investigated in the future, as this knowledge may expand our understanding of bacterial gene expression.
MATERIALS AND METHODS
Media and growth conditions.
E. coli TOP10 cells were grown in 5 ml LB medium (1% tryptone, 0.5% yeast extract, and 1% NaCl) at 37°C with shaking. Bifidobacterium longum strains were grown in 12 ml MRS medium at 37°C under anaerobic conditions. Solid agar medium was used at a concentration of 1.5% (wt/vol). Spectinomycin (Sp) supplementation was provided at 75 μg/ml for transformant cultivation.
Bacterial strains and plasmid construction.
Strains, plasmids, and oligo-DNA primers used in this study are listed in Table 1 and Table S1 and S2 in the supplemental material. Thirteen plasmids were constructed for the CAT reporter assay. A control plasmid (pKO403-TPCTcon) based on pKO403 (62) was constructed using the Golden Gate method (63, 64). This control plasmid has two consensus motifs, namely, TTGTGC and TACAAT, separated by a 17-bp spacer. Using this plasmid as a template, we designed 36 mutant plasmids with single base alterations in both boxes. Each promoter region was PCR-amplified using phosphorylated primers and then the amplified fragments were self-ligated and introduced into E. coli TOP10 cells. Transformants were selected on LB (Sp) agar plates. Transformant plasmids were extracted, nucleotide sequences confirmed, and then plasmids introduced into B. longum NCC2705 via electroporation (MicroPulser; Bio-Rad, CA). The transformants were then selected on MRS agar plates (Sp).
Promoter assay using the CAT reporter.
CAT is a resistance protein for chloramphenicol that catalyzes the acetylation of chloramphenicol with acetyl coenzyme A (acetyl-CoA). The sulfhydryl (SH) group, in the resulting CoA-SH, reacts with 5, 5-dithiobis (2-nitrobenzoic acid) (DTNB; λmax = 325 nm) to form 5-mercapto-2-nitrobenzoic acid (TNB; λmax = 412 nm). For the CAT reporter promoter assay, transformants were cultured in MRS medium (Sp) at 37°C until the absorbance at 660 nm reached 0.6. Cells were then washed twice with phosphate-buffered saline (PBS) by centrifugation (15,000 × g, 5 min, 4°C), sonicated for 2 min, and then centrifuged again to remove debris (15,000 × g, 15 min, 4°C). A 5-μl aliquot of supernatant was then mixed with 295 μl of CAT assay solution (100 mM Tris-HCl [pH 8.0], 2.5 mM DTNB, 5.0 mM acetyl-CoA, and 0.3% [wt/vol] chloramphenicol) in a 96-well plate and incubated at 37°C for 5 min, and then the absorbance was measured at 414 nm. CAT assays were performed on extracts of three independent bacterial cultures.
RNA-Seq experiments.
To prepare total RNA, a 48-ml culture was centrifuged (5,000 × g, 10 min, 4°C), and the resulting pellet was resuspended in 500 μl of Tris-EDTA (TE) buffer (10 mM Tris-HCl [pH 8.0] and 1.0 mM EDTA). The RNAprotect bacterial reagent (1 ml; Qiagen, Hilden, Germany) was added to the sample and then vortexed and centrifuged (5,000 × g, 10 min). After the supernatant was removed, the cells were suspended in a cell wall lysis solution called Labiase (2 mg/ml in 10 mM sodium citrate [pH 4.0]; OZEKI, Hyogo, Japan) and proteinase K (10 mg/ml in TE buffer, Wako Pure Chem, Osaka, Japan), incubated for 10 min at room temperature, mixed with 1 ml TRIzol (Thermo Fisher Scientific, MA), vortexed for 1 min, incubated for 3 min at room temperature, and then mixed with 200 μl chloroform. The mixture was vortexed for 15 s, incubated for 5 min at room temperature, and then centrifuged (12,000 × g, 15 min, 4°C). The supernatant was transferred to a new tube, mixed with 0.5 ml 2-propanol, incubated for 10 min at room temperature, and then centrifuged (12,000 × g, 10 min, 4°C) to prepare RNA. The pellet was rinsed with 75% ethanol, centrifuged (7,500 × g, 5 min, 4°C), and then dissolved in 50 μl RNase-free water.
The total RNA preparation was then treated with the Ribo-Zero rRNA removal kit (Illumina, CA) to deplete the rRNA and then reverse transcribed. RNA-Seq was performed using a MiSeq instrument (Illumina) The fragment depth graph was used to assign the transcription start sites (TSSs).
