Biofilms are an important mode of growth in many settings. Here, we looked at small differences in the genomes of the bacterium Listeria monocytogenes isolate EGDe and used them to find out how biofilms form. This important fundamental information may help new treatments to be developed and also highlights the fact that isolates of the same identity often diverge.
KEYWORDS: Listeria monocytogenes, biofilm formation, sigma B, rhamnose, cell wall teichoic acid, biofilms, genome analysis
ABSTRACT
Listeria monocytogenes is a Gram-positive firmicute that causes foodborne infections, in part due to its ability to use multiple strategies, including biofilm formation, to survive adverse growth conditions. As a potential way to screen for genes required for biofilm formation, we harnessed the ability of bacteria to accumulate mutations in the genome over time, diverging the properties of seemingly identical strains. By sequencing the genomes of four laboratory reference strains of the commonly used L. monocytogenes EGDe, we showed that each isolate contains single nucleotide polymorphisms (SNPs) compared with the reference genome. We discovered that two SNPs, contained in two independent genes within one of the isolates, impacted biofilm formation. Using bacterial genetics and phenotypic assays, we confirmed that rsbU and rmlA influence biofilm formation. RsbU is the upstream regulator of the alternative sigma factor SigB, and mutation of either rsbU or sigB increased biofilm formation. In contrast, deletion of rmlA, which encodes the first enzyme for TDP-l-rhamnose biosynthesis, resulted in a reduction in the amount of biofilm formed. Further analysis of biofilm formation in a strain that still produces TDP-l-rhamnose but which cannot decorate the wall teichoic acid with rhamnose (rmlT mutant) showed that it is the decorated wall teichoic acid that is required for adhesion of the cells to surfaces. Together, these data uncover novel routes by which biofilm formation by L. monocytogenes can be impacted.
IMPORTANCE Biofilms are an important mode of growth in many settings. Here, we looked at small differences in the genomes of the bacterium Listeria monocytogenes isolate EGDe and used them to find out how biofilms form. This important fundamental information may help new treatments to be developed and also highlights the fact that isolates of the same identity often diverge.
INTRODUCTION
Biofilms are complex communities of microbial cells that are encased within a self-produced extracellular matrix. The biofilm matrix provides protection from environmental insults, increasing the tolerance of cells to antimicrobial agents and biocides (1). Listeria monocytogenes is a Gram-positive bacterium that causes the foodborne infection listeriosis. In susceptible individuals (e.g., people who are immunocompromised), the mortality rate of Listeria infections has been estimated to be up to 30%. Biofilms of L. monocytogenes can form on machinery in food-processing plants, contributing to food contamination (2) and potentially leading to the closure of manufacturing facilities for deep-clean processes (3). Thus, routes to inhibit or disrupt biofilm formation by L. monocytogenes could present one means of reducing Listeria infections. It is currently known that biofilm formation by L. monocytogenes is dependent on an active flagellum (4). Moreover, two major transcription factors, SigB and PrfA, and the virulence factor ActA have been shown to contribute to biofilm formation (5–7). However, there are still many unanswered questions regarding the molecular processes underpinning L. monocytogenes biofilm formation.
Reference strains of bacteria are widely used in laboratories as research models for the study of bacterial behavior and physiology (8). However, mutations can be inadvertently introduced into the genome during routine culture, modifying the strains derived from the designated laboratory reference strain (9). Diverging mutations within laboratory reference strains can contribute to differences in observed phenotypic behavior between different research groups. For example, Bacillus subtilis laboratory reference strain 168 was identified as a nonrugose biofilm-forming strain (10); however, it has been shown that some variants can form biofilms (11). By sequencing a collection of 12 sublines of strain 168, it was revealed that the epsC gene, which is essential for biofilms, carried point mutations in the nonrugose biofilm isolates. L. monocytogenes EGDe, serovar 1/2a, is widely used for molecular and cellular studies as the model organism (12), and we chose to use this isolate in our studies. We predicted that if we were able to identify genomic variations between L. monocytogenes isolates used by different laboratories, this could potentially shed light on the underlying genetics of biofilm formation. Using a comparative sequencing approach, we identified and connected genomic variations in L. monocytogenes EGDe isolates with differences in biofilm formation. More specifically, our bioinformatic analysis and experimental approaches revealed two genes, rsbU and rmlA, involved in biofilm formation. This work contributes to our understanding of biofilm formation by an important human pathogen.
(Data included in this article have been published in Chih-Yu Hsu’s doctoral thesis [53].)
RESULTS
Assessing growth and flagellum-based motility.
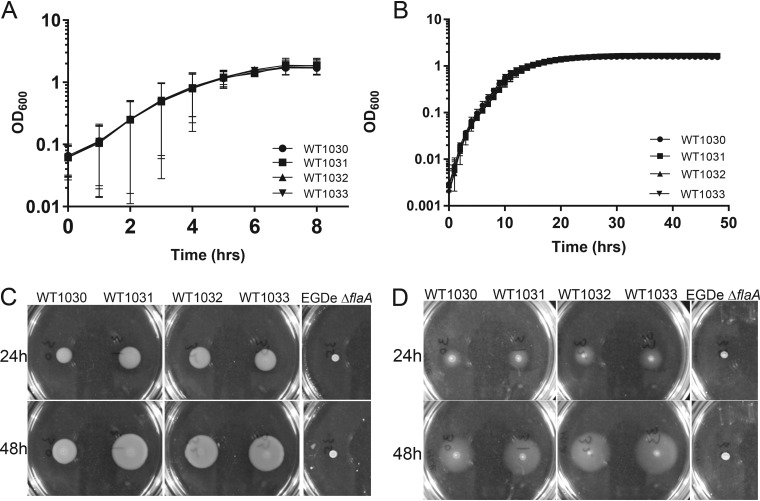
Four different L. monocytogenes EGDe isolates were obtained for this study and are here referred to as WT1030, WT1031, WT1032, and WT1033 (Table 1). The designation of the bacteria used in the study as EGDe was initially based on information obtained from the source supplying them and was later confirmed by whole-genome sequencing. We first compared the growth rates and yields of all strains and assayed motility. Growth was monitored under shaking culture conditions using brain heart infusion (BHI) medium and under static culture conditions using modified Welshimer’s broth (MWB). We did not identify any statistically significant differences in the growth rates or final yields of the four isolates for either condition (Fig. 1A and B). Next, we assessed flagellum-based motility by quantifying the ability of the cells to spread on semisolid agar plates, using EGDe ΔflaA as a negative control (13). The four EGDe isolates and EGDe ΔflaA were spotted onto BHI- and MWB-based semisolid agar plates that were incubated at 30°C, the permissive temperature for motility by L. monocytogenes (13, 14). As expected, the EGDe ΔflaA strain did not spread from the inoculation point (Fig. 1C and D). In contrast, the four EGDe stocks spread from the seeding point over time, although WT1030 showed reduced motility on BHI agar by comparison to the other three isolates (Fig. 1C and D; see Fig. S1 in the supplemental material). These data indicate that any differences in biofilm formation observed are not due to impaired growth or mutation of the flagellar genes.
TABLE 1.
The Listeria monocytogenes EGDe isolates used in this study
| Strain | Referencea | Origin |
|---|---|---|
| WT1030 | ANG882 | Carmen Buchrieser via Angelika Gründling |
| WT1031 | ANG873 | Martin Loessner via Angelika Gründling |
| WT1032 | EGDe | University College Cork |
| WT1033 | BAA-679 | Carmen Buchrieser via ATCC |
The strain name used in the originating lab.
FIG 1.
Growth and motility of the four L. monocytogenes EGDe isolates. (A) Growth in BHI medium under shaking conditions at 37°C. (B) Growth in MWB under static conditions at 30°C. The values presented in panels A and B are the means from 2 independent experiments, and the error bars represent the standard deviations. (C and D) Motility of the four isolates assessed after 24 and 48 h at 30°C using BHI (C) or MWB (D) soft agar. The EGDe ΔflaA strain was used as a negative control. Representative images are presented.
Differences in chitinase activity.
