Bacterial cell division is essential and requires the recruitment and regulation of a complex network of proteins needed to initiate and guide constriction and cytokinesis. FtsZ serves as a master regulator for this process, and its function is highly dependent on both its assembly into the canonical Z ring and interactions with protein binding partners, all of which results in the activation of enzymes that remodel the cell wall to drive constriction. Using mutants of FtsZ, we have elaborated on the role of its C-terminal linker domain in regulating Z-ring stability and dynamics, as well as the requirement for its conserved C-terminal domain and interaction with the membrane-anchoring protein FtsA for regulating the process of cell wall remodeling for constriction.
KEYWORDS: Caulobacter crescentus, FtsA, FtsZ, cell division, cell wall, peptidoglycan
ABSTRACT
Bacterial cell division requires the assembly of a multiprotein division machinery, or divisome, that remodels the cell envelope to cause constriction. The cytoskeletal protein FtsZ forms a ringlike scaffold for the divisome at the incipient division site. FtsZ has three major regions: a conserved GTPase domain that polymerizes into protofilaments on binding GTP, a C-terminal conserved peptide (CTC) required for binding membrane-anchoring proteins, and a C-terminal linker (CTL) region of varied length and low sequence conservation. Recently, we demonstrated that the CTL regulates FtsZ polymerization properties in vitro and Z-ring structure and cell wall metabolism in vivo. In Caulobacter crescentus, an FtsZ variant lacking the CTL (designated ΔCTL) can recruit all known divisome members and drive local cell wall synthesis but has dominant lethal effects on cell wall metabolism. To understand the underlying mechanism of the CTL-dependent regulation of cell wall metabolism, we expressed chimeras of FtsZ domains from C. crescentus and Escherichia coli and observed that the E. coli GTPase domain fused to the C. crescentus CTC phenocopies C. crescentus ΔCTL. By investigating the contributions of FtsZ-binding partners, we identified variants of FtsA, a known membrane anchor for FtsZ, that delay or exacerbate the ΔCTL phenotype. Additionally, we observed that the ΔCTL protein forms extended helical structures in vivo upon FtsA overproduction. We propose that misregulation downstream of defective ΔCTL assembly is propagated through the interaction between the CTC and FtsA. Overall, our study provides mechanistic insights into the CTL-dependent regulation of cell wall enzymes downstream of FtsZ polymerization.
IMPORTANCE Bacterial cell division is essential and requires the recruitment and regulation of a complex network of proteins needed to initiate and guide constriction and cytokinesis. FtsZ serves as a master regulator for this process, and its function is highly dependent on both its assembly into the canonical Z ring and interactions with protein binding partners, all of which results in the activation of enzymes that remodel the cell wall to drive constriction. Using mutants of FtsZ, we have elaborated on the role of its C-terminal linker domain in regulating Z-ring stability and dynamics, as well as the requirement for its conserved C-terminal domain and interaction with the membrane-anchoring protein FtsA for regulating the process of cell wall remodeling for constriction.
INTRODUCTION
Bacterial cell division requires spatially and temporally coordinated remodeling of the cell envelope to cause constriction. To this end, a multiprotein machinery, called the divisome, is assembled at the incipient site of division. FtsZ, the most conserved protein of the divisome and the first to arrive at midcell prior to division, is a tubulin homolog that polymerizes into a discontinuous ringlike scaffold, or Z ring, for the recruitment of other members of the divisome (1). Over two dozen proteins are directly or indirectly recruited to the divisome. In Caulobacter crescentus, these include FtsZ-binding proteins that regulate Z-ring structure (ZapA, ZauP, and FzlA), membrane-anchoring proteins of FtsZ (FtsA, FzlC, and FtsEX), peptidoglycan (PG) cell wall enzymes (DipM, AmiC, FtsI/Pbp3, and FtsW) and their regulators (FtsN and FtsQLB), outer membrane remodeling proteins (Tol-Pal complex), pole remodeling proteins (TipN), and factors involved in chromosome segregation and translocation (e.g., FtsK) (2–3). While most of the essential members of the divisome have likely been identified, the interactions among these proteins and the regulation of their organization and function are unclear.
In addition to serving as a scaffold for the divisome, FtsZ actively regulates the activity of the divisome. The existence of FtsZ mutants that can assemble into Z rings, recruit the divisome, and drive local cell wall synthesis but are incapable of cell division suggests that cell division requires additional FtsZ-dependent regulation of organization or activity of the divisome (4). Recent studies have shown that clusters of FtsZ protofilaments in the Z ring undergo directional treadmilling motion that drives the movement of cell wall enzymes (5–6). Thus, Z-ring assembly properties are directly relevant for the regulation of local cell wall remodeling. However, the pathways downstream of Z-ring assembly that regulate cell wall enzymes are largely unknown.
FtsZ has three regions: (i) a conserved GTPase domain, (ii) a C-terminal linker (CTL), and (iii) a C-terminal conserved peptide (CTC) (7). The GTPase domain is structurally similar to that of eukaryotic tubulin (8–9) and is sufficient for polymerization on binding GTP (4, 10). Mutations in the GTPase domain affect Z-ring dynamics, organization, and regulation of cell wall synthetic enzymes, at least in some bacteria (5, 6, 11–13). FtsZ-binding proteins such as FtsZ-localized protein A (FzlA) bind the GTPase domain (4). Overproduction, depletion, and mutation of these FtsZ-binding proteins have been observed to influence Z-ring structure through unclear mechanisms. The CTC is composed of a conserved peptide that is required for FtsZ’s interactions with membrane-anchoring proteins such as FtsA across multiple species of bacteria (14–19) and FzlC in C. crescentus (20).
The CTL is an intrinsically disordered region that connects the GTPase domain to the CTC and varies in length and sequence across species. While there are no known binding partners for the CTL, changes in the length and sequence of the CTL affect polymer turnover and lateral interactions between FtsZ protofilaments, at least in the cases of FtsZ from Caulobacter crescentus (CcFtsZ), Bacillus subtilis, and Agrobacterium tumefaciens (10, 21–23). Surprisingly, large modifications of CTL sequence are tolerated in B. subtilis and Escherichia coli cells as long as flexibility of the CTL and a length range of ±50% of wild-type (WT) length are maintained (21, 24). Conversely, in C. crescentus, large truncations of the CTL are tolerated to some extent, but significant changes to the CTL sequence impact protein stability and, therefore, cell division (4). Complete deletion of the CTL causes dominant lethal defects in Z-ring assembly and cell lysis, at least in C. crescentus and B. subtilis (4, 21). Identifying the contributions of the CTL to FtsZ function is thus essential to understanding the communication between Z-ring structure and cell wall enzyme activities.
We previously showed that the expression of FtsZ lacking its CTL (designated ΔCTL, wherein the GTPase domain is fused directly to the CTC) in the alphaproteobacterium C. crescentus causes misregulation of cell wall enzymes, resulting in the formation of envelope bulges at the sites of ΔCTL assembly and rapid cell lysis (4). Using a fluorescent fusion to ZapA, a protein that binds FtsZ’s GTPase domain, we found that Z-ring superstructure is affected in cells producing ΔCTL: ΔCTL forms large, amorphous assemblies instead of focused rings (4). FtsZ with a minimal CTL of 14 amino acids (designated L14) exhibits WT-like Z-ring shape and does not lead to bulging and lysis. In vitro, ΔCTL polymerizes into straight multifilament bundles that are significantly longer than the curved protofilaments observed for WT FtsZ or L14 by electron microscopy (4, 10). Moreover, ΔCTL exhibits lower GTP hydrolysis rates than WT FtsZ in vitro (4, 10). The effects of the loss of the CTL on polymer assembly and superstructure result in the formation of stable networks of protofilaments of ΔCTL on supported lipid bilayer membranes in contrast to smaller dynamic clusters formed by WT FtsZ, as observed by total internal reflection fluorescence microscopy in vitro (25). Unlike the CTL, the CTC does not obviously contribute to polymer structure or dynamics for C. crescentus FtsZ; polymer structure, observed by electron microscopy (EM), or GTP hydrolysis rates of FtsZ lacking its CTC (ΔCTC) are comparable to those of WT FtsZ (10). Moreover, in vivo, ΔCTC assembles into rings and broad bands but is incapable of cytokinesis (4).
Determining how CTL-dependent changes in FtsZ polymerization are communicated to the divisome is essential for understanding how Z-ring structure and dynamics regulate cell wall metabolism. We hypothesize that there are specific pathways downstream of the aberrant ΔCTL superstructures in vivo that contribute to the misregulation of cell wall enzymes. In the current study, we examined the contributions of the GTPase and CTC domains to the ΔCTL phenotype. By expressing chimeric FtsZ variants bearing domains from E. coli FtsZ (EcFtsZ) and/or C. crescentus FtsZ in C. crescentus cells, we found that a chimeric FtsZ with the E. coli GTPase domain and C. crescentus CTC causes bulging and lysis but only in the absence of a CTL from either organism. We also tested the effects of candidate division proteins on the lethal cell wall metabolic defects downstream of ΔCTL. Of all the division proteins tested, only FtsA appears to be required for ΔCTL-induced bulging and lysis. Specifically, we observed that variants of FtsA are able to reduce or enhance the toxicity of ΔCTL, and an overabundance of FtsA leads to profound distortion of the superstructures of WT FtsZ and ΔCTL Z rings in vivo. Together, our results suggest that whereas the CTL is required to prevent defective Z-ring assembly, the interaction between the CTC and FtsA is required for CTL-dependent signaling from the Z ring to the regulation of cell wall enzymes in cells.