Bioinformatics analysis.
The 55-bp-long upstream sequences were extracted from 130 TSSs and then analyzed by HMM using the pattern discovery function of CLC Genomics Workbench v. 5.1 (Qiagen) to identify the promoter motifs. The upper 30 bp and the lower 30 bp of extracted sequences were used to analyze –35 and –10 box sequences, respectively (Fig. 3A).
The motif, shown in Fig. 2 and 3 and in Fig. S4, was searched for in sequences of the entire genome, including 150-bp upstream regions of all genes and upstream regions of TSSs in 18 members of the Bifidobacterium genus (Table S2) and other bacterial strains. Genomic sequences and coding sequences were obtained from the NCBI nucleotide database. Spacer length was analyzed using an original Perl script (see the supplemental text).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by JSPS KAKENHI (grant number 16H04896) and a research grant from The Skylark Food Science Institute.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Hawley DK, McClure WR. 1983. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res 11:2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harley CB, Reynolds RP. 1987. Analysis of E. coli promoter sequences. Nucleic Acids Res 15:2343–2361. doi: 10.1093/nar/15.5.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shine J, Dalgarno L. 1974. The 3’-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A 71:1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shine J, Dalgarno L. 1975. Determinant of cistron specificity in bacterial ribosomes. Nature 254:34–38. doi: 10.1038/254034a0. [DOI] [PubMed] [Google Scholar]
- 5.Murakami KS, Darst SA. 2003. Bacterial RNA polymerases: the wholo story. Curr Opin Struct Biol 13:31–39. doi: 10.1016/s0959-440x(02)00005-2. [DOI] [PubMed] [Google Scholar]
- 6.He J, Sakaguchi K, Suzuki T. 2012. Determination of the ribosome-binding sequence and spacer length between binding site and initiation codon for efficient protein expression in Bifidobacterium longum 105-A. J Biosci Bioeng 113:442–444. doi: 10.1016/j.jbiosc.2011.11.019. [DOI] [PubMed] [Google Scholar]
- 7.Wang Y, Kim JY, Park MS, Ji GE. 2012. Novel Bifidobacterium promoters selected through microarray analysis lead to constitutive high-level gene expression. J Microbiol 50:638–643. doi: 10.1007/s12275-012-1591-x. [DOI] [PubMed] [Google Scholar]
- 8.Sun Z, Westermann C, Yuan J, Riedel CU. 2014. Experimental determination and characterization of the gap promoter of Bifidobacterium bifidum S17. Bioengineered 5:371–377. doi: 10.4161/bioe.34423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sakanaka M, Tamai S, Hirayama Y, Onodera A, Koguchi H, Kano Y, Yokota A, Fukiya S. 2014. Functional analysis of bifidobacterial promoters in Bifidobacterium longum and Escherichia coli using the α-galactosidase gene as a reporter. J Biosci Bioeng 118:489–495. doi: 10.1016/j.jbiosc.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 10.Bottacini F, Zomer A, Milani C, Ferrario C, Lugli GA, Egan M, Ventura M, van Sinderen D. 2017. Global transcriptional landscape and promoter mapping of the gut commensal Bifidobacterium breve UCC2003. BMC Genomics 18:991. doi: 10.1186/s12864-017-4387-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pfeifer-Sancar K, Mentz A, Rückert C, Kalinowski J. 2013. Comprehensive analysis of the Corynebacterium glutamicum transcriptome using an improved RNAseq technique. BMC Genomics 14:888. doi: 10.1186/1471-2164-14-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li X, Mei H, Chen F, Tang Q, Yu Z, Cao X, Andongma BT, Chou S-H, He J. 2017. Transcriptome landscape of Mycobacterium smegmatis. Front Microbiol 8:2505. doi: 10.3389/fmicb.2017.02505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shell SS, Wang J, Lapierre P, Mir M, Chase MR, Pyle MM, Gawande R, Ahmad R, Sarracino DA, Ioerger TR, Fortune SM, Derbyshire KM, Wade JT, Gray TA. 2015. Leaderless transcripts and small proteins are common features of the mycobacterial translational