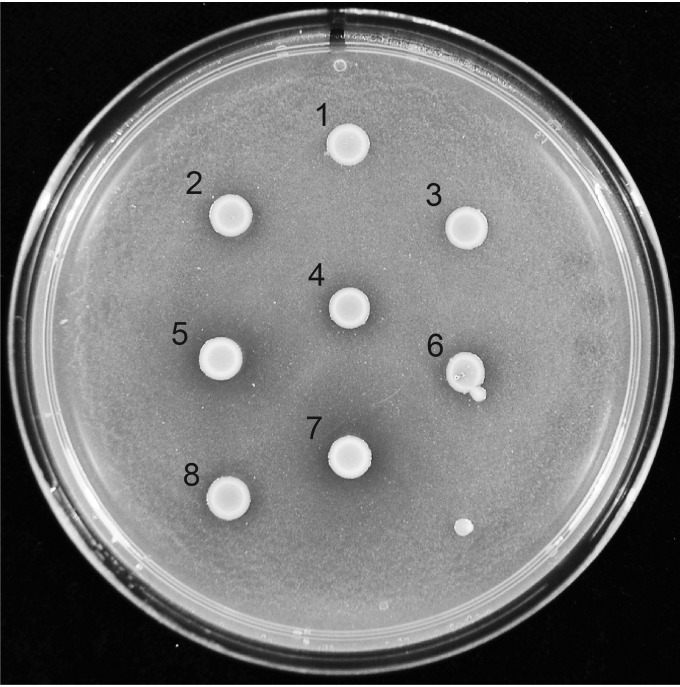
Certain regions of the L. monocytogenes genome are prone to incorporating mutations during growth (15), including rsbS, rsbU, and rsbV (16). The products of these genes comprise part of the complex regulatory system that activates the alternative sigma factor sigma B (SigB) (17). In turn, SigB controls a large regulon in L. monocytogenes that includes the genes chiA and chiB, which encode extracellular chitinases (18). Thus, to test if SigB regulation was disrupted due to mutations in the rsb genes, we examined chitinase activity (16). After spotting the four EGDe isolates onto chitin-rich agar, we noted that two of the isolates displayed clear evidence of chitinolytic activity: WT1031 and WT1032. In contrast, colonies formed by WT1030 and WT1033 had less distinct clearance zones, suggesting altered expression of members of the SigB regulon (Fig. 2). These gross phenotypic differences are indicative of genomic variations existing between the four EGDe isolates.
FIG 2.
Chitinase activity of the four L. monocytogenes EGDe isolates. Chitinolytic activity assessed using LB agar containing 2% (wt/vol) chitin. Incubation was at 30°C for 120 h. The genotypes of the strains tested are as follows: 1 and 3, 1031 ΔsigB; 2 and 4, 1031 ΔrsbU; 5, WT1031; 6, WT1030; 7, WT1032; 8, WT1033. The 1031 ΔsigB and 1031 ΔrsbU strains were used as controls.
Whole-genome sequencing.
We next sequenced the genomes of the EGDe strains using Illumina next-generation technologies. The reads were mapped to the published wild-type EGDe reference genome (NC_003210), and single nucleotide polymorphisms (SNPs) were identified in each of the four strains by using variant detection (Table 2). Some of the SNPs initially identified (not shown in Table 2) in the WT1032 genome were close to the prophage A118 integration site; further bioinformatic analysis revealed that these were caused by excision of the prophage from the chromosome, restoring a functional copy of comK (19, 20). Isolates WT1030 and WT1033 both contained a nonsense SNP in rsbU; this is consistent with the chitinase analyses which showed that these isolates generated a less distinct clearance zone on chitin-containing growth medium. WT1031 contained the fewest SNPs, all of which were identified in the other EGDe isolates, and so was designated the parental “wild-type” strain. These findings support the conclusion that variations in the genome have emerged between the EGDe isolates obtained from different sources.
TABLE 2.
Analysis of single nucleotide polymorphisms using whole-genome sequencing data
| Relative position in genomea | Gene | Refb | SNP |
Alteration of amino acidc | Type of mutationd | ||||
|---|---|---|---|---|---|---|---|---|---|
| WT 1030 | WT 1031 | WT 1032 | WT 1033 | EGDe ΔflaA | |||||
| 188308 | lmo0184 | G | T | —e | — | T | — | 148 E to stop codon | Nonsense |
| 189757 | lmo0185 | C | A | — | — | — | — | — | Synonymous |
| 264578 | lmo0247 | G | T | T | T | T | T | — | Synonymous |
| 280225 | rpoC | C | G | — | — | — | — | 1166 I to M | Missense |
| 435968 | Intergenic | C | A | A | A | A | A | Intergenic | Intergenic |
| 929469 | rsbU | C | CTT | — | — | CTT | — | 245 L to F, frameshift | Nonsense |
| 1116367 | lmo1081 (rmlA) | G | T | — | — | — | — | 241 E to stop codon | Nonsense |
| 1442124 | Intergenic | C | A | A | A | A | A | Intergenic | Intergenic |
| 1890030 | lmo1814 | C | A | — | — | A | — | 82 G to W | Missense |
| 2003900 | aroF | C | A | — | — | — | — | 138 V to F | Missense |
| 2207164 | lmo2125 | T | G | — | — | — | — | 400 Q to P | Missense |
| 2734614 | lmo2660 | C | A | — | — | — | — | 211 G to V | Missense |
| 2836724 | lmo2757 | G | — | — | A | — | — | 354 R to C | Missense |
| 2849710 | lmo2769 | G | — | — | — | T | — | 247 Y to stop codon | Nonsense |
| 2943565 | Intergenic | G | T | T | T | T | T | Intergenic | Intergenic |
The relative locations of the SNPs present in the strains are compared with NC_003210.
Ref, nucleotide present at the corresponding relative position in NC_003210.
The codons of the coding sequences with SNPs were analyzed by ExPASy translation tool followed by BLAST with the original amino acid sequences.
SNPs were categorized as intergenic, synonymous, missense, or nonsense.
—, no difference from the reference genome.
Biofilm formation by the L. monocytogenes EGDe stocks.
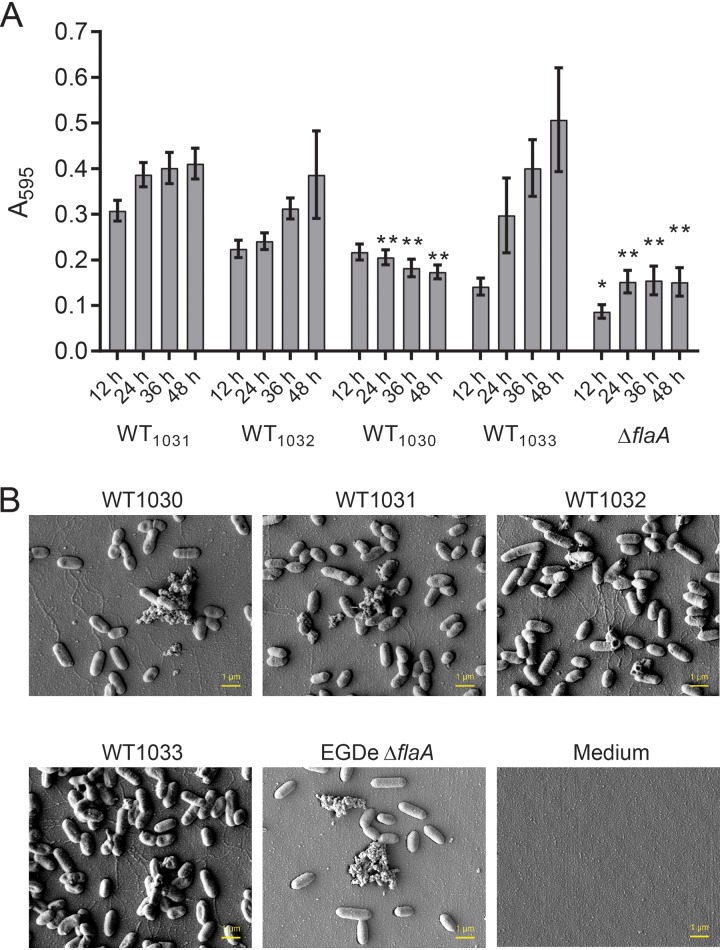
Having identified that the genomes of the four EGDe isolates were nonidentical, we assessed biofilm formation. The four EGDe stocks were inoculated in a 96-well microtiter plate platform where polystyrene pegs protruded from the lid into the well (this is also known as a Calgary biofilm device [21]). The EGDe ΔflaA strain, which was previously shown to be impeded in biofilm formation (4), was included as a negative control. The cultures were incubated statically at 30°C, and the biomass of each biofilm was measured every 12 h for a 48-h period. As expected, EGDe ΔflaA exhibited lower A595 readings then those of the four EGDe stocks (Fig. 3A), indicative of biofilm formation being reduced. Using the data from WT1031 as a baseline, the profiles of biofilm biomasses measured for the other three EGDe isolates were found to differ (Fig. 3A). Overall, the biomass of WT1030 was lower at all time points (Fig. 3A), whereas the biomass of WT1033 started at a lower point than WT1031 but ended with higher measurements at later time points (Fig. 3A). The statistical analysis revealed the measurements for WT1032 to be comparable to those of the reference WT1031 (Fig. 3A). The findings indicate that excision of prophage A118 does not impact biofilm formation as assessed here.