RESULTS
E. coli GTPase domain fused to C. crescentus CTC is sufficient to cause bulging and lysis.
We first sought to determine the contributions of each region of FtsZ to the ΔCTL-induced bulging and lysis phenotype by making chimeric FtsZ variants using the GTPase domain, CTL, and/or CTC regions of E. coli and/or C. crescentus FtsZ (Fig. 1; see also Fig. S1 in the supplemental material). We reasoned that, due to the low sequence homology, FtsZ-binding partners specific to C. crescentus would not be able to bind and regulate the evolutionarily distant E. coli FtsZ. We expressed chimeric FtsZ mutants in C. crescentus cells depleted of WT FtsZ using a strain wherein the only copy of ftsZ is under vanillate-driven expression via the PvanA promoter and wherein the ftsZ chimera is under xylose-inducible expression via the PxylX promoter and followed their effects on cell morphology. Additionally, we imaged the incorporation of fluorescently labeled d-alanine (HADA) (26) to visualize regions of active cell wall metabolism.
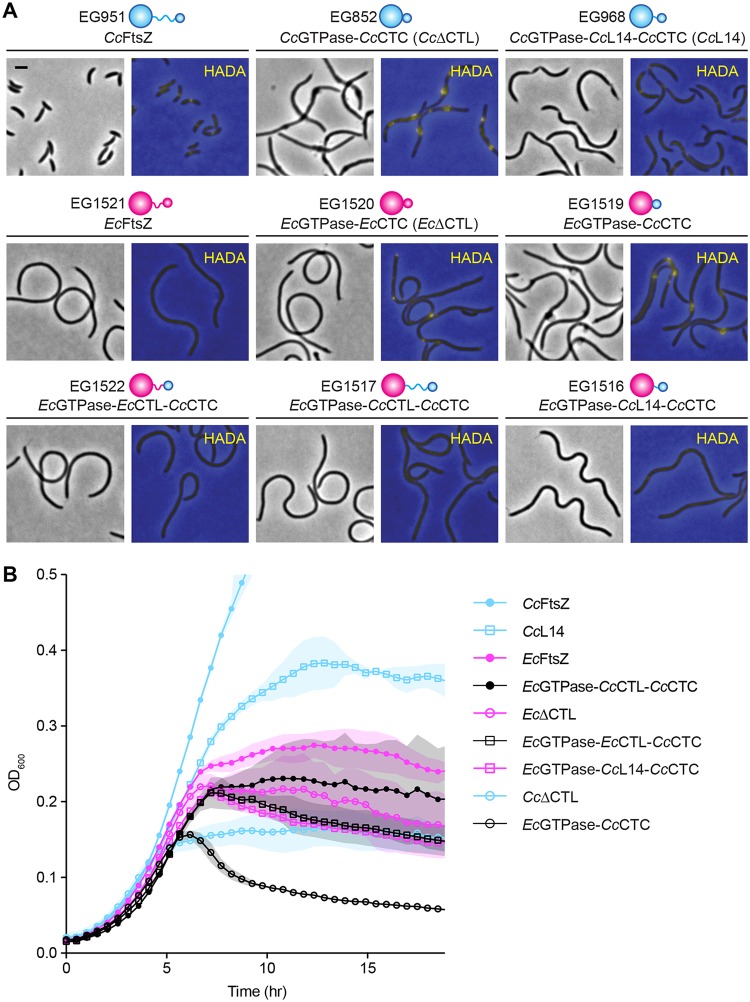
FIG 1.
EcGTPase-CcCTC can cause bulging and lysis similar to that with CcΔCTL. (A) Phase-contrast images of morphology and merged epifluorescence images showing HADA incorporation (yellow) overlaid on phase-contrast images (blue) corresponding to cells depleted of FtsZ and simultaneously induced for xylose-dependent production of C. crescentus FtsZ (cyan), E. coli FtsZ (magenta), CTL truncations, or their chimeric variants. Phase-contrast images were acquired after 5 h of induction of FtsZ variant. HADA fluorescence images were acquired after 4.5 h of induction of FtsZ variant. Scale bar, 2 μm. (B) Growth characteristics of strains shown in panel A represented as absorbance at OD600 over time. Shaded regions represent standard deviations of three technical replicates at each point. Strains are as indicated, and all are depleted of native CcFtsZ.
In C. crescentus, FtsZ drives the majority of cell wall synthesis at midcell (Fig. 1A, CcFtsZ), and depletion of WT FtsZ in C. crescentus cells caused diffuse cell wall synthesis (27). Xylose-induced CcΔCTL production resulted in filamentation and bulges, which corresponded to sites of active cell wall metabolism (Fig. 1A). On the other hand, xylose-induced production of CcL14, an FtsZ variant with a CTL consisting of only 14 amino acids that is incapable of cytokinesis but does not cause bulging and lysis (4), could direct initiation of constriction and drive cell wall metabolism at multiple sites along filamentous cells. Production of EcFtsZ in C. crescentus cells did not result in any constriction or localized cell wall metabolism, consistent with the expectation that EcFtsZ cannot efficiently engage the C. crescentus division or PG metabolic machinery. When we expressed EcΔCTL (EcGTPase-EcCTC), we did not observe any constriction initiation, bulging, or lysis. However, this mutant was surprisingly able to drive limited local cell wall metabolism. The localization of HADA fluorescence appeared diffuse, with occasional asymmetrically distributed foci along the short axis of the cells or at the cell pole.
Strikingly, when we expressed a chimera wherein the EcGTPase domain is fused to CcCTC (EcGTPase-CcCTC), we observed cell envelope bulges similar to those resulting from CcΔCTL production. Once again, similar to observations of CcΔCTL, bulges were the primary sites of cell wall metabolism in these cells, and expression of EcGTPase-CcCTC resulted in rapid cell lysis (Fig. 1B). The toxic effects of EcGTPase-CcCTC were not observed when we introduced CcCTL, EcCTL, or CcL14 back into this chimera; xylose-induced expression of EcGTPase-CcCTL-CcCTC, EcGTPase-EcCTL-CcCTC, or EcGTPase-CcL14-CcCTC resulted in smooth filamentous cells with diffuse cell wall synthesis, similar to cells with EcFtsZ. We confirmed by probing with antibodies against both C. crescentus and E. coli FtsZ that there were no significant differences in the expression levels of these chimeras that could account for the differences in phenotypes observed (Fig. S2).
We hypothesized that the localization (or lack thereof) of HADA signal in cells producing different FtsZ variants would reflect the localization of the corresponding FtsZ variant. Indeed, CcFtsZ with an N-terminal monomeric-neon green (mNG) (28) fusion localized to midcell rings, as expected, while mNG-CcΔCTL and mNG-EcGTPase-CcCTC showed similar localizations in bulges (Fig. S3), recapitulating what we observed previously using a tagged variant of an FtsZ-binding protein (ZapA-Venus) (4). mNG-EcGTPase-CcL14-CcCTC and mNG-EcΔCTL had diffuse localizations as well with foci along the cell body and, in the case of mNG-EcΔCTL, at cell polls (Fig. S3), reflecting a localization pattern similar to that of HADA in the strains bearing the untagged variants (Fig. 1A). In contrast to EcΔCTL, mNG-EcFtsZ in cells depleted of CcFtsZ exhibited diffuse localization with limited formation of foci.
Our finding that EcGTPase-CcCTC induces bulging and lysis while EcΔCTL fails to do so implies that partners that bind the CTC, but not the GTPase domain, are important for downstream regulation of PG metabolism. To confirm that C. crescentus proteins that bind the GTPase domain of CcFtsZ do not bind the GTPase domain of EcFtsZ, we performed in vitro copelleting assays with the FtsZ-binding proteins FzlA and MipZ, regulators of constriction activation and division site positioning, respectively. As expected, purified FzlA and MipZ were each efficiently recruited to the pellet by polymers of CcFtsZ or CcΔCTL after high-speed centrifugation, indicating direct binding. In contrast, FtsZ variants including the EcGTPase domain (EcFtsZ and EcGTPase-CcCTC) failed to recruit either FzlA or MipZ to the pellet, suggesting that these proteins are unable to interact strongly with the GTPase domain of EcFtsZ (Fig. S4).
Finally, to gain insight into why EcFtsZ and EcΔCTL would exhibit different localization patterns when expressed in C. crescentus, we analyzed their in vitro assemblies, along with those of CcFtsZ, CcΔCTL, and EcGTPase-CcCTC, with right-angle light scatter assays using limiting amounts of GTP (Fig. S5). In line with previous results (10), CcΔCTL exhibited a greater degree of light scatter for a longer period of time after addition of GTP than CcFtsZ, in agreement with a higher degree of interfilament bundling and slower turnover. On the other hand, EcΔCTL exhibited a similar degree of light scatter, albeit for a longer period of time than EcFtsZ, after addition of GTP. This is consistent with the idea that the CTL contributes to polymer turnover in EcFtsZ as it does in CcFtsZ (4, 10). The low level of light scatter for EcΔCTL suggests that it does not exhibit substantially increased bundling, at least compared to that of CcΔCTL. The slowed polymer dynamics observed in vitro for EcΔCTL compared to that of EcFtsZ could potentially explain the difference in their abilities to form foci in C. crescentus cells (Fig. S3). Interestingly, we also observed that the EcGTPase-CcCTC variant exhibits a higher degree of light scatter than EcFtsZ and EcΔCTL and for a longer period of time than EcFtsZ (Fig. S5), suggesting that in addition to slower turnover, this variant is able to form interfilament bundles. This bundling behavior may be mediated in part by the presence of CcCTC, suggesting that this domain contributes to the bundling propensity of the EcGTPase domain.