landscape. PLoS Genet 11:e1005641. doi: 10.1371/journal.pgen.1005641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jeong Y, Kim J-N, Kim MW, Bucca G, Cho S, Yoon YJ, Kim B-G, Roe J-H, Kim SC, Smith CP, Cho B-K. 2016. The dynamic transcriptional and translational landscape of the model antibiotic producer Streptomyces coelicolor A3(2). Nat Commun 7:11605. doi: 10.1038/ncomms11605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Irla M, Neshat A, Brautaset T, Rückert C, Kalinowski J, Wendisch VF. 2015. Transcriptome analysis of thermophilic methylotrophic Bacillus methanolicus MGA3 using RNA-sequencing provides detailed insights into its previously uncharted transcriptional landscape. BMC Genomics 16:73. doi: 10.1186/s12864-015-1239-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Innocenti N, Golumbeanu M, Fouquier d'Hérouël A, Lacoux C, Bonnin RA, Kennedy SP, Wessner F, Serror P, Bouloc P, Repoila F, Aurell E. 2015. Whole-genome mapping of 5′ RNA ends in bacteria by tagged sequencing: a comprehensive view in Enterococcus faecalis. RNA 21:1018–1030. doi: 10.1261/rna.048470.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu Z, Wu C, Shao J, Zhu Z, Wang W, Zhang W, Tang M, Pei N, Fan H, Li J, Yao H, Gu H, Xu X, Lu C. 2014. The Streptococcus suis transcriptional landscape reveals adaptation mechanisms in pig blood and cerebrospinal fluid. RNA 20:882–898. doi: 10.1261/rna.041822.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van der Meulen SB, de Jong A, Kok J. 2016. Transcriptome landscape of Lactococcus lactis reveals many novel RNAs including a small regulatory RNA involved in carbon uptake and metabolism. RNA Biol 13:353–366. doi: 10.1080/15476286.2016.1146855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mitschke J, Vioque A, Haas F, Hess WR, Muro-Pastor AM. 2011. Dynamics of transcriptional start site selection during nitrogen stress-induced cell differentiation in Anabaena sp. PCC7120. Proc Natl Acad Sci U S A 108:20130–20135. doi: 10.1073/pnas.1112724108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitschke J, Georg J, Scholz I, Sharma CM, Dienst D, Bantscheff J, Voss B, Steglich C, Wilde A, Vogel J, Hess WR. 2011. An experimentally anchored map of transcriptional start sites in the model cyanobacterium Synechocystis sp. PCC6803. Proc Natl Acad Sci U S A 108:2124–2129. doi: 10.1073/pnas.1015154108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qiu Y, Cho B-K, Park YS, Lovley D, Palsson BO, Zengler K. 2010. Structural and operational complexity of the Geobacter sulfurreducens genome. Genome Res 20:1304–1311. doi: 10.1101/gr.107540.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Porcelli I, Reuter M, Pearson BM, Wilhelm T, van Vliet A. 2013. Parallel evolution of genome structure and transcriptional landscape in the Epsilonproteobacteria. BMC Genomics 14:616. doi: 10.1186/1471-2164-14-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sharma CM, Hoffmann S, Darfeuille F, Reignier J, Findeiss S, Sittka A, Chabas S, Reiche K, Hackermüller J, Reinhardt R, Stadler PF, Vogel J. 2010. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature 464:250–255. doi: 10.1038/nature08756. [DOI] [PubMed] [Google Scholar]
- 24.Čuklina J, Hahn J, Imakaev M, Omasits U, Förstner KU, Ljubimov N, Goebel M, Pessi G, Fischer H-M, Ahrens CH, Gelfand MS, Evguenieva-Hackenberg E. 2016. Genome-wide transcription start site mapping of Bradyrhizobium japonicum grown free-living or in symbiosis—a rich resource to identify new transcripts, proteins and to study gene regulation. BMC Genomics 17:302. doi: 10.1186/s12864-016-2602-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilms I, Overlöper A, Nowrousian M, Sharma CM, Narberhaus F. 2012. Deep sequencing uncovers numerous small RNAs on all four replicons of the plant pathogen Agrobacterium tumefaciens. RNA Biol 9:446–457. doi: 10.4161/rna.17212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schlüter J-P, Reinkensmeier J, Barnett MJ, Lang C, Krol E, Giegerich