FIG 3.
Biofilm formation of the four L. monocytogenes EGDe isolates. (A) The biomasses of the four EGDe isolates adherent to the substratum were quantified over time when incubated at 30°C. The EGDe ΔflaA strain was used as a negative control. The values presented are the means from 29 independent experiments for the EGDe isolates and 4 experiments for the ΔflaA strain. The error bars are the standard errors of the means. The data were analyzed by one-way ANOVA comparing with WT1031. *, P ≤ 0.05; **, P ≤ 0.01. (B) The biomass adherent to the substratum was imaged using scanning electron microscopy after 48 h of incubation. The representative images shown were taken at the midpoint of the peg.
We next imaged the adherent cells by using scanning electron microscopy (Fig. 3B). This analysis was conducted at 30°C after biofilms were grown for 48 h. Five regions of interest (ROI) were chosen for each sample that covered the top (liquid surface) to near to the bottom of the peg (Fig. S2A). We first compared the overall cell morphology of the EGDe isolates and concluded that there were no discernible differences (Fig. 3B). We next counted the individual cells per field of view (FOV), and in doing so, we noticed that dense aggregates of cells encased in extracellular material were only encountered infrequently for all of the strains. The biomass produced by WT1031 contained on average ∼810 ± 320 (mean ± standard deviation [SD]) cells per FOV (Fig. S2B). Moreover, consistent with the measurements derived from crystal violet staining, the number of cells per FOV calculated for WT1032 did not significantly vary from those measured for WT1031. In contrast, fewer cells were counted per FOV for WT1030, while considerably more cells were detected in the WT1033 samples (1,255 ± 539). It is worth noting that in some cases, the cell density per FOV seemed to change with the location on the peg; the region of the peg that was closer to the bottom of the well had a higher number of cells than an equivalent region nearer the liquid-air interface (Fig. S2C). This gradient of cell attachment was most apparent for the biofilms formed by WT1033 (Fig. S2C). In summary, the biomasses measured using crystal violet and by counting the adherent cells per FOV correlate well.
Linking genotype and biofilm formation.
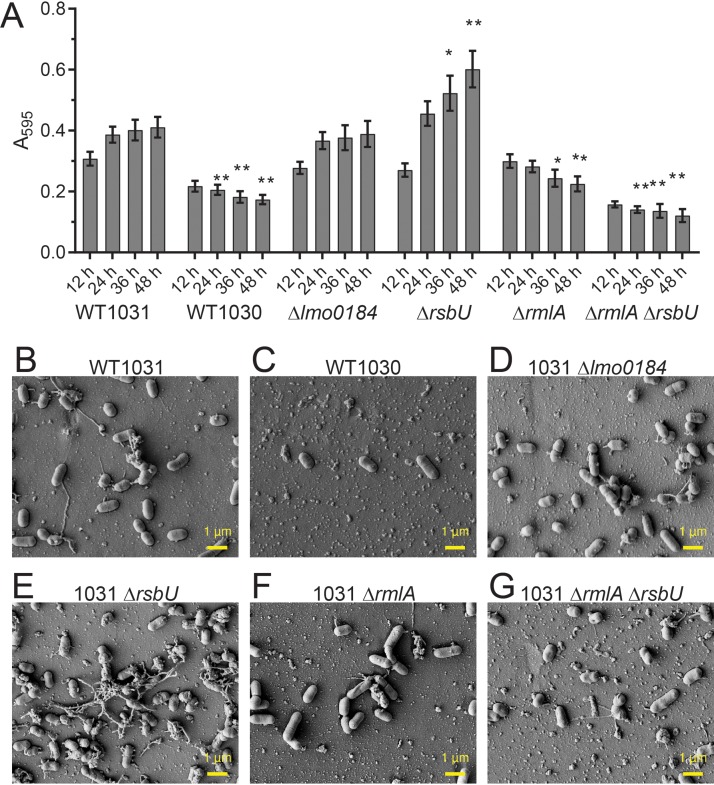
Our data suggest that WT1030 is impeded in biofilm formation by comparison with WT1031, a phenotype that is a consequence of fewer cells attaching to the substratum. As detailed in Table 2, the WT1030 genome contains 6 missense SNPs and three nonsense SNPs. To identify which of these mutations was responsible for reducing cell attachment, we constructed single gene deletions in the coding regions that contained nonsense SNPs, lmo0184, rmlA (lmo1081), and rsbU, using WT1031 as the parent. We reasoned that the nonsense SNPs were more likely to have a significant impact on protein function than the missense SNPs and, additionally, links to biofilm formation can be made for both rsbU and rmlA (22).
We checked if planktonic growth of the deletion strains was different from that of the parental strain WT1031 (Fig. S3). No significant differences were detected. Next, we measured the biomass adhered to the pegs of the Calgary biofilm device for the deletion strains by using crystal violet staining. We discovered that deletion of lmo0184 did not impact biofilm formation compared with that of WT1031 (Fig. 4A). In contrast, deletion of either rsbU or rmlA produced differences in the levels of crystal violet staining measured over time. For the rsbU deletion strain, the biomass was higher than that of WT1031 at 36 and 48 h (Fig. 4A). For the rmlA mutant strain, biofilm formation was reduced at 36 and 48 h (Fig. 4A). These findings were in agreement with the average number of cells adherent per FOV that were visualized (Fig. 4B to G) and quantified following scanning electron microscopy (Fig. S4A and B). Therefore, we concluded that two genes that impact biofilm formation are mutated in WT1030: rsbU and rmlA. By constructing a double rsbU rmlA deletion strain in the WT1031 background, we established that the impact of the rmlA mutation dominated the moderate increase in biofilm observed when rsbU was deleted alone (Fig. 4A, E, and G).
FIG 4.
RmlA and RsbU influence biofilm formation by L. monocytogenes EGDe. (A) The biomasses of WT1031, WT1030, WT1031 Δlmo0184 (LSW1024), WT1031 ΔrsbU (LSW1028), WT1031 ΔrmlA (LSW1040), and WT1031 ΔrmlA ΔrsbU (LSW1051) strains that were adherent to the substratum were quantified. The samples were incubated at 30°C for the time points indicated. The values presented for WT1031 and WT1030 are reproduced from Fig. 3. The means from ≥4 experiments are presented for the remaining strains. The error bars are the standard errors of the means. The data were analyzed by one-way ANOVA, comparing with WT1031. *, P ≤ 0.05; **, P ≤ 0.01. The biomasses adherent to the substratum were imaged using scanning electron microscopy for WT1031 (B), WT1030 (C), WT1031 Δlmo0184 (D), WT1031 ΔrsbU (E), WT1031 ΔrmlA (F), and WT1031 ΔrmlA ΔrsbU (G). The representative images shown were taken at the midpoint of the peg after 48 h of incubation.