Taken together, our results suggest that the misregulation of PG metabolism downstream of CcΔCTL assembly requires mainly three factors: (i) a polymerizing GTPase domain (4), (ii) absence of a minimal CTL, and (iii) the CcCTC. Moreover, since CcΔCTL (CcGTPase-CcCTC) and EcGTPase-CcCTC cause almost identical effects on cell morphology and cell wall integrity, since the CcGTPase domain alone was previously shown to be insufficient to cause bulges (4), and since the EcGTPase domain does not efficiently bind CcFtsZ partners (Fig. S4), we conclude that differences in the interactions of FtsZ-binding proteins with the GTPase domain are unlikely to be responsible for the CTL-dependent misregulation of cell wall synthesis.
FtsA is implicated in ΔCTL-induced bulging.
The ability of EcGTPase-CcCTC to cause bulging and lysis suggests that divisome proteins that bind to the CTC are critical for CTL-dependent regulation of cell wall metabolism, whereas those that interact with the GTPase domain of FtsZ are likely not required. To address this hypothesis, we asked if xylose-inducible expression of ΔCTL can cause bulging and lysis in cells deleted of nonessential members of the divisome that bind FtsZ at the GTPase domain (zapA, zauP, and both zapA and zauP), the CTC (fzlC), or at an unknown site (ftsE). We observed filamentation, bulging, and lysis in strains lacking each of these factors (Fig. S6), suggesting that CTL-dependent regulation of cell wall metabolism does not require these divisome proteins.
FtsA is an essential membrane-anchoring protein for FtsZ that binds to the CTC (14–19). Since FtsA is essential, we expressed ΔCTL in a previously described ftsA temperature-sensitive [ftsA(Ts)] strain background (EG1776) (29). At the restrictive temperature of 37˚C, the ftsA(Ts) strain exhibited filamentation in the presence or absence of ΔCTL production (Fig. S7A and B). Upon induction of ΔCTL, the ftsA(Ts) strain failed to exhibit characteristic bulging (Fig. S7A), and lysis was delayed (Fig. S7C), although the ftsA(Ts) strain was unable to suppress the viability defect associated with ΔCTL production (Fig. S7D). However, the ftsA(Ts) strain also suppressed bulging upon ΔCTL induction at the permissive temperature. This prompted us to sequence the ftsA mutation (I275N) and introduce it into a clean genetic background with xylose-inducible ΔCTL (EG2805). This strain exhibited a filamentous morphology, low growth rate, and reduced viability at 30°C, characteristics which were exacerbated at 37°C without induction of ΔCTL (Fig. S7A, C, and D), implying that there are additional mutations in the original temperature-sensitive strain that suppress filamentation at 30°C. We did not observe bulging in a strain expressing an I-to-N change at position 275 encoded by ftsA (ftsAI275N strain) upon ΔCTL expression at either temperature (Fig. S7A), although increased lysis and the viability defect associated with ΔCTL were still present (Fig. S7C and D). Collectively, the above data implicate FtsA in the dominant lethal bulging effect of ΔCTL and suggest that it likely plays a key role in CTL-dependent regulation of cell wall metabolism.
Given that cells bearing the ftsAI275N mutation are very sick even at 30°C without ΔCTL production, we sought additional genetic evidence implicating FtsA in ΔCTL-mediated phenotypes. Prior studies implicated FtsA’s polymerization state in its function in vivo (30–35), so we endeavored to generate mutants of ftsA that would produce variants predicted to be impaired for self-interaction based on analogous mutations in E. coli ftsA (Fig. S8, adapted from PDB accession number 4A2B) (32). Importantly, these residues (Fig. S8, shown in red) are located at FtsA’s self-interaction interface and are not in proximity to either the ATP- or FtsZ-binding region (shown in cyan and blue, respectively). In the absence of ΔCTL, strains expressing these mutants as the only copy of ftsA were similar to the wild type in morphology, growth, and viability, although the R371A variant resulted in slightly elongated cells (Fig. 2A to G). However, upon induction of ΔCTL (Fig. 2H), the mutants displayed distinct phenotypes. Two mutants (R195C and A295M) appeared to delay the ΔCTL-associated filamentation, bulging, and lysis phenotypes (Fig. 2B to F). Interestingly, both of these strains exhibited late-stage division defects when ΔCTL was induced, as indicated by the presence of cells with nearly complete constriction, resulting in a chaining morphology (Fig. 2B and C, white arrowheads). Additionally, HADA incorporation nearly resembled that of uninduced cells, further suggesting that these mutants can minimize or delay the signaling defects imparted by ΔCTL. Neither mutant was able to suppress ΔCTL-induced viability defects (Fig. 2G).
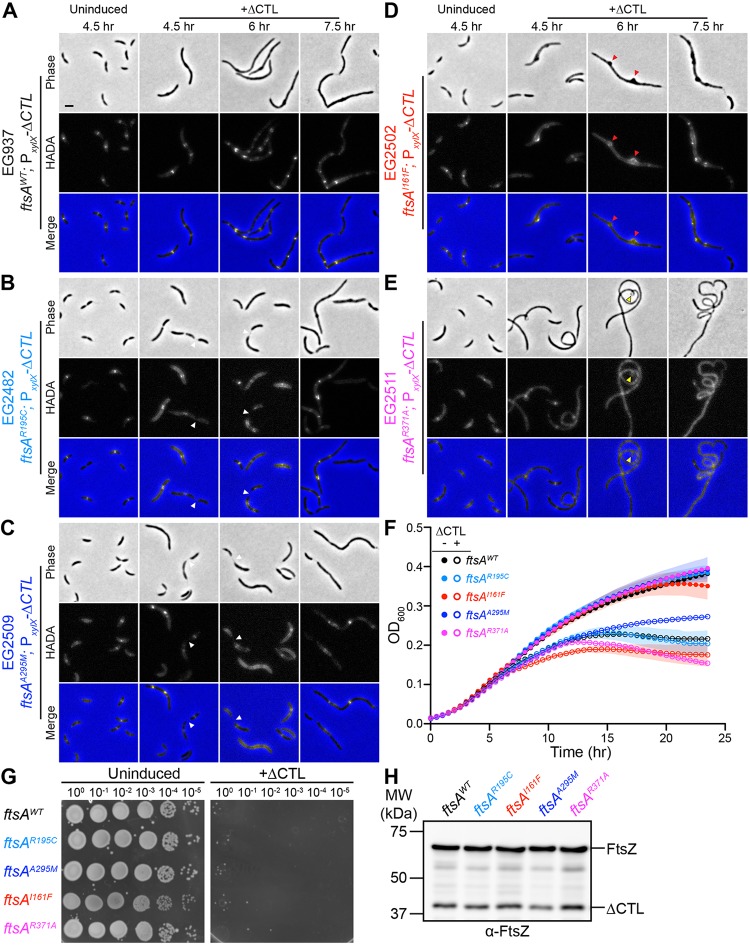
FIG 2.
ftsA point mutants can delay or exacerbate ΔCTL-induced bulging, filamentation, and lysis. (A to E) Phase-contrast, epifluorescence, and merged images of cells with ftsA WT (A), ftsAR195C (B), ftsAA295M (C), ftsAI161F (D), or ftsAR371A (E) as the only copy of ftsA uninduced with glucose or induced with xylose to drive expression of ΔCTL from PxylX promoter for indicated amounts of time prior to imaging. White arrowheads indicate cells with nearly complete constriction, resulting in a chaining morphology. Red arrowheads indicate bulges that are asymmetric about the long axis of the cell. Yellow arrowheads indicate regions of hypercurvature. (F) Growth characteristics of cells shown in panel A, uninduced (closed circles) or induced (open circles) for ΔCTL expression, represented as absorbance at OD600 over time. Shaded regions represent standard deviations of three technical replicates at each point. (G) Spot dilutions of strains shown in panel A showing growth of cells uninduced with glucose or induced with xylose (+ΔCTL) for ΔCTL expression. Cells in log phase were diluted to an OD600 of 0.05, serially diluted, and spotted onto PYE agar plates with indicated inducer (glucose or xylose). Plates were incubated at 30°C for 48 h before imaging. (H) Immunoblot using anti-FtsZ antibody showing protein levels of ΔCTL and WT FtsZ corresponding to the experiments shown in panel A at 6.5 h of incubation with xylose. Strains are as indicated, and all strains have xylose-inducible ΔCTL.
The other two mutants (I161F and R371A) displayed distinct morphologies in the presence of ΔCTL (Fig. 2D and E), as well as more rapid lysis than that of ΔCTL in an ftsA WT background (Fig. 2F). Cells bearing the I161F mutant exhibited severe, asymmetric bulging (Fig. 2D, arrowheads), which is remarkable since ΔCTL typically results in bulges symmetric about the longitudinal axis. The R371A mutant in combination with ΔCTL, however, yielded highly filamentous cells with diffuse HADA signal and regions of hypercurvature (Fig. 2E, yellow arrowheads), suggesting that division is immediately and completely inhibited upon ΔCTL induction in this background. Although further work is needed to elucidate the exact effects these point mutations impart on FtsA function, it is clear that they are sufficient to affect ΔCTL-mediated phenotypes. Collectively, our data thus far indicate that the CTC is required for ΔCTL-mediated signaling through FtsA to misregulate cell wall metabolism but that all known nonessential FtsZ-binding proteins are dispensable for this signaling.
Overproduction of FtsA exacerbates ΔCTL-induced cell wall defects.