R, Long SR, Becker A. 2013. Global mapping of transcription start sites and promoter motifs in the symbiotic α-proteobacterium Sinorhizobium meliloti 1021. BMC Genomics 14:156. doi: 10.1186/1471-2164-14-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kroger C, Dillon SC, Cameron ADS, Papenfort K, Sivasankaran SK, Hokamp K, Chao Y, Sittka A, Hebrard M, Handler K, Colgan A, Leekitcharoenphon P, Langridge GC, Lohan AJ, Loftus B, Lucchini S, Ussery DW, Dorman CJ, Thomson NR, Vogel J, Hinton J. 2012. The transcriptional landscape and small RNAs of Salmonella enterica serovar Typhimurium. Proc Natl Acad Sci U S A 109:E1277–E1286. doi: 10.1073/pnas.1201061109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim D, Hong J-J, Qiu Y, Nagarajan H, Seo J-H, Cho B-K, Tsai S-F, Palsson BØ. 2012. Comparative analysis of regulatory elements between Escherichia coli and Klebsiella pneumoniae by genome-wide transcription start site profiling. PLoS Genet 8:e1002867. doi: 10.1371/journal.pgen.1002867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Plágaro AH, Pearman PB, Kaberdin VR. 2019. Defining the transcription landscape of the Gram-negative marine bacterium Vibrio harveyi. Genomics 111:1547–1556. doi: 10.1016/j.ygeno.2018.10.013. [DOI] [PubMed] [Google Scholar]
- 30.Campanaro S, De Pascale F, Telatin A, Schiavon R, Bartlett DH, Valle G. 2012. The transcriptional landscape of the deep-sea bacterium Photobacterium profundum in both a toxR mutant and its parental strain. BMC Genomics 13:567. doi: 10.1186/1471-2164-13-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmidtke C, Findeiss S, Sharma CM, Kuhfuss J, Hoffmann S, Vogel J, Stadler PF, Bonas U. 2012. Genome-wide transcriptome analysis of the plant pathogen Xanthomonas identifies sRNAs with putative virulence functions. Nucleic Acids Res 40:2020–2031. doi: 10.1093/nar/gkr904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Remmele CW, Xian Y, Albrecht M, Faulstich M, Fraunholz M, Heinrichs E, Dittrich MT, Müller T, Reinhardt R, Rudel T. 2014. Transcriptional landscape and essential genes of Neisseria gonorrhoeae. Nucleic Acids Res 42:10579–10595. doi: 10.1093/nar/gku762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Albrecht M, Sharma CM, Dittrich MT, Müller T, Reinhardt R, Vogel J, Rudel T. 2011. The transcriptional landscape of Chlamydia pneumoniae. Genome Biol 12:R98. doi: 10.1186/gb-2011-12-10-r98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schell MA, Karmirantzou M, Snel B, Vilanova D, Berger B, Pessi G, Zwahlen M-C, Desiere F, Bork P, Delley M, Pridmore RD, Arigoni F. 2002. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc Natl Acad Sci U S A 99:14422–14427. doi: 10.1073/pnas.212527599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen X, Zhiguo E, Gu D, Lv L, Li Y. 2015. Complete genome sequence of Bifidobacterium actinocoloniiforme type strain DSM 22766T, isolated from bumblebee digestive tracts. Genome Announc 3:e01084-15. doi: 10.1128/genomeA.01084-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Milani C, Lugli GA, Duranti S, Turroni F, Bottacini F, Mangifesta M, Sanchez B, Viappiani A, Mancabelli L, Taminiau B, Delcenserie V, Barrangou R, Margolles A, van Sinderen D, Ventura M. 2014. Genomic encyclopedia of type strains of the genus Bifidobacterium. Appl Environ Microbiol 80:6290–6302. doi: 10.1128/AEM.02308-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Toh H, Hayashi J, Oshima K, Nakano A, Takayama Y, Takanashi K, Morita H, Hattori M. 2015. Complete genome sequence of Bifidobacterium dentium strain JCM 1195T, isolated from human dental caries. Genome Announc 3:e00284-15. doi: 10.1128/genomeA.00284-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vazquez-Gutierrez P, Lacroix C, Chassard C, Klumpp J, Jans C, Stevens M. 2015. Complete and assembled genome sequence of Bifidobacterium kashiwanohense PV20-2, isolated from the feces of an anemic Kenyan infant. Genome Announc 3:e01467-14. doi: 10.1128/genomeA.01467-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morita H, Toh H, Oshima K, Nakano A, Arakawa K, Takayama Y, Kurokawa R, Takanashi K, Honda K, Hattori M. 2015. Complete genome sequence of Bifidobacterium pseudocatenulatum JCM 1200T isolated from infant feces. J Biotechnol 210:68–69. doi: 10.1016/j.jbiotec.2015.06.416. [DOI] [PubMed] [Google Scholar]