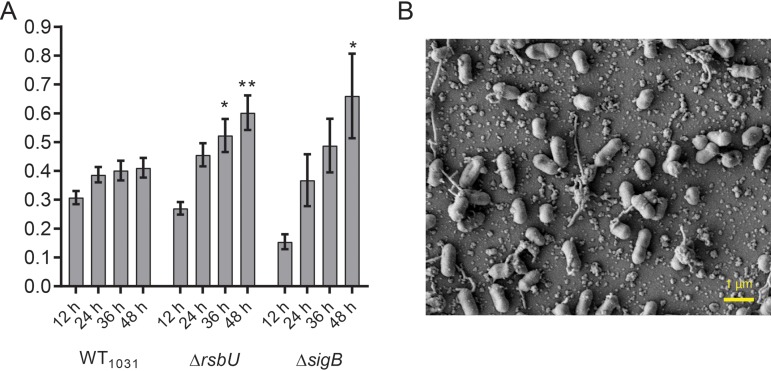
When sigma B is inactive, cell adherence increases.
Deletion of rsbU enhances biofilm formation, whereas deletion of rmlA decreases biofilm formation. RsbU is an upstream positive regulator of SigB (23); therefore, one possible interpretation of our data is that deletion of rsbU decreases transcription of the SigB regulon, leading to an increase in rmlA transcription. While an effect of SigB on transcription of rmlA has not been reported, this hypothesis would explain the enhanced biofilm capability of the rsbU mutant and decreased biofilm levels in the double rsbU rmlA strain and in the rmlA single mutant. Therefore, we first tested if the impact of mutating rsbU on biofilm formation manifests as a consequence of SigB inactivation. If our hypothesis was correct, then deletion of sigB should phenocopy the rsbU mutation.
We constructed a sigB deletion in WT1031, examined the level of chitinase activity, and assessed the impact on biofilm formation. As expected, the sigB deletion strain did not display chitinolytic activity (Fig. 2) (16). During biofilm formation, the sigB deletion strain was initially observed to have a lower level of biomass adherent to the pegs than the parental WT1031 strain. However, the value surpassed that of the parental strain at later time points (Fig. 5A). As suggested by the crystal violet staining in Fig. 5A, the sigB and rsbU strains were shown to have similar numbers of cells attached per FOV when the samples were imaged by scanning electron microscopy (SEM) (Fig. 5B; Fig. S4A and B). Together, these findings are consistent with the conclusion that the impact of the SNP in rsbU on biofilm formation was due to a reduction in sigB activity.
FIG 5.
SigB influences biofilm formation by L. monocytogenes EGDe. (A) The biomasses of WT1031, WT1031 ΔrsbU (LSW1028), and WT1031 ΔsigB (LSW1026) strains that were adherent to the substratum were quantified. The samples were incubated at 30°C for the time points indicated. The values presented for WT1031 and WT1031 ΔrsbU are reproduced from Fig. 3 and 4. The means from ≥4 experiments are presented for the WT1031 ΔsigB strain. The error bars are the standard errors of the means. The data were analyzed by one-way ANOVA comparing with WT1031. *, P ≤ 0.05; **, P ≤ 0.01. (B) The biomass adherent to the substratum was imaged using scanning electron microscopy for WT1031 ΔsigB. The representative image shown was taken at the midpoint of the peg after 48 h of incubation.
We next reasoned that if the reduction of SigB activity in the rsbU mutant impacted rmlA transcription, this would manifest as an alteration in l-rhamnose decoration of the wall teichoic acid (WTA). This is because RmlA is an enzyme in the TDP-l-rhamnose pathway. TDP-l-rhamnose is used for the synthesis of cell wall carbohydrates (24–26) and for the decoration of WTA in L. monocytogenes (27, 28). Therefore, we extracted WTA from the sigB and rsbU mutants and compared the apparent molecular mass with the WTA extracted from the rmlA mutant. These analyses showed there was no gross difference in the apparent molecular mass of WTA produced by the sigB and rsbU strains compared with that of the parental strain, at either a lower or higher position, as would be expected for material with fewer or more rhamnose moieties, respectively. In contrast, for the rmlA mutant, the molecular mass of WTA extracted was lower than that observed for WT1031. The mobility of the WTA extracted from the rmlA mutant was comparable to that of the WTA extracted from EGDe isolate WT1030 (Fig. S5). Therefore, taking these data together, it is unlikely that rsbU or sigB is mediating the impact on biofilm formation via rmlA and its impact on WTA decoration.
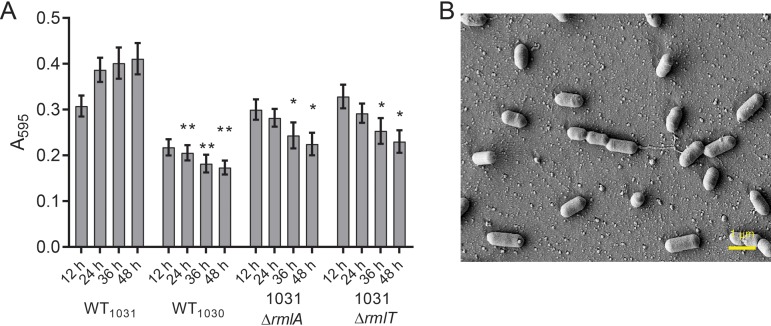
Sugar decoration of wall teichoic acids alters adhesion properties of L. monocytogenes.
RmlA is the first enzyme in the pathway that catalyzes the conversion of d-glucose-1-phosphate into TDP-l-rhamnose (27). We wanted to confirm if deletion of rmlA had an impact on biofilm formation due to the lack of the l-rhamnose moiety on WTA or if TDP-l-rhamnose was used in the synthesis of a different polymer. To do this, we constructed a derivative of WT1031 that still produced TDP-l-rhamnose but lacked the glycosyltransferase, RmlT, which is responsible for the transfer of TDP-l-rhamnose onto ribitol phosphate (27). Biofilm formation was measured for the 1031 ΔrmlT strain every 12 h and found to be more comparable to that of the 1031 ΔrmlA strain than to that of the WT1031 strain (Fig. 6A). Using SEM imaging to visualize the attached biomass, the 1031 ΔrmlA and 1031 ΔrmlT strains were shown to have similar numbers of cells attached per FOV (Fig. 6B; Fig. S4A and B). Therefore, as presence of the TDP-l-rhamnose pool in the rmlT mutant strain was not sufficient to allow biofilm formation, these findings suggest that decoration of WTA with l-rhamnose is needed for cell adhesion to the substratum. It is also possible that decoration of WTA with l-rhamnose is needed to promote the formation of clusters of the bacteria, but further analysis would be needed to determine this conclusively.
FIG 6.
Decoration of the wall teichoic acid with L-rhamnose is needed for cell adhesion by L. monocytogenes EGDe. (A) The biomasses of WT1031, WT1030, WT1031 ΔrmlA (LSW1040), and WT1031 ΔrmlT (LSW1039) strains that were adherent to the substratum were quantified. The samples were incubated at 30°C for the time points indicated. The values presented for WT1031, WT1030, and WT1031 ΔrmlA are reproduced from Fig. 3 and 4. The means from ≥4 experiments are presented for the WT1031 ΔrmlT strain. The error bars are the standard errors of the means. The data were analyzed by one-way ANOVA comparing with WT1031. *, P ≤ 0.05; **, P ≤ 0.01. (B) The biomass adherent to the substratum was imaged using scanning electron microscopy for WT1031 ΔrmlT after 48 h of incubation. The representative image shown was taken at the midpoint of the peg.