Next, we investigated whether overproducing FtsZ-binding proteins could impact the dominant lethal effects of ΔCTL expression. To this end, we overproduced FzlA, ZapA, ZauP, both ZapA and ZauP, FtsA, FtsE, both FtsE and FtsX, FzlC, or MipZ in cells inducibly expressing ΔCTL. We found that overproduction of either ZapA (without ZauP) or FzlC suppresses the formation of bulges up to 10 h postinduction and allows initiation of constriction in cells producing ΔCTL (Fig. 3). Overproduction of ZapA or FzlC in the absence of ΔCTL causes a minimal increase in cell length (20, 36). In cells overproducing ZapA or FzlC and producing ΔCTL, the region of constriction appeared elongated, and many cells were chained. Overproduction of FtsE or FtsEX in the absence of ΔCTL expression caused cell elongation and ectopic pole formation (37). We observed a similar phenotype in cells overproducing FtsE or FtsEX and producing ΔCTL, with no envelope bulges, suggesting that the effects of ftsEX overexpression are dominant to ΔCTL-induced toxicity (Fig. 3A).
FIG 3.
FtsA overproduction exacerbates formation of ΔCTL-induced bulges. (A) Phase-contrast images of cells in the absence or presence of inducer (xylose) for the expression of ΔCTL and overproduction of FtsZ binding proteins (or empty vector control). Scale bar, 2 μm. (B) Phase-contrast images of cells with xylose-inducible expression of ΔCTL and vanillate-inducible overexpression of fzlC, mipZ, or empty vector control. Scale bar, 2 μm. All strains have xylose-inducible ΔCTL.
MipZ is a negative regulator of FtsZ polymerization, and overproduction inhibits assembly of Z rings. Overexpression of mipZ in cells expressing ΔCTL prevented the formation of envelope bulges up to 10 h postinduction (Fig. 3B). Since the ability of ΔCTL to form polymers is required for inducing bulging and lysis (4), overproducing MipZ could affect ΔCTL-induced bulging by limiting ΔCTL assembly. Although overproduction of ZapA, FzlC, FtsE, FtsEX, or MipZ is sufficient to suppress ΔCTL-induced bulging, none of the proteins, when overproduced, were able to support growth, as demonstrated by a spot dilution assay (Fig. S9). We confirmed that the effects of overproducing these proteins on ΔCTL-induced bulging were not due to differences in expression of ΔCTL via immunoblot probing for FtsZ/ΔCTL (Fig. S10).
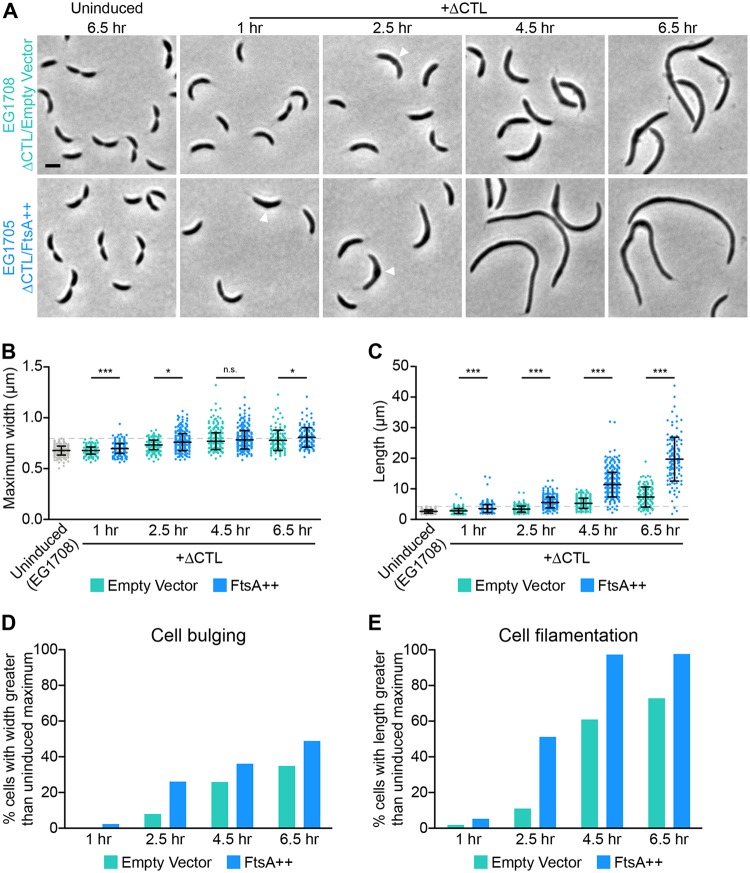
The ratio of FtsZ to FtsA has been shown to affect cell division and morphology in several species (38–41), so we predicted that increasing FtsA levels would have an effect on cells producing ΔCTL. Indeed, in contrast to the effects of overproducing ZapA, FzlC, or MipZ, overproduction of FtsA exacerbated the effects of ΔCTL production: envelope bulges were larger, less symmetric, and appeared earlier in ΔCTL-producing cells overexpressing ftsA than in those not overexpressing ftsA (empty vector control) (Fig. 3A). We confirmed these observations using a time course that included earlier time points to gain insight into when these bulges begin to appear (Fig. 4A, arrowheads). Quantification of the maximum widths of cells revealed that ftsA overexpression causes larger and/or more frequent ΔCTL-induced envelope bulges than empty vector controls at all time points except 4.5 h postinduction (Fig. 4B). Moreover, the proportion of cells with bulges (quantified as cells with maximum cell widths greater than the width of the widest ΔCTL-uninduced cell of the same strain) was higher for FtsA-overproducing cells than for empty vector controls at all time points (Fig. 4D), further suggesting that FtsA overproduction accelerates ΔCTL-induced bulging. As expected, FtsA-overproducing cells were also longer than empty vector controls at all time points, and the proportion of filamentous cells (quantified as cells with lengths greater than the length of the longest ΔCTL-uninduced cell of the same strain) increased rapidly on ftsA overexpression until virtually the entire population was filamentous (Fig. 4C and E). Collectively, our genetic interaction analyses implicate FtsA as a major mediator of signaling downstream of FtsZ in CTL-dependent regulation of PG metabolism.
FIG 4.
FtsA overproduction induces earlier bulge formation and greater filamentation in cells producing ΔCTL. (A) Phase-contrast images of cells uninduced or induced with xylose to drive expression of mNG-ΔCTL and empty vector (turquoise) or ftsA (blue) from PxylX promoter for the indicated amounts of time prior to imaging. Arrowheads indicate bulges at early time points. (B and C) Quantification of maximum width or length of cells from strains shown in panel A at indicated time points. The maximum value of the uninduced EG1708 strain is denoted by a dashed line in each. Bars represent standard deviations (n = 379 cells per strain for 1-, 2.5-, and 4.5-h induced and 6.5-h uninduced time points; n = 179 cells per strain for the 6.5-h induced time point). *, P ≤ 0.05; ***, P ≤ 0.001; n.s., not significant. (D and E) Proportions of cells from strains shown in panel A at indicated time points with maximum width (as a readout of bulging) or length (as a readout of filamentation) greater than that of the maximum value for uninduced cells of the same strain. Strains are as indicated, and all strains have xylose-inducible ΔCTL.
The FtsZ CTL impacts Z-ring superstructure in vivo.
ΔCTL has significantly altered polymerization properties compared to those of WT FtsZ (4, 10, 25) and altered Z-ring morphology in cells (4) (Fig. S3), implicating FtsZ dynamics and superstructure in PG misregulation downstream of ΔCTL. To investigate whether FtsZ-binding proteins affect Z-ring structure to modulate ΔCTL toxicity, we first more comprehensively compared Z-ring structures of ΔCTL and other variants in vivo. We found that mNG-ΔCTL produced from PxylX in the presence or absence of WT FtsZ causes filamentation and local cell wall bulges at 5 h of induction, indicating that mNG-ΔCTL is dominant lethal, similar to untagged ΔCTL (Fig. 5A; also Fig. S11 and S14). This allowed us to compare structures formed by FtsZ or ΔCTL in vivo to identify CTL-dependent differences in the Z ring that might correlate with bulging and lysis. In our analysis, we also included as controls mNG fusions to L14 and an FtsZ variant with the CcCTL sequence replaced with the CTL from Hyphomonas neptunium FtsZ (HnCTL) (Fig. S1) that causes inefficient cytokinesis (4).
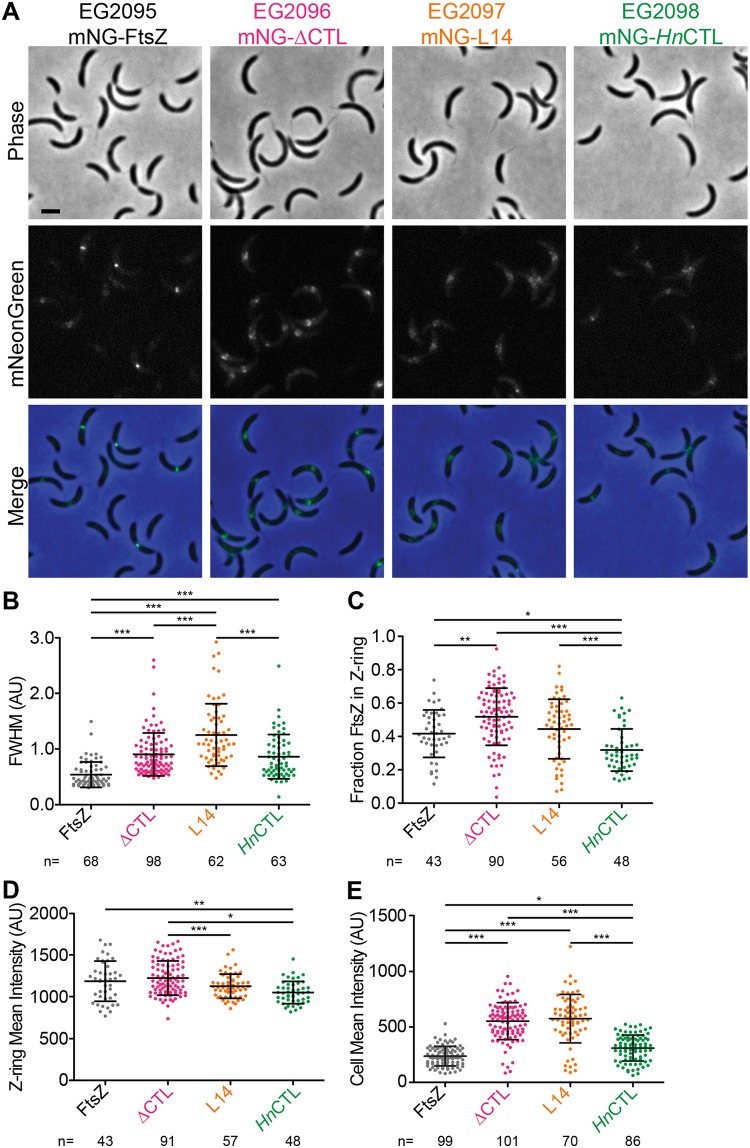
FIG 5.