- 40.Vazquez-Gutierrez P, Lacroix C, Chassard C, Klumpp J, Stevens MJA, Jans C. 2015. Bifidobacterium pseudolongum strain PV8-2, isolated from a stool sample of an anemic Kenyan infant. Genome Announc 3:e01469-14. doi: 10.1128/genomeA.01469-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Toh H, Oshima K, Nakano A, Yamashita N, Iioka E, Kurokawa R, Morita H, Hattori M. 2015. Complete genome sequence of Bifidobacterium scardovii strain JCM 12489T, isolated from human blood. Genome Announc 3:e00285-15. doi: 10.1128/genomeA.00285-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jans C, Lacroix C, Follador R, Stevens M. 2013. Complete genome sequence of the probiotic Bifidobacterium thermophilum strain RBL67. Genome Announc 1:e00191-13. doi: 10.1128/genomeA.00191-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yasui K, Tabata M, Yamada S, Abe T, Ikemura T, Osawa R, Suzuki T. 2009. Intra-species diversity between seven Bifidobacterium adolescentis strains identified by genome-wide tiling array analysis. Biosci Biotechnol Biochem 73:1422–1424. doi: 10.1271/bbb.80843. [DOI] [PubMed] [Google Scholar]
- 44.Morita H, Toh H, Oshima K, Nakano A, Kiuchi M, Kuroyanagi H, Arakawa K, Suda W, Honda K, Hattori M. 2015. Complete genome sequence of Bifidobacterium angulatum JCM 7096T isolated from human feces. J Biotechnol 211:10–11. doi: 10.1016/j.jbiotec.2015.06.412. [DOI] [PubMed] [Google Scholar]
- 45.Liu L, Qin Y, Wang Y, Li H, Shang N, Li P. 2014. Complete genome sequence of Bifidobacterium animalis RH, a probiotic bacterium producing exopolysaccharides. J Biotechnol 189:86–87. doi: 10.1016/j.jbiotec.2014.08.041. [DOI] [PubMed] [Google Scholar]
- 46.Bottacini F, Milani C, Turroni F, Sánchez B, Foroni E, Duranti S, Serafini F, Viappiani A, Strati F, Ferrarini A, Delledonne M, Henrissat B, Coutinho P, Fitzgerald GF, Margolles A, van Sinderen D, Ventura M. 2012. Bifidobacterium asteroides PRL2011 genome analysis reveals clues for colonization of the insect gut. PLoS One 7:e44229. doi: 10.1371/journal.pone.0044229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhurina D, Zomer A, Gleinser M, Brancaccio VF, Auchter M, Waidmann MS, Westermann C, van Sinderen D, Riedel CU. 2011. Complete genome sequence of Bifidobacterium bifidum S17. J Bacteriol 193:301–302. doi: 10.1128/JB.01180-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.O'Connell Motherway M, Zomer A, Leahy SC, Reunanen J, Bottacini F, Claesson MJ, O'Brien F, Flynn K, Casey PG, Munoz JAM, Kearney B, Houston AM, O'Mahony C, Higgins DG, Shanahan F, Palva A, de Vos WM, Fitzgerald GF, Ventura M, O'Toole PW, van Sinderen D. 2011. Functional genome analysis of Bifidobacterium breve UCC2003 reveals type IVb tight adherence (Tad) pili as an essential and conserved host-colonization factor. Proc Natl Acad Sci U S A 108:11217–11222. doi: 10.1073/pnas.1105380108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morita H, Toh H, Oshima K, Nakano A, Yamashita N, Iioka E, Arakawa K, Suda W, Honda K, Hattori M. 2015. Complete genome sequence of Bifidobacterium catenulatum JCM 1194T isolated from human feces. J Biotechnol 210:25–26. doi: 10.1016/j.jbiotec.2015.06.415. [DOI] [PubMed] [Google Scholar]
- 50.Jung D-H, Chung W-H, Seo D-H, Nam Y-D, Yoon S, Park C-S. 2018. Complete genome sequence of Bifidobacterium choerinum FMB-1, a resistant starch-degrading bacterium. J Biotechnol 274:28–32. doi: 10.1016/j.jbiotec.2018.03.009. [DOI] [PubMed] [Google Scholar]