DISCUSSION
To study biofilm formation by the Gram-positive pathogen Listeria monocytogenes, we chose an approach that was based on the hypothesis that diverged stocks of the EGDe wild type may contain mutations that could impact biofilm formation. We proposed that identifying the mutations would allow us to link genotype with phenotype and thereby gain insights into the mechanisms underpinning biofilm formation in this pathogen. We sourced four EGDe isolates, checked planktonic growth, and identified differences in chitinase activity. Using next-generation sequencing technologies, we sequenced the genomes of the four isolates and identified genomic variations. Some features of the genomic sequencing data were able to be readily connected to phenotypic differences displayed by the four EGDe isolates. For example, RsbU is an upstream regulator of SigB activity (17, 29, 30), and in WT1030 and WT1033, a frameshift mutation that leads to the premature termination of translation is contained within rsbU (lmo0892) (Table 2). A consequence of the rsbU mutation may be that SigB is not activated, and transcription of the genes in its regulon will not be triggered (30); although, there is evidence showing that SigB retains partial activity in an RsbV mutant background (31). The presence of the SNP in rsbU correlated with the reduction of chitinolytic activity observed for WT1030 and WT1033 (Fig. 2). Additionally, WT1030 contains a nonsense SNP within the rmlA (lmo1081) coding region. RmlA is the first enzyme in a four-step reaction resulting in the synthesis of TDP-l-rhamnose (27), which is a substrate to transfer l-rhamnose onto the ribitol phosphate backbone of wall teichoic acid. The nonsense SNP in rmlA is predicted to disrupt TDP-l-rhamnose production, resulting in a strain that carries WTA without the l-rhamnose decoration. The presence of this mutation correlates with the lower molecular weight of the WTA extracted from WT1030.
We adapted and implemented a robust method of assessing biofilm formation by the four EGDe isolates. The biofilm formed under these conditions did not typically appear to generate an obvious extracellular matrix; when viewed by microscopy, the biomass appeared to be isolated cells or small clusters that were adherent to the surface. This is different from the honeycomb arrangement of L. monocytogenes cells seen in some biofilms (32) but comparable to that in other studies where cells have been observed as an attached monolayer (33). Through our analysis, we identified one strain (named WT1030) that displayed a defect in biofilm formation. Having ruled out that differences in growth or motility caused the differences in the biofilm formation we observed, we used the details from the next-generation sequencing analysis to link rmlA to surface adhesion and biofilm formation. As detailed above, RmlA is needed for TDP-l-rhamnose production, and through assessing biofilm formation lacking RmlT, we were able to determine that the lack of l-rhamnose decoration of wall teichoic acid was the factor influencing biofilm formation rather than the loss of TDP-l-rhamnose production per se. The defect in biofilm formation appeared to be due to reduced cell surface adhesion. Our findings are consistent with data derived from a global transposon screen of L. monocytogenes isolate 568 which identified lmo1080 (rmlT) as needed for biofilm formation at low temperature (34). In addition, they are in line with experiments that uncovered wall teichoic acids as a major polysaccharide present in the L. monocytogenes biofilm matrix (35). However, exactly how the l-rhamnose decorated wall teichoic acid aids cell surface interaction remains unknown.
We also strengthened the already identified connection between sigB and biofilm formation and in so doing, reinforced the need to obtain dynamic data when analyzing biofilm formation using a microtiter plate-based assay (7, 36). SigB was previously found to promote biofilm formation (37, 38). However, here, for the sigB deletion strain, a defect in biofilm formation at early time points culminated in an enhanced level of biofilm produced at later time points. We therefore conclude that SigB appears to suppress transcription of genes involved in biofilm formation, perhaps those directly linked with matrix synthesis, as deletion resulted in greater adhesion and more extracellular material being deposited and visible by SEM analysis.
Concluding comments.
The use of laboratory reference strains was initially focused on allowing the cooperation of research groups around the world (8). It provides a baseline of commonality to compare observations and accelerate the progression of research. Although this goal has been accomplished, the approach also allows seemingly identical isolates of bacteria to independently evolve in different laboratories (9, 11). Using a comparative sequencing approach, we have uncovered variations in the genomes of EGDe isolates used in laboratories across the world. Moreover, we have reinforced the importance and necessity of obtaining whole-genome sequencing data to ensure that strains do not contain inadvertent mutations when a new isolate is used in research settings.
MATERIALS AND METHODS
Growth media and additives.
Brain heart infusion (BHI) medium (237500; BD Biosciences) was used for propagating L. monocytogenes strains. Strains were routinely grown either in liquid BHI medium, on BHI medium solidified with 1.5% (wt/vol) select agar, or in liquid modified Welshimer’s broth (MWB) (6.56 g/liter KH2PO4, 16.39 g/liter Na2HPO4, 0.41 g/liter MgSO4·7H2O, 10 g/liter glucose, 0.088 g/liter ferric citrate, 0.1 g/liter leucine, 0.1 g/liter isoleucine, 0.1 g/liter valine, 0.1 g/liter methionine, 0.1 g/liter arginine, 0.1 g/liter cysteine, 0.6 g/liter glutamine, 0.5 mg/liter riboflavin, 1.0 mg/liter thiamine, 0.5 mg/liter biotin, and 0.005 mg/liter lipoic acid). Starter cultures were prepared by inoculating a single colony of L. monocytogenes grown on BHI agar into 5 ml of BHI medium, which was grown with shaking. The growth medium was supplemented with selective antibiotics (100 μg/ml ampicillin [Amp], 5 μg/ml erythromycin [Ery], or 50 μg/ml X-Gal [5-bromo-4-chloro-3-indolyl-d-galactopyranoside]) during cloning and the construction of mutant strains as required.
Strains, plasmids, and primers.
Complete details of the strains, plasmids and primers used in this study are provided in Tables S1 to S3 in the supplemental material.
Growth measurement.
To follow the growth of L. monocytogenes strains, starter cultures were grown at 37°C for ∼20 h and inoculated in 100 ml of BHI medium at a starting optical density at 600 nm (OD600) of 0.05. The cultures were incubated in a water bath with shaking at 200 rpm, and the OD600 was measured every hour. Alternatively, growth over time was monitored using a plate reader (Synergy 2; BioTek Instruments). The starting cultures were subcultured in MWB at an initial OD600 of 0.01 in 200 μl per well in a round-bottom polystyrene 96-well plate. The OD600 was measured every hour during incubation at 30°C for 48 h without shaking.
Motility.
Semisolid (0.3% [wt/vol]) agar was prepared in BHI medium or MWB. Starter cultures for each strain were grown at 30°C for up to 48 h. To seed the strains, the OD600 of starting cultures was normalized to 1.0, and 1 μl of the normalized culture was stabbed into the center of a semisolid agar plate. A negative control, the nonmotile strain EGDe ΔflaA (13), was included. The seeded semisolid agar plates were incubated at 30°C, and after 24 and 48 h of incubation, images were captured using a digital single-lens reflex (DSLR) camera (Nikon D3200 with Nikkor 18- to 55-mm lens). Quantification of motility was performed by measuring the diameter of the zone occupied by the cells. For each sample, the diameter of the swarm was measured at two positions. The average of the two values was used for further statistical analysis.
Chitinase activity.
Chitinase activity was tested as described previously (18). Starter cultures were grown at 37°C for ∼20 h. The cultures of the strains were normalized to an OD600 of 1.0, and 10 μl was spotted onto an LB agar plate supplemented with colloidal chitin at a final concentration of 2% (wt/vol). The plates were then incubated at 30°C for 24 and 48 h prior to imaging using a DSLR camera (Nikon D3200 with Nikkor 18- to 55-mm lens).
Cell wall teichoic acid analysis.