ΔCTL assembles into large asymmetric superstructures at sites of cell wall bulging in cells depleted of WT FtsZ. (A) Phase-contrast, epifluorescence, and merged images of cells induced with xylose to drive expression of mNG-FtsZ, mNG-ΔCTL, mNG-L14, or mNG-HnCTL from the PxylX promoter for 1 h while simultaneously depleting WT FtsZ. Scale bar, 2 μm. (B to E) Quantification of epifluorescence images of cells 3 to 5 μm long indicating the full width at half-maximum (FWHM) values of Z-ring intensity (B), fraction of mNG-FtsZ or variants in the Z ring (C), and mean epifluorescence intensity of the whole cells (D) or the Z ring (E) in an FtsZ depletion background. Bars represent standard deviations. Numbers of cells per strain are indicated for each measurement. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001. Strains are indicated in panel A. AU, arbitrary units.
We expressed mNG fusions to FtsZ or CTL variants using the xylose-inducible PxylX promoter while depleting WT FtsZ simultaneously using strains wherein the only copy of ftsZ is under the control of the vanillate-inducible PvanA promoter. As we observed earlier (Fig. S3), mNG-FtsZ formed ringlike structures that appeared as a band or two closely spaced foci aligned along the short axis or a single focus per dividing cell after 1 h of induction (Fig. 5A). At longer induction times, mNG-FtsZ Z-ring structure was maintained despite cell filamentation due to depletion of WT FtsZ (Fig. S11). Unlike WT FtsZ, within 1 h of induction, mNG-ΔCTL formed brighter, wider, and fewer ringlike structures. These structures increased in size and intensity over time. Frequently, we observed these structures to be asymmetrically distributed along the short axis (Fig. 5A; Fig. S11). mNG-L14 assembled into apparently less dense, diffuse structures after 1 h of induction (Fig. 5A), which became more diffuse and scattered at longer induction times (Fig. S11). mNG-HnCTL structures appeared predominantly as faint rings or foci or more dispersed structures, similar to those of mNG-L14, and did not change appreciably with longer induction or cell filamentation (Fig. 5A; Fig. S11).
We quantitatively analyzed Z-ring structures using MicrobeJ (42) and Oufti (43). To avoid potential effects of cell length on Z-ring organization, we focused on cells 3 to 5 μm long, which we determined from demographs (Fig. S12) to have stable Z rings after induction of mNG-FtsZ or CTL variants for 1 h. We calculated the full width at half-maximum (FWHM) value for mNG intensity along the longitudinal axis for each variant as a measure of the degree of focusing of the Z ring. Z rings formed by mNG-ΔCTL, mNG-L14, and mNG-HnCTL were wider than those formed by mNG-FtsZ (Fig. 5B). We asked if these altered Z-ring structures formed by CTL variants could be explained by differences in the fraction of FtsZ present in the Z ring by determining relative enrichment of fluorescence signal at the Z ring compared to that of the rest of the cell. Indeed, cells expressing mNG-ΔCTL had a significantly greater proportion of fluorescence signal in the Z ring than those expressing mNG-FtsZ (Fig. 5C). The fraction of mNG-HnCTL was lower than that of each of the other CTL variants, suggesting a lower tendency to assemble into polymers at the Z ring, while the fraction of mNG-L14 was similar to that of WT FtsZ (Fig. 5C). Moreover, mNG-L14 and mNG-HnCTL each formed less dense structures, as measured by mean intensity of the Z ring, than mNG-FtsZ or mNG-ΔCTL (Fig. 5D), consistent with their apparent dimness in the images.
We next determined the relative amount of each tagged FtsZ variant per cell in each strain to address whether protein levels are affected. Mean fluorescence intensity values for the whole cell were significantly increased in cells expressing mNG-ΔCTL or mNG-L14 compared to levels in those expressing mNG-FtsZ or mNG-HnCTL (Fig. 5E), suggesting increased protein levels of these two variants. Using quantitative immunoblotting, we found that, indeed, mNG-ΔCTL and mNG-L14 levels were ∼5-fold higher than those of mNG-FtsZ and mNG-HnCTL and that these levels increased relative to the level of mNG-FtsZ over time (Fig. S13). mNG-L14 was present at higher levels than mNG-ΔCTL, whereas mNG-HnCTL levels were nearly equivalent to those of mNG-FtsZ (Fig. S13). Since all mNG fusions were expressed using identical induction conditions, increased protein levels are likely due to differences in posttranslational stability.
Finally, we tested if the structures formed by the CTL variants are influenced by the presence of WT FtsZ by expressing mNG fusions in an otherwise WT strain (i.e., without depleting WT FtsZ). All four variants formed ringlike structures at 1 h of induction, with mNG-ΔCTL forming slightly wider and brighter rings (Fig. S14A). However, after 5 h of induction, mNG-ΔCTL structures became less ringlike and more asymmetric, while the structures formed by the other CTL variants resembled those formed by mNG-FtsZ (Fig. S14B). This observation agrees with our prior observations that the presence of WT FtsZ delays the onset of ΔCTL-induced bulging and lysis (4) and that ΔCTL and WT FtsZ can form long, bundled copolymers in vitro (10). In all cells expressing ΔCTL, the appearance of aberrant structures preceded the appearance of cell envelope bulges, suggesting that aberrant Z-ring morphology in the absence of CTL is not a result of altered cell geometry but, rather, is inherent to the assembly properties of ΔCTL. Quantitation of Z-ring structures formed by and protein levels of each CTL variant in the presence of WT FtsZ after 1 h of induction revealed trends similar to those observed in cells depleted of WT FtsZ, with the exception that the mean Z-ring intensity for mNG-L14 was similar to that of mNG-ΔCTL in the presence of WT FtsZ (Fig. S14C to F and S15).
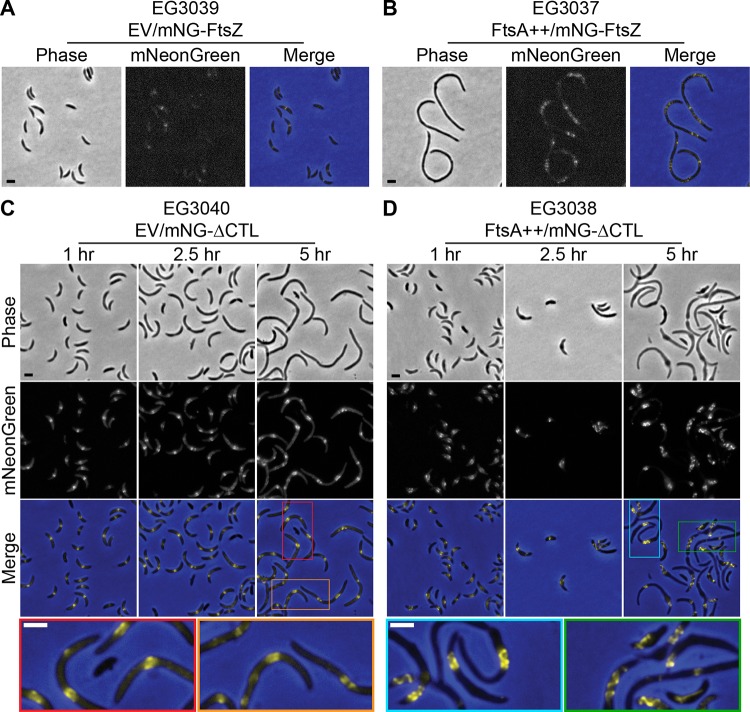
FtsA overproduction promotes formation of long, helical ΔCTL assemblies.
Since we observed a correlation between effects of the CTL on FtsZ assembly properties and in vivo phenotype (4, 10, 25) and since FtsA has a drastic effect on the ΔCTL phenotype (Fig. 2 and 3), we reasoned that perhaps FtsA alters the structure of Z rings in vivo. We investigated this hypothesis by comparing mNG fusions of FtsZ and ΔCTL in strains with native ftsA expression to those overexpressing ftsA from the PxylX promoter. In cells producing mNG-FtsZ, FtsA overproduction yielded both cell filamentation and a patchy, dispersed mNG-FtsZ signal compared to results in empty vector control cells (Fig. 6A and B), similar to what has been observed in previous studies (20) and suggesting that increased FtsA levels interfere with FtsZ localization. Intriguingly, FtsA overproduction in cells producing mNG-ΔCTL resulted in the formation of what appeared to be large-scale ΔCTL helices that stretch for multiple micrometers along the inner membrane of the cell in contrast to amorphous ring morphologies observed following production of mNG-ΔCTL with native levels of FtsA (Fig. 6C and D). We compared these findings to the localization of mNG-FtsZ and mNG-ΔCTL in cells overexpressing zapA, mipZ, and fzlC, which were shown to moderately reduce envelope bulging. We observed that Z-ring structure and FtsZ/ΔCTL localization remain largely unaffected, with the exception of a lack of diffuse mNG-ΔCTL signal in the presence of overabundant MipZ (Fig. S16).