- 51.Kanamori-Katayama M, Itoh M, Kawaji H, Lassmann T, Katayama S, Kojima M, Bertin N, Kaiho A, Ninomiya N, Daub CO, Carninci P, Forrest ARR, Hayashizaki Y. 2011. Unamplified cap analysis of gene expression on a single-molecule sequencer. Genome Res 21:1150–1159. doi: 10.1101/gr.115469.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu D-Q, Liu F, Sun Y, Yang L-M, Xin L, Meng X-C. 2015. Genome-wide identification of small RNAs in Bifidobacterium animalis subsp. lactis KLDS 2.0603 and their regulation role in the adaption to gastrointestinal environment. PLoS One 10:e0117373. doi: 10.1371/journal.pone.0117373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nudler E, Mironov AS. 2004. The riboswitch control of bacterial metabolism. Trends Biochem Sci 29:11–17. doi: 10.1016/j.tibs.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 54.Ross W, Gosink K, Salomon J, Igarashi K, Zou C, Ishihama A, Severinov K, Gourse R. 1993. A third recognition element in bacterial promoters: DNA binding by the alpha subunit of RNA polymerase. Science 262:1407–1413. doi: 10.1126/science.8248780. [DOI] [PubMed] [Google Scholar]
- 55.Mitchell JE, Zheng D, Busby SJW, Minchin SD. 2003. Identification and analysis of “extended -10” promoters in Escherichia coli. Nucleic Acids Res 31:4689–4695. doi: 10.1093/nar/gkg694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Blattner FR, Plunkett G, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, Gregor J, Davis NW, Kirkpatrick HA, Goeden MA, Rose DJ, Mau B, Shao Y. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 57.Kunst F, Ogasawara N, Moszer I, Albertini AM, Alloni G, Azevedo V, Bertero MG, Bessières P, Bolotin A, Borchert S, Borriss R, Boursier L, Brans A, Braun M, Brignell SC, Bron S, Brouillet S, Bruschi CV, Caldwell B, Capuano V, Carter NM, Choi S-K, Codani J-J, Connerton IF, Cummings NJ, Daniel RA, Denizot F, Devine KM, Düsterhöft A, Ehrlich SD, Emmerson PT, Entian KD, Errington J, Fabret C, Ferrari E, Foulger D, Fritz C, Fujita M, Fujita Y, Fuma S, Galizzi A, Galleron N, Ghim S-Y, Glaser P, Goffeau A, Golightly EJ, Grandi G, Guiseppi G, Guy BJ, Haga K, et al. 1997. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature 390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 58.Vassylyev DG, Sekine S, Laptenko O, Lee J, Vassylyeva MN, Borukhov S, Yokoyama S. 2002. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 Å resolution. Nature 417:712–719. doi: 10.1038/nature752. [DOI] [PubMed] [Google Scholar]
- 59.Campbell EA, Muzzin O, Chlenov M, Sun JL, Olson CA, Weinman O, Trester-Zedlitz ML, Darst SA. 2002. Structure of the bacterial RNA polymerase promoter specificity σ subunit. Mol Cell 9:527–539. doi: 10.1016/s1097-2765(02)00470-7. [DOI] [PubMed] [Google Scholar]
- 60.Feklistov A, Darst SA. 2011. Structural basis for promoter −10 element recognition by the bacterial RNA polymerase σ subunit. Cell 147:1257–1269. doi: 10.1016/j.cell.2011.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Narayanan A, Vago FS, Li K, Qayyum MZ, Yernool D, Jiang W, Murakami KS. 2018. Cryo-EM structure of Escherichia coli σ 70 RNA polymerase and promoter DNA complex revealed a role of σ non-conserved region during the open complex formation. J Biol Chem 293:7367–7375. doi: 10.1074/jbc.RA118.002161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sakaguchi K, He J, Tani S, Kano Y, Suzuki T. 2012. A targeted gene knockout method using a newly constructed temperature-sensitive plasmid mediated homologous recombination in Bifidobacterium longum. Appl Microbiol Biotechnol 95:499–509. doi: 10.1007/s00253-012-4090-4. [DOI] [PubMed] [Google Scholar]
- 63.Engler C, Gruetzner R, Kandzia R, Marillonnet S. 2009. Golden Gate shuffling: a one-pot DNA shuffling method based on type iis restriction enzymes. PLoS One 4:e5553. doi: 10.1371/journal.pone.0005553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Altaib H, Ozaki Y, Kozakai T, Badr Y, Nomura I, Suzuki T. 2019. A new Escherichia coli entry vector series (pIIS18) for seamless gene cloning using type iis restriction enzymes. Microbiol Resour Announc 8:e00323-19. doi: 10.1128/MRA.00323-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.