Extraction of the cell wall teichoic acids from L. monocytogenes was performed as described previously (39). Starter cultures were grown at 37°C for ∼8 h and inoculated in 50 ml of MWB at an initial OD600 of 0.01, which was incubated at 30°C for ∼17 h with shaking at 200 rpm. The cells were harvested by centrifugation at 3,800 × g for 10 min. The cell pellet was washed with 20 ml of MES buffer [50 mM 2-(N-morpholino)ethanesulfonic acid, pH 6.5] and centrifuged at 3,800 × g for 10 min. The cell pellet was resuspended in 1 ml of MES buffer supplemented with 4% (wt/vol) SDS and boiled at 99°C for 1 h. The SDS-treated cells were harvested by centrifugation at 17,000 × g for 10 min. The cell pellets were washed twice with MES buffer containing 2% (wt/vol) NaCl, rinsed with MES buffer, and resuspended in 1 ml of MES buffer with 0.4 g acid-washed glass beads (≤106 μm, catalog number G4649-500G; Sigma-Aldrich) per sample. The cells were lysed by vortexing at the highest speed for 10 min with the tube lying horizontally. The glass beads were discarded after centrifugation at 1,000 × g for 5 min, and the cell lysate was harvested for the following steps. The proteins in the samples were digested with 20 μg/ml proteinase K (03508811103; Roche) in 20 mM Tris-HCl (pH 8.0) at 50°C for 2 h. After centrifuging at 17,000 × g for 10 min, the pellet was treated with 1 ml of 0.1 M NaOH for 17 h with shaking at 1,200 rpm at 25°C on Thermomixer R (Eppendorf). The supernatant was harvested by centrifugation at 14,000 × g for 15 min, and 0.1 ml of 1 M HCl was added to each sample. The liquid was dialyzed into Milli-Q water using a 1-kDa dialysis membrane (132105; Spectrum). The dialyzed samples were dried with a SpeedVac (RVC2-25 with CT02-05; Christ). Each sample was resuspended with 100 μl of WTA loading buffer (20 mM Tris-HCl, 20 mM Tricine, 10% [vol/vol] glycerol) for further analysis by native polyacrylamide gel electrophoresis. The gel was rinsed with Milli-Q water and stained with alcian blue staining solution (5% [vol/vol] acetic acid, 30% [vol/vol] ethanol, and 1 mg/ml alcian blue 8GX) for 1 h. An image of the stained gel was taken after incubation in destaining solution (5% [vol/vol] acetic acid, 30% [vol/vol] ethanol) for 20 min.
Biofilm formation.
Starter cultures were grown at 37°C for ∼20 h, and the OD600 was normalized to 0.01 in MWB. One hundred fifty microliters of the diluted cultures was subcultured in the Calgary biofilm device (catalog number 445497 for the lid, and catalog number 262162 for the plate; Nunc, Thermo Scientific) and incubated at 30°C for 12 to 48 h. The biomass of the biofilm formed was determined by crystal violet staining. The cultures were discarded by aspiration, each well was rinsed three times with 1.2 volumes of 1× phosphate-buffered saline (PBS; 8 g/liter NaCl, 0.2 g/liter KCl, 2.56 g/liter Na2HPO4·7H2O, 0.2 g/liter KH2PO4, pH 7.4), and cells were incubated with 1.3 volumes of 0.1% (wt/vol) crystal violet (diluted from 2.3% solution in Milli-Q water, HT901-8FOZ; Sigma-Aldrich) for 1 h at room temperature. The staining solution was aspirated, and the peg was washed with 1.5 volumes of 1× PBS three times. The biofilm was destained by incubating with 30% (vol/vol) acetic acid for 30 min at room temperature. The absorbance of the stained 30% (vol/vol) acetic acid was measured at a wavelength of 595 nm. For each replicate, the A595 of a medium-only control was used as the background reading.
Scanning electron microscopy.
Biofilms formed on the pegs of the Calgary biofilm device were fixed for scanning electron microscopy (SEM) largely as described previously (40). The protocol involved two different stages of fixation, critical-point drying and sputter coating with platinum prior to final imaging. The biofilm-coated pegs were first rinsed with 1× PBS three times and fixed with 200 μl per well of primary fixative for 2 h at room temperature. The primary fixative comprised 2.5% (vol/vol) glutaraldehyde, 4% (wt/vol) paraformaldehyde, 75 mM l-lysine, and 0.075% (wt/vol) alcian blue in 1× PBS. Next, the pegs were removed from the Calgary biofilm device by using diagonal pliers. A secondary fixation step was included after a brief wash with 1× PBS. The secondary fixative was composed of 1% (wt/vol) osmium tetroxide (diluted from 4% stock, 75632; Sigma-Aldrich). After 1 h of secondary fixation, the biofilms were treated with a gradient ethanol series (50%, 70%, 90%, and 99.9% [vol/vol]). The biofilm-coated pegs were transferred into a chamber to be critical-point dried. Biofilm-coated pegs were stuck onto a 25-mm sample stub (AGG3023; Agar Scientific) with carbon stickers (AGG3303; Agar Scientific) and conductive carbon double-sided tape (AGG3939; Agar Scientific). The sample stub carrying the biofilm-coated pegs was sputter coated with 25-nm-thick platinum to create a conductive surface. The biofilms were imaged with field emission SEM (JSM-7400f; Jeol). All images were taken with 5 kV detected by a lower secondary electron (LEI) detector. The cells in each image were counted manually with a cell counter plug-in in ImageJ.
Electrocompetent cells.
To insert plasmids into L. monocytogenes strains, electrocompetent cells were prepared as described previously (41). Plasmid DNA (1 μg) was gently mixed with 50 μl of electrocompetent cells before incubating them on ice for 10 min. The cells were transferred into a chilled electroporation cuvette (1652089; Bio-Rad) and electroporated at 10 kV/cm, 400 Ω, and 25 μF. A recovery medium, 1 ml of 0.5 M sucrose-supplemented BHI medium, was gently added to each electroporation reaction. Following incubation at 30°C for 90 min without shaking, 150 μl of the cell suspension was plated on a BHI agar plate supplemented with antibiotics as required.
Construction of deletion strains.
In-frame deletions of protein-coding regions on the chromosome were introduced by the pMAD-based approach (42). First, the pMAD-based plasmid was modified such that it could be used for allelic exchange. Both upstream and downstream regions of the gene to be deleted were either amplified and fused with a KpnI restriction enzyme site using PCR or were synthesized commercially. The modified DNA sequences were first inserted into an intermediate cloning vector, pUC19 or pUC57, prior to ligation into pMAD. The pMAD vector containing the required insert was introduced into the desired parental strain. The recovered cells were spread on BHI agar plates supplied with 5 μg/ml Ery and 50 μg/ml X-Gal and incubated at 30°C for 72 h. The resultant colonies were collected, inoculated in BHI medium containing 5 μg/ml Ery, and incubated at 39°C with shaking at 200 rpm for 17 h. The cultures were serially diluted to a factor of 10−6 and isolated on 5 μg/ml Ery- and 50 μg/ml X-Gal-supplemented BHI agar plates that were incubated at 39°C for 48 h. Blue-colored colonies were used to inoculate liquid BHI medium, and the cells were incubated at 30°C for 17 h without shaking and then for 4 h with shaking at 200 rpm. The cultures were serially diluted to a factor of 10−6 and isolated on 50 μg/ml X-Gal-supplemented tryptic soy agar plates. The plates were incubated at 37°C for 72 h to allow the formation of white colonies. Each white colony was inoculated in 5 ml of BHI medium and incubated at 37°C with shaking at 200 rpm for ∼17 h. Deletions were confirmed using PCR and DNA sequencing.
Statistical analysis.
GraphPad Prism 7 was used to generate plots and analyze data. Statistical analyses of the data were performed by one-way analyses of variance (ANOVAs) with Dunnett’s multiple-comparison tests.
Genome sequencing.
Genomic DNA was extracted from starter cultures incubated at 37°C for ∼17 to 20 h. The cells were harvested by centrifugation at 3,500 × g for 10 min, and the cell pellet suspended in 180 μl of enzymatic lysis buffer (20 mM Tris-HCl, 2 mM EDTA [pH 8.0], 1.2% [vol/vol] Triton X-100 containing 20 mg/ml lysozyme). The cells were lysed at 37°C for 30 min after which the cell lysate was applied to the DNeasy blood and tissue kit (69504; Qiagen). The final product was eluted in water and stored at −20°C.
Illumina next-generation sequencing was performed by the Genome Sequencing Unit at the Tayside Centre for Genome Analysis. The DNA was quantified using the QuBit 2.0 DNA kit, and 1 μg of DNA was sheared into 300-bp fragments using a Covaris M220 focused ultrasonicator. Paired-end libraries were generated using the Illumina TruSeq DNA sample preparation guide and sequenced using the Illumina MiSeq reagent kit v2 on the Illumina MiSeq platform.
Sequence analysis.