FIG 6.
FtsA overproduction affects the structure of Z rings formed by mNG-FtsZ and mNG-ΔCTL. (A and B) Phase-contrast, epifluorescence, and merged images of cells induced with xylose to drive expression of mNG-FtsZ and empty vector (EV) or ftsA from the PxylX promoter for 6.5 h prior to imaging. (C and D) Phase-contrast, epifluorescence, and merged images of cells induced with xylose to drive expression of mNG-ΔCTL and empty vector (EV) or ftsA from the PxylX promoter for indicated times prior to imaging. Colored insets in merged images from 5-h time points are expanded below to increase visibility of Z-ring structures. Scale bars, 2 μm. Strains are as indicated.
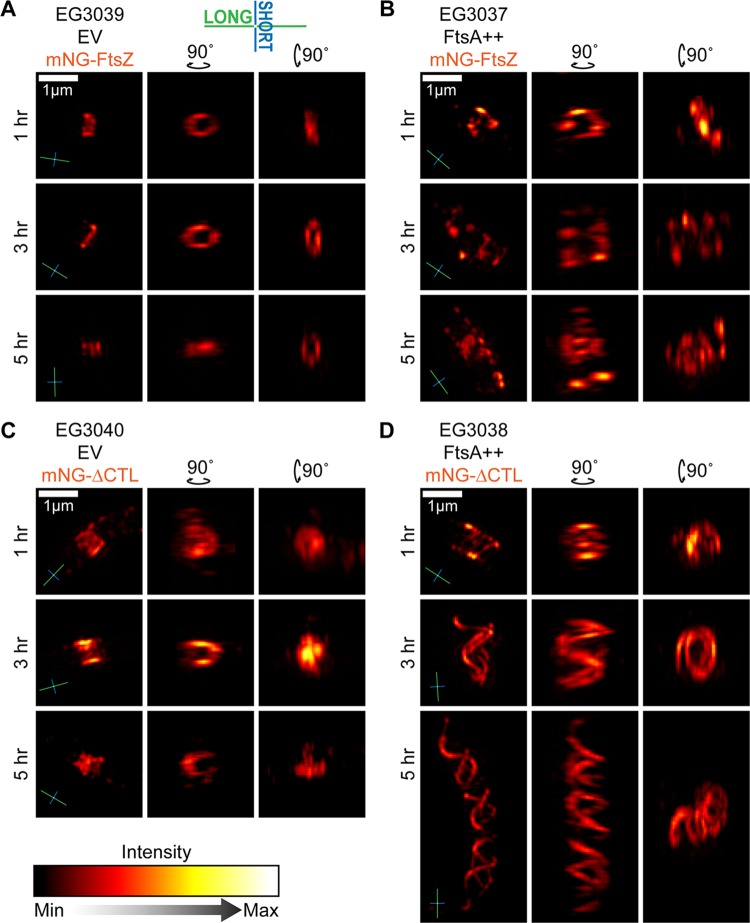
To gain more detailed insight into the mNG-ΔCTL structures formed in cells overproducing FtsA, we imaged the same strains at multiple time points following induction using three-dimensional structured-illumination microscopy (3D-SIM). mNG-FtsZ formed relatively narrow, discontinuous rings at midcell at all time points (Fig. 7A; also Fig. S17A and Movie S1 in the supplemental material) that exhibited a range of diameters corresponding to progression through the cell cycle and cytokinesis (Fig. S18A), observations that agree with WT Z-ring structures previously observed in vivo in C. crescentus (36, 44). ftsA overexpression resulted in more patchy and discontinuous Z rings that stretched farther along the length of the cell over time. At longer inductions of ftsA overexpression, we observed multiple, loosely ringlike structures along the length of the cell (Fig. 7B; Fig. S17B and S18B and Movie S2). Although these cells are filamentous, this FtsZ localization pattern differs from others observed upon filamentation caused by depletion of WT FtsZ (Fig. S11B, first column) and other perturbations (37, 45), suggesting that FtsA has a direct effect on FtsZ organization.
FIG 7.
FtsA overproduction interferes with Z-ring assembly and causes mNG-ΔCTL to form long helical structures in vivo. Shown are orthogonal views of 3D projections of image stacks acquired by 3D-SIM visualization of Z rings in cells induced with xylose to drive expression of empty vector and mNG-FtsZ (A), ftsA and mNG-FtsZ (B), empty vector and mNG-ΔCTL (C), or ftsA and mNG-ΔCTL (D) from the PxylX promoter for the indicated amounts of time. Compasses in the lower left of images indicate the orientation of long (green) and short (blue) axes of cells. Scale bar, 1 μm.
mNG-ΔCTL formed amorphous structures that were wider along the long axis of the cell than those of mNG-FtsZ (Fig. 7C; Fig. S17C and S18C and Movie S3), even after 1 h of induction, consistent with our FWHM calculations (Fig. 4B). Notably, as suggested from epifluorescence imaging, FtsA overproduction induced formation of striking, large-scale mNG-ΔCTL helices at 3 h postinduction and beyond (Fig. 7D; Fig. S17D and S18D and Movie S4). Helix shape was pleomorphic throughout the population. Helix length varied largely among the cells observed, and the pitch was variable not only from cell to cell but also within helices themselves, resulting in structures that were nearly ringlike at one end and highly extended at the other (Fig. 7D, 3-h time point; Fig. S17D). Virtually every cell in each population exhibited a categorically similar structure (e.g., all mNG-ΔCTL/FtsA cells contained helices and all mNG-FtsZ cells contained rings or foci), as demonstrated by three-dimensional z-projections made using a large field of view at the 5-h time point (Fig. S18B to D). Based on these results, we conclude that FtsA overproduction both exacerbates the deleterious effects of ΔCTL on cell morphology and profoundly alters the morphology of polymeric structures formed by both WT FtsZ and ΔCTL, highlighting a crucial role for FtsA in FtsZ-mediated regulation of PG metabolism during constriction.
DISCUSSION
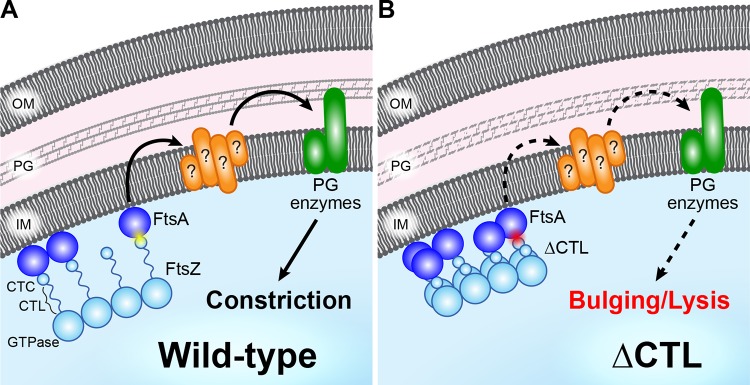
FtsZ serves both as a lynchpin for assembly of the division machinery and, at least in some bacteria, as a master regulator of cell wall metabolic activity at the division site through unclear mechanisms. The unique lethal effects of ΔCTL production in C. crescentus provides a powerful system to characterize the signaling pathway(s) between FtsZ and cell wall enzymes. Here, we refined the molecular determinants of the dominant lethal effects of ΔCTL (Fig. 8). We determined that ΔCTL-mediated bulging and lysis require (i) a polymerizing GTPase domain fused to (ii) the C. crescentus CTC with (iii) a CTL of less than 14 amino acids (Fig. 1). Importantly, our data indicate that the primary role of the CTC in CTL-dependent cell wall regulation lies in its ability to bind FtsA. Mutations to or overexpression of ftsA impacts the severity of the ΔCTL phenotype, and overproduction of FtsA profoundly alters ΔCTL morphology in cells. Collectively, our data indicate that signals from the Z ring are propagated in a CTL-dependent manner through FtsA to regulate cell wall metabolic activities for constriction (Fig. 8). Our data implicate the superstructure and dynamics of the Z ring in this proposed signaling pathway.
FIG 8.
FtsA mediates CTL-dependent regulation of PG enzyme activity during division. (A) In wild-type cells, FtsZ (light blue) directs activity of PG enzymes (green) through FtsA (dark blue) and other divisome components (orange) via a mechanism requiring interaction between the FtsZ C terminus (CTC) and FtsA (yellow flash), resulting in constriction. (B) FtsZ lacking its C-terminal linker (ΔCTL) exhibits a higher degree of filament lateral interaction and has a disrupted interaction with FtsA (red flash), resulting in aberrant regulation of PG enzymes (dashed arrows) that culminates in bulging and lysis.