The list of the single nucleotide polymorphisms (SNPs) was acquired by aligning the reads to the published genome (NC_003210). The sequence data were analyzed using MiSeq Reporter, alignment to the reference genome was conducted using Burrows-Wheeler Aligner (43), and variant calling to identify SNPs was performed using the Genome Analysis Toolkit UnifiedGenotyper (44). To determine if the A118 prophage was integrated, genome assemblies of strains WT1030 and WT1032 were carried out using the BugBuilder (45) pipeline, using SPAdes (46) for contig assembly (version 3.13.1, coverage cutoff = 5, kmer size = auto, and “careful” mode enabled). Scaffolding was carried out with the Mauve Contig Mover (47) (version 2.4.0) with NC_003210.1 as a reference sequence, followed by automated gap closure using Pilon 1.23 (48). Annotation of the assembled sequences was carried out using Prokka 1.13.4 (49). Assembled genomes were aligned against NC_003210.1 using pairwise comparisons with NCBI BLAST (50) (blastn version 2.7.1, E value cutoff = 0.01), and alignments were visualized using the Artemis comparison tool (51).
Additional bioinformatics analysis performed in this study used CLC Main Workbench 8 to organize the DNA sequences. Basic Local Alignment Search Tool (BLAST) was used to align sequences of nucleic acids (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The ExPASy translation tool was used to assess the impact of the mutations on the protein sequences (https://web.expasy.org/translate/). The alignment of the protein sequences was generated by Clustal Omega (52).
Data availability.
Sequence data have been deposited in the European Nucleotide Archive under study accession numbers PRJEB35200 and ERZ1188925.
Supplementary Material
ACKNOWLEDGMENTS
C.-Y.H. received funding from BeautyHsiao Biotechnology Inc. (registered in Taiwan). N.R.S.-W. received funding from Tenovus Scotland. L.C. was the recipient of a Wellcome Trust Ph.D. studentship (093714/Z/10/Z). C.O. and N.R.S.-W. received funding from the European Union’s Horizon 2020 research and innovation program under the Marie Sklodowska-Curie grant (agreement number 721456).
We thank Claire Gorby for constructing the strain 1031 rmlA and rmlT mutations and Angelika Gründling for kindly providing two of the EGDe variants and the flaA deletion strain. We acknowledge the Dundee Imaging Facility, Dundee, United Kingdom, which is supported by the Wellcome Trust Technology Platform award (097945/B/11/Z) and the MRC Next Generation Optical Microscopy award (MR/K015869/1). We also thank Yongchang Fanin for assistance.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Flemming HC, Wingender J, Szewzyk U, Steinberg P, Rice SA, Kjelleberg S. 2016. Biofilms: an emergent form of bacterial life. Nat Rev Microbiol 14:563–575. doi: 10.1038/nrmicro.2016.94. [DOI] [PubMed] [Google Scholar]
- 2.Colagiorgi A, Bruini I, Di Ciccio PA, Zanardi E, Ghidini S, Ianieri A. 2017. Listeria monocytogenes biofilms in the wonderland of food industry. Pathogens 6:41. doi: 10.3390/pathogens6030041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fagerlund A, Moretro T, Heir E, Briandet R, Langsrud S. 2017. Cleaning and disinfection of biofilms composed of Listeria monocytogenes and background microbiota from meat processing surfaces. Appl Environ Microbiol 83:e01046-17. doi: 10.1128/AEM.01046-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lemon KP, Higgins DE, Kolter R. 2007. Flagellar motility is critical for Listeria monocytogenes biofilm formation. J Bacteriol 189:4418–4424. doi: 10.1128/JB.01967-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Travier L, Guadagnini S, Gouin E, Dufour A, Chenal-Francisque V, Cossart P, Olivo-Marin JC, Ghigo JM, Disson O, Lecuit M. 2013. ActA promotes Listeria monocytogenes aggregation, intestinal colonization and carriage. PLoS Pathog 9:e1003131. doi: 10.1371/journal.ppat.1003131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Price R, Jayeola V, Niedermeyer J, Parsons C, Kathariou S. 2018. The Listeria monocytogenes key virulence determinants hly and prfa are involved in biofilm formation and aggregation but not colonization of fresh produce. Pathogens 7:E18. doi: 10.3390/pathogens7010018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van der Veen S, Abee T. 2010. Importance of SigB for Listeria monocytogenes static and continuous-flow biofilm formation and disinfectant resistance. Appl Environ Microbiol 76:7854–7860. doi: 10.1128/AEM.01519-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fux CA, Shirtliff M, Stoodley P, Costerton JW. 2005. Can laboratory reference strains mirror “real-world” pathogenesis? Trends Microbiol 13:58–63. doi: 10.1016/j.tim.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 9.Read TD, Massey RC. 2014. Characterizing the genetic basis of bacterial phenotypes using genome-wide association studies: a new direction for bacteriology. Genome Med 6:109. doi: 10.1186/s13073-014-0109-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McLoon AL, Guttenplan SB, Kearns DB, Kolter R, Losick R. 2011. Tracing the domestication of a biofilm-forming bacterium. J Bacteriol 193:2027–2034. doi: 10.1128/JB.01542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gallegos-Monterrosa R, Mhatre E, Kovács ÁT. 2016. Specific Bacillus subtilis 168 variants form biofilms on nutrient-rich medium. Microbiology 162:1922–1932. doi: 10.1099/mic.0.000371. [DOI] [PubMed] [Google Scholar]
- 12.Bécavin C, Bouchier C, Lechat P, Archambaud C, Creno S, Gouin E, Wu Z, Kühbacher A, Brisse S, Pucciarelli MG, García-del Portillo F, Hain T, Portnoy DA, Chakraborty T, Lecuit M, Pizarro-Cerdá J, Moszer I, Bierne H, Cossart P. 2014. Comparison of widely used Listeria monocytogenes strains EGD, 10403S, and EGD-e highlights genomic variations underlying differences in pathogenicity. mBio 5:e00969-14. doi: 10.1128/mBio.00969-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grundling A, Burrack LS, Bouwer HG, Higgins DE. 2004. Listeria monocytogenes regulates flagellar motility gene expression through MogR, a transcriptional repressor required for virulence. Proc Natl Acad Sci U S A 101:12318–12323. doi: 10.1073/pnas.0404924101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peel M, Donachie W, Shaw A. 1988. Temperature-dependent expression of flagella of Listeria monocytogenes studied by electron microscopy, SDS-PAGE and western blotting. J Gen Microbiol 134:2171–2178. doi: 10.1099/00221287-134-8-2171. [DOI] [PubMed] [Google Scholar]
- 15.Orsi RH, Borowsky ML, Lauer P, Young SK, Nusbaum C, Galagan JE, Birren BW, Ivy RA, Sun Q, Graves LM, Swaminathan B, Wiedmann M. 2008. Short-term genome evolution of Listeria monocytogenes in a non-controlled environment. BMC Genomics 9:539. doi: 10.1186/1471-2164-9-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Quereda JJ, Pucciarelli MG, Botello-Morte L, Calvo E, Carvalho F, Bouchier C, Vieira A, Mariscotti JF, Chakraborty T, Cossart P, Hain T, Cabanes D, García-Del Portillo F. 2013. Occurrence of mutations impairing sigma factor B (SigB) function upon inactivation of Listeria monocytogenes genes encoding surface proteins. Microbiology 159:1328–1339. doi: 10.1099/mic.0.067744-0. [DOI] [PubMed] [Google Scholar]
- 17.Tiensuu T, Guerreiro DN, Oliveira AH, O’Byrne C, Johansson J. 2019. Flick of a switch: regulatory mechanisms allowing Listeria monocytogenes to transition from a saprophyte to a killer. Microbiology 165:819–833. doi: 10.1099/mic.0.000808. [DOI] [PubMed] [Google Scholar]
- 18.Larsen MH, Leisner JJ, Ingmer H. 2010. The chitinolytic activity of Listeria monocytogenes EGD is regulated by carbohydrates but also by the virulence regulator PrfA. Appl Environ Microbiol 76:6470–6476. doi: 10.1128/AEM.00297-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rabinovich L, Sigal N, Borovok I, Nir-Paz R, Herskovits AA. 2012. Prophage excision activates Listeria competence genes that promote phagosomal escape and virulence. Cell 150:792–802. doi: 10.1016/j.cell.2012.06.036. [DOI] [PubMed] [Google Scholar]
- 20.Loessner MJ, Inman RB, Lauer P, Calendar R. 2000. Complete nucleotide sequence, molecular analysis and genome structure of bacteriophage A118 of Listeria monocytogenes: implications for phage evolution. Mol Microbiol 35:324–340. doi: 10.1046/j.1365-2958.2000.01720.x. [DOI] [PubMed] [Google Scholar]
- 21.Ceri H, Olson ME, Stremick C, Read RR, Morck D, Buret A. 1999. The Calgary Biofilm Device: new technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J Clin Microbiol 37:1771–1776. doi: 10.1128/JCM.37.6.1771-1776.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Griffiths A, Miller J, Suzuki D. 2000. An introduction to genetic analysis. 7th edition W. H. Freeman, New York, NY. [Google Scholar]
- 23.Delumeau O, Dutta S, Brigulla M, Kuhnke G, Hardwick SW, Volker U, Yudkin MD, Lewis RJ. 2004. Functional and structural characterization of RsbU, a stress signaling protein phosphatase 2C. J Biol Chem 279:40927–40937. doi: 10.1074/jbc.M405464200. [DOI] [PubMed] [Google Scholar]
- 24.Ma Y, Pan F, McNeil M. 2002. Formation of dTDP-rhamnose is essential for growth of mycobacteria. J Bacteriol 184:3392–3395. doi: 10.1128/jb.184.12.3392-3395.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsukioka Y, Yamashita Y, Oho T, Nakano Y, Koga T. 1997. Biological function of the dTDP-rhamnose synthesis pathway in Streptococcus mutans. J Bacteriol 179:1126–1134. doi: 10.1128/jb.179.4.1126-1134.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van der Beek SL, Zorzoli A, Canak E, Chapman RN, Lucas K, Meyer BH, Evangelopoulos D, de Carvalho LPS, Boons GJ, Dorfmueller HC, van Sorge NM. 2019. Streptococcal dTDP-l-rhamnose biosynthesis enzymes: functional characterization and lead compound identification. Mol Microbiol 111:951–964. doi: 10.1111/mmi.14197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carvalho F, Atilano ML, Pombinho R, Covas G, Gallo RL, Filipe SR, Sousa S, Cabanes D. 2015. l-Rhamnosylation of Listeria monocytogenes wall teichoic acids promotes resistance to antimicrobial peptides by delaying interaction with the membrane. PLoS Pathog 11:e1004919. doi: 10.1371/journal.ppat.1004919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kamisango K, Fujii H, Okumura H, Saiki I, Araki Y, Yamamura Y, Azuma I. 1983. Structural and immunochemical studies of teichoic acid of Listeria monocytogenes. J Biochem 93:1401–1409. doi: 10.1093/oxfordjournals.jbchem.a134275. [DOI] [PubMed] [Google Scholar]
- 29.Wiedmann M, Arvik TJ, Hurley RJ, Boor KJ. 1998. General stress transcription factor sigmaB and its role in acid tolerance and virulence of Listeria monocytogenes. J Bacteriol 180:3650–3656. doi: 10.1128/JB.180.14.3650-3656.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shin JH, Brody MS, Price CW. 2010. Physical and antibiotic stresses require activation of the RsbU phosphatase to induce the general stress response in Listeria monocytogenes. Microbiology 156:2660–2669. doi: 10.1099/mic.0.041202-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Utratna M, Cosgrave E, Baustian C, Ceredig RH, Byrne CP. 2014. Effects of growth phase and temperature on σB activity within a Listeria monocytogenes population: evidence for RsbV-independent activation of σB at refrigeration temperatures. BioMed Res Int 2014:641647. doi: 10.1155/2014/641647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guilbaud M, Piveteau P, Desvaux M, Brisse S, Briandet R. 2015. Exploring the diversity of Listeria monocytogenes biofilm architecture by high-throughput confocal laser scanning microscopy and the predominance of the honeycomb-like morphotype. Appl Environ Microbiol 81:1813–1819. doi: 10.1128/AEM.03173-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Renier S, Hebraud M, Desvaux M. 2011. Molecular biology of surface colonization by Listeria monocytogenes: an additional facet of an opportunistic Gram-positive foodborne pathogen. Environ Microbiol 13:835–850. doi: 10.1111/j.1462-2920.2010.02378.x. [DOI] [PubMed] [Google Scholar]
- 34.Piercey MJ, Hingston PA, Hansen LT. 2016. Genes involved in Listeria monocytogenes biofilm formation at a simulated food processing plant temperature of 15°C. Int J Food Microbiol 223:63–74. doi: 10.1016/j.ijfoodmicro.2016.02.009. [DOI] [PubMed] [Google Scholar]
- 35.Brauge T, Sadovskaya I, Faille C, Benezech T, Maes E, Guerardel Y, Midelet-Bourdin G. 2016. Teichoic acid is the major polysaccharide present in the Listeria monocytogenes biofilm matrix. FEMS Microbiol Lett 363:fnv229. doi: 10.1093/femsle/fnv229. [DOI] [PubMed] [Google Scholar]
- 36.Azeredo J, Azevedo NF, Briandet R, Cerca N, Coenye T, Costa AR, Desvaux M, Di Bonaventura G, Hebraud M, Jaglic Z, Kacaniova M, Knochel S, Lourenco A, Mergulhao F, Meyer RL, Nychas G, Simoes M, Tresse O, Sternberg C. 2017. Critical review on biofilm methods. Crit Rev Microbiol 43:313–351. doi: 10.1080/1040841X.2016.1208146. [DOI] [PubMed] [Google Scholar]
- 37.Lee JJ, Lee G, Shin JH. 2014. sigma(B) affects biofilm formation under the dual stress conditions imposed by adding salt and low temperature in Listeria monocytogenes. J Microbiol 52:849–855. doi: 10.1007/s12275-014-4369-5. [DOI] [PubMed] [Google Scholar]
- 38.Lee T, Jun SH, Choi CW, Kim SI, Lee JC, Shin JH. 2018. Salt stress affects global protein expression profiles of extracellular membrane-derived vesicles of Listeria monocytogenes. Microb Pathog 115:272–279. doi: 10.1016/j.micpath.2017.12.071. [DOI] [PubMed] [Google Scholar]
- 39.Carvalho F, Pucciarelli MG, Garcia-del Portillo F, Cabanes D, Cossart P. 2013. Extraction of cell wall-bound teichoic acids and surface proteins from Listeria monocytogenes. Methods Mol Biol 966:289–308. doi: 10.1007/978-1-62703-245-2_18. [DOI] [PubMed] [Google Scholar]
- 40.Fischer ER, Hansen BT, Nair V, Hoyt FH, Dorward DW. 2012. Scanning electron microscopy. Curr Protoc Microbiol Chapter 2:Unit 2B.2. doi: 10.1002/9780471729259.mc02b02s25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Monk IR, Gahan CG, Hill C. 2008. Tools for functional postgenomic analysis of Listeria monocytogenes. Appl Environ Microbiol 74:3921–3934. doi: 10.1128/AEM.00314-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arnaud M, Chastanet A, Debarbouille M. 2004. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl Environ Microbiol 70:6887–6891. doi: 10.1128/AEM.70.11.6887-6891.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abbott JC. 2017. BugBuilder - an automated microbial genome assembly and analysis pipeline. bioRxiv doi: 10.1101/148783. [DOI]
- 46.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Darling AE, Mau B, Perna NT. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, Earl AM. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 50.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carver TJ, Rutherford KM, Berriman M, Rajandream M-A, Barrell BG, Parkhill J. 2005. ACT: the Artemis comparison tool. Bioinformatics 21:3422–3423. doi: 10.1093/bioinformatics/bti553. [DOI] [PubMed] [Google Scholar]
- 52.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hsu C-Y. 2019. Investigation of molecular mechanisms of biofilm formation by Listeria monocytogenes. PhD thesis. University of Dundee, Dundee, UK. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data have been deposited in the European Nucleotide Archive under study accession numbers PRJEB35200 and ERZ1188925.