Altered Z-ring architecture has been correlated with defects in cell wall metabolism and cell division in C. crescentus (4, 20). Our in-depth characterization of Z-ring morphologies and HADA incorporation for different CTL variants allows further correlation between in vitro polymerization properties, Z-ring assembly in cells, and cell wall metabolism. Notably, we observed that, compared to WT FtsZ, ΔCTL forms amorphous structures, whereas two other variants, L14 and HnCTL, form dispersed, faint rings (Fig. 5; see also Fig. S11 and S14 in the supplemental material). We previously observed that L14 and HnCTL form sparse, unbundled filaments in vitro while ΔCTL forms bundled, more stable filaments than WT FtsZ (4, 10). We therefore postulate that the dispersed, less dense structures formed by mNG-L14 and mNG-HnCTL result from their reduced polymerization propensity while the aberrant, nonring structures formed by ΔCTL are a consequence of its increased tendency to form bundles. The increased fraction of ΔCTL in the Z ring likely reflects the hyperstability of ΔCTL polymers, as observed in vitro (10, 25) (Fig. 5C; Fig. S14D). The absence of asymmetric, nonring structures for mNG-FtsZ or mNG-HnCTL, even in filamentous cells, suggests that the CTL-dependent effects we observed on Z-ring structure are not due to cell length or morphology but are specific to the assembly properties of each FtsZ variant. In addition, the finding that total mNG-ΔCTL and mNG-L14 protein levels were increased compared to the level of mNG-FtsZ suggests that ΔCTL and L14 variants exhibit increased protein stability (Fig. 5E; Fig. S13, S14F, and S15). However, increased protein concentration is not sufficient to explain the altered Z-ring morphology or function observed for different CTL variants in cells as mNG-ΔCTL and mNG-L14 are present at similar levels but form distinct structures. The increased fraction of mNG-ΔCTL in the Z ring, increased mean Z-ring intensity in the absence of WT FtsZ, and its capacity to form persistently aberrant structures in the presence of WT FtsZ distinguish it from the L14 variant, implicating these characteristics in the downstream misregulation of cell wall metabolism specific to ΔCTL.
FtsA appears to be a critical factor in signaling from ΔCTL to misregulation of PG metabolism. Two mutants of ftsA proposed to interfere with FtsA’s self-interaction delayed the toxic effects of ΔCTL, while two others enhanced or altered the phenotype in a deleterious manner (Fig. 2). While it is unclear if FtsA forms functional polymeric structures in vivo, these results suggest that FtsA self-interaction may be relevant for cell wall misregulation downstream of ΔCTL. Notably, FtsA and FtsZ polymers formed in vitro exhibit a subunit length mismatch (30–31), wherein the monomer-monomer distance in an FtsZ polymer is shorter than that in an FtsA polymer. The CTL of FtsZ could function as a flexible linker between FtsA and FtsZ filaments to accommodate this mismatch. Consistent with this idea, increased FtsA levels significantly alter the superstructure of ΔCTL rings in vivo (Fig. 6 and 7; Fig. S17 and S18), leading to formation of large-scale ΔCTL helices. These ΔCTL structures appear to be constrained to the membrane in a way that forces them into a helical conformation rather than a ring. FtsA has been shown to affect the GTPase rate and filament structure of FtsZ in vitro in a concentration-dependent manner (46–50), and mutants of ftsA predicted to have altered self-interaction characteristics have previously been implicated in affecting Z-ring structure in vivo (33–34). We propose that when the CTL is shorter than ∼14 amino acids, aberrant FtsZ-FtsA coassemblies form as a result of altered intrinsic FtsZ polymerization properties and an inability of FtsZ and FtsA polymers to interact appropriately, leading to misregulation of downstream pathways (Fig. 8). Further in vitro and in vivo efforts to characterize FtsA self-interaction and polymerization state, as well as their effects on Z-ring architecture and constriction, are required to resolve the contributions of FtsA to the CTL-dependent regulation of FtsZ function in cell wall metabolism and constriction, in general.
The present study highlights the role of the CTL in bacterial cell division and provides evidence for the necessity of both the CTC and FtsA in signaling for the regulation of cell wall metabolism by FtsZ. Our results emphasize the need to investigate the role(s) FtsA plays in mediating/modulating FtsZ’s signaling capabilities in vivo and open the door for further exploration of how FtsA affects Z-ring structure.
MATERIALS AND METHODS
Caulobacter crescentus and Escherichia coli growth media and conditions.
C. crescentus NA1000 cells were grown at 30°C in peptone-yeast extract (PYE) medium. Antibiotic concentrations used in liquid (solid) medium for C. crescentus were as follows: gentamicin, 1 (5) μg ml−1; kanamycin, 5 (25) μg ml−1; spectinomycin, 25 (100) μg ml−1. Streptomycin was used at 5 μg ml−1 in solid medium. For experiments with inducible expression of genes, inducer concentrations used were as follows: xylose, 0.3% (wt/vol); vanillate, 0.5 mM; glucose, 0.2% (wt/vol). E. coli Rosetta(DE3)/pLysS cells were grown at either 30°C or 37°C in Luria-Bertani (LB) medium. Antibiotic concentrations used in liquid (solid) media for E. coli were as follows: kanamycin, 30 (50) μg ml−1; ampicillin, 50 (100) μg ml−1.
Epifluorescence and phase-contrast microscopy and image analysis.
Cells were immobilized on 1% agarose pads and imaged using a Nikon Eclipse Ti inverted microscope through a Nikon Plan Fluor 100× (numeric aperture, 1.30) oil Ph3 objective with a Photometrics CoolSNAP HQ2 cooled charge-coupled-device (CCD) camera. Images were prepared for figure presentation in Adobe Photoshop by adjusting the fluorescence channel of each image to the same levels across samples in a given experiment (without saturating pixels or losing data) and merging on top of the corresponding phase-contrast image (Fig. 5A, in blue). Length and maximum width analyses were performed using meshes generated from phase-contrast images in the MicrobeJ plugin for Fiji (42). Prior to analyzing the images shown in Fig. 5A, the background was subtracted from raw fluorescence images by finding the average value of a rectangular region of interest where no cells were present and subtracting that value from the value for the whole image. Images were input into either Oufti (43) for demographs and FWHM or the MicrobeJ for Z-ring fraction and intensity. Cells were then outlined with meshes using phase-contrast images, and fluorescence signal was analyzed. FWHM calculations of midcell mNG signal from Oufti output were performed using a custom MATLAB script (51) that fit the normalized signal output from Oufti into an eighth-term Fourier series model and determined the width of the fluorescence curve at 50% of maximum intensity. A one-way analysis of variance (ANOVA) Kruskal-Wallis test with Dunn’s posttest was used to compare pairs of groups within each data set indicated in Fig. 5 and Fig. S14 in the supplemental material and in the associated figure legends and determine significance.
Three-dimensional structured-illumination microscopy (3D-SIM) and image processing.
Liquid cultures of log-phase C. crescentus cells induced with xylose (0.3%, wt/vol) and grown for indicated amounts of times before being diluted to an optical density at 600 nm (OD600) of 0.2 to 0.3 and immobilized onto 1% agarose pads. Cells were imaged with a Deltavision OMX-SR microscope (General Electric) using a 60×/1.42-numerical-aperture UPlanApo oil objective. Thirteen to 17 images were taken at 0.125-μm z-step intervals (three angles and five phases) with a 488-nm laser for each field of view. Image stacks were reconstructed using softWoRx (version 7.0.0) software with a Weiner filter of 0.001. Specific features from image stacks were duplicated in Fiji and used to generate interpolated 3D projections. Orthogonal views were selected for each image. The z-projections were generated from full wide-field images. A Red Hot look-up table (LUT) was applied to all images, and brightness and contrast were adjusted to enable optimal viewing.
Spot dilution assay.
Cells were grown without inducer until they reached log phase (absorption at 600 nm of 0.1 to 0.7). Then, cultures were diluted to an OD600 of 0.05 and serially diluted up to 10−6 before being spotted onto PYE plates containing glucose (0.2% [wt/vol]), xylose (0.3% [wt/vol]), and/or vanillate (0.5 mM) as indicated (along with antibiotics corresponding to the resistance of each strain). The plates were then incubated at 30°C until the appearance of colonies at the lowest dilution in the control strain in the glucose plates (48 h).
Growth rate measurement.
Cells were grown until they reached log phase. They were then diluted to an OD600 of 0.05, and inducer (or glucose control) was added at the beginning of growth measurements. OD600 values of three technical replicates for each culture were measured every 30 min for 24 h in 96-well plates using a Tecan Infinite 200 Pro plate reader with intermittent shaking and incubation at 30°C.
Immunoblotting.
Immunoblotting for FtsZ and ΔCTL were performed using standard procedures. For anti-FtsZ blots, CcFtsZ antiserum was used at a 1:20,000 dilution (4) to determine levels of WT FtsZ, ΔCTL, and other variants of FtsZ in lysates collected at the specified time points. Additionally, EcFtsZ antiserum (a gift from Harold Erickson) was used at 1:1,000 to recognize E. coli FtsZ variants or variants containing parts of E. coli FtsZ. SpmX antiserum was used as a control for loading concentration at a 1:50,000 dilution (52). For anti-mNeonGreen blots, anti-mNeonGreen antibody (ChromoTek) was used at a 1:1,000 dilution to determine levels of mNG-FtsZ variants indicated in Fig. S13 and S15 and in the associated figure legends at specified time points. Anti-Huβ antibody was used as a loading control at a 1:50,000 dilution (53). Anti-rabbit or anti-mouse secondary antibodies conjugated to horseradish peroxidase were used at 1:10,00 dilution (PerkinElmer). Immunoblots were developed using PerkinElmer Western Lightning Plus-ECL and imaged with a GE Healthcare Amersham Imager 600. Quantification of immunoblots was completed in Image Lab, version 6.0 (Bio Rad), by manually finding bands, detecting the total volume within the region, and subtracting the background volume.
HADA labeling.
To image cell wall metabolism patterning, cells were incubated with 0.82 mM HADA for 5 min with shaking at 30°C. Following incubation, cells were removed, washed twice in phosphate-buffered saline (PBS), and resuspended in PBS before imaging. Alternately, when cells were not imaged immediately, they were fixed by resuspension in 100% ethanol and incubated on ice (at 4°C) until imaging and were pelleted and resuspended in PBS before imaging.
Protein purification.
FtsZ and ΔCTL from C. crescentus and E. coli, as well as EcGTPase-CcCTC, were purified using the protocol described for CcFtsZ in Sundararajan and Goley (10). Briefly, FtsZ (or variant) expression was induced from pET21a or pET43.1a vectors in E. coli Rosetta(DE3)/pLysS using 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 37°C for 3 h after the uninduced cultures reached an OD600 of 1.0. Cells were then pelleted and resuspended in lysis buffer (50 mM Tris-HCl [pH 8.0], 50 mM KCl, 1 mM EDTA, 10% glycerol, DNase I, 1 mM β-mercaptoethanol, 2 mM phenylmethylsulfonyl fluoride [PMSF], 1 cOmplete mini, EDTA-free protease inhibitor tablet [Roche]). Resuspended cell pellets were lysed by incubation with 1 mg ml−1 lysozyme for 1 h, followed by sonication. FtsZ variants were then purified from the lysate using an anion exchange chromatography column (HiTrap Q HP, 5 ml; GE Life Sciences), followed by ammonium sulfate precipitation (at 20 to 30% ammonium sulfate saturation). The ammonium sulfate pellet was resuspended in FtsZ storage buffer (50 mM HEPES-KOH [pH 7.2], 50 mM KCl, 0.1 mM EDTA, 1 mM β-mercaptoethanol, 10% glycerol) and was subjected to size exclusion chromatography (Superdex 200 10/300 GL column; GE Life Sciences) to further purify the protein and snap-frozen in liquid nitrogen and stored long-term in FtsZ storage buffer at –80°C.
His6-FzlA was purified using the protocol described for His6-FzlA in Sundararajan et al. (4). A pET28c vector bearing fzlA (pEG327) was transformed into Rosetta(DE3)/pLysS E. coli cells, which were grown at 30°C to an OD600 of 0.5, at which point His6-fzlA expression was induced with 0.5 mM IPTG for 4 h. Cells were then pelleted, resuspended in FzlA lysis buffer (50 mM Tris-HCl [pH 8.0], 300 mM NaCl, 20 mM imidazole, 10% glycerol), snap-frozen in liquid nitrogen, and stored at −80°C until purification. Thawed cells were lysed with 1 mg ml−1 lysozyme with one cOmplete Mini protease inhibitor tablet per 30 ml (Roche), 2.5 mM MgCl2, and 2 units ml−1 DNase I (New England Biolabs). Cells were incubated for 45 min at room temperature before sonication, and lysates were spun for 30 min at 15,000 × g at 4°C. Supernatant was filtered and run through two tandem HisTrap FF columns (1 ml each) (GE Life Sciences). The columns were equilibrated and washed in FzlA lysis buffer, and His6-FzlA was eluted with FzlA elution buffer (FzlA lysis buffer with 300 mM imidazole). Peak fractions were combined, His6-FzlA was dialyzed into FzlA storage buffer (50 mM Tris-HCl [pH 8.0], 300 mM NaCl, 5% glycerol), and aliquots were snap-frozen in liquid nitrogen and stored long-term at −80°C.
His6-MipZ was purified using the protocol described for His6-MipZ by Sundararajan et al. (4). A pET21a vector bearing mipZ-His6 was transformed into Rosetta(DE3)/pLysS E. coli cells, which were grown at 37°C to an OD600 of 0.6, at which point mipZ-His6 expression was induced with 0.5 mM IPTG for 3.25 h. Cells were then pelleted, washed with PBS, snap-frozen in liquid nitrogen, and stored at −80°C until purification. Upon thawing, cells were resuspended in Ni lysis buffer (50 mM Tris-HCl [pH 8.0], 300 mM NaCl, 10 mM imidazole, 1 mM EDTA, 10% glycerol) with one cOmplete Mini Protease inhibitor tablet per 30 ml (Roche), 2 mM PMSF, and 2 units ml−1 DNase I (New England Biolabs). Cells were lysed by two passages through a French press at 15,000 lb/in2, and lysates were spun for 30 min at 15,000 × g at 4°C. Lysates were run through a Ni-nitrilotriacetic acid (NTA)–agarose column (Qiagen) equilibrated with Ni lysis buffer. The column was washed with Ni wash buffer (Ni lysis buffer with 20 mM imidazole), and His6-MipZ was eluted using Ni elution buffer (Ni lysis buffer with 300 mM imidazole). Peak fractions were concentrated and run through a Superdex 200 10/300 GL column (GE Life Sciences) equilibrated in MipZ storage buffer (50 mM HEPES-NaOH [pH 7.2], 50 mM NaCl, 0.1 mM EDTA, 1 mM β-mercaptoethanol, 10% glycerol). Peak fractions were combined, concentrated, snap-frozen in liquid nitrogen, and stored long-term at −80°C.
Copelleting assay.
Each protein (5 μM) in the indicated combinations was incubated in HEK50 buffer (50 mM HEPES-KOH [pH 7.2], 0.1 mM EDTA, 50 mM KCl) with 2 mM GTP, 0.05% Triton X-100, and 10 mM MgCl2 (and 2 mM ATP for MipZ) at 25˚C for 15 min and spun at 25˚C for 15 min at 250,000 × g. Supernatants and resuspended pellets were resolved by SDS-PAGE and visualized with Coomassie stain to determine relative levels of FzlA or MipZ in the supernatant and pellet for each FtsZ variant.
Polymerization kinetic assay.
A polymerization kinetic assay was performed, similar to those described by Sundararajan and Goley (10). Briefly, 4 μM each FtsZ variant was added to HEK300 buffer (50 mM HEPES-KOH [pH 7.2], 0.1 mM EDTA, 300 mM KCl) with 10 mM MgCl2. Following addition of a limiting concentration of GTP (0.5 mM), polymerization was measured using a Fluoromax-3 spectrofluorometer (Jobin Yvon, Inc.) to measure right-angle light scatter (excitation and emission at 350 nm, 2-nm slits). Measurements were taken every 10 s.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Goley lab for helpful discussions throughout this work and feedback on the manuscript. We are grateful to Patrick Viollier, Martin Thanbichler, Lucy Shapiro, and Harold Erickson for antibodies and strains and to Caren Freel Meyers and Amer Al-khouja for synthesis of HADA. We also thank Jason Lyu and the Microscopy Facility of Johns Hopkins School of Medicine for assistance with the 3D-SIM imaging.
This work was funded in part by the National Institutes of Health, National Institute of General Medical Sciences, through R01GM108640 (E.D.G.) and T32GM007445 (training grant support of J.M.B.).
J.M.B., K.S., and E.D.G. conceived the study, designed and carried out experiments, analyzed data, and contributed to writing and editing the manuscript. A.B. wrote the custom MATLAB script for analyzing fluorescence FWHM data.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Erickson HP, Anderson DE, Osawa M. 2010. FtsZ in bacterial cytokinesis: cytoskeleton and force generator all in one. Microbiol Mol Biol Rev 74:504–528. doi: 10.1128/MMBR.00021-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goley ED, Dye NA, Werner JN, Gitai Z, Shapiro L. 2010. Imaging-based identification of a critical regulator of ftsz protofilament curvature in Caulobacter. Mol Cell 39:975–987. doi: 10.1016/j.molcel.2010.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goley ED, Yeh Y-C, Hong S-H, Fero MJ, Abeliuk E, McAdams HH, Shapiro L. 2011. Assembly of the Caulobacter cell division machine. Mol Microbiol 80:1680–1698. doi: 10.1111/j.1365-2958.2011.07677.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sundararajan K, Miguel A, Desmarais SM, Meier EL, Huang KC, Goley ED. 2015. The bacterial tubulin FtsZ requires its intrinsically disordered linker to direct robust cell wall construction. Nat Commun 6:7281. doi: 10.1038/ncomms8281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang X, Lyu Z, Miguel A, McQuillen R, Huang KC, Xiao J. 2017. GTPase activity-coupled treadmilling of the bacterial tubulin FtsZ organizes septal cell wall synthesis. Science 355:744–747. doi: 10.1126/science.aak9995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bisson-Filho AW, Hsu Y-P, Squyres GR, Kuru E, Wu F, Jukes C, Sun Y, Dekker C, Holden S, VanNieuwenhze MS, Brun YV, Garner EC. 2017. Treadmilling by FtsZ filaments drives peptidoglycan synthesis and bacterial cell division. Science 355:739–743. doi: 10.1126/science.aak9973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vaughan S, Wickstead B, Gull K, Addinall SG. 2004. Molecular evolution of FtsZ protein sequences encoded within the genomes of archaea, bacteria, and eukaryota. J Mol Evol 58:19–29. doi: 10.1007/s00239-003-2523-5. [DOI] [PubMed] [Google Scholar]
- 8.Nogales E, Wolf SG, Downing KH. 1998. Structure of the αβ tubulin dimer by electron crystallography. Nature 391:199–203. doi: 10.1038/34465. [DOI] [PubMed] [Google Scholar]
- 9.Löwe J, Amos LA. 1998. Crystal structure of the bacterial cell-division protein FtsZ. Nature 391:203–206. doi: 10.1038/34472. [DOI] [PubMed] [Google Scholar]
- 10.Sundararajan K, Goley ED. 2017. The intrinsically disordered C-terminal linker of FtsZ regulates protofilament dynamics and superstructure in vitro. J Biol Chem 292:20509–20527. doi: 10.1074/jbc.M117.809939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stricker J, Maddox P, Salmon ED, Erickson HP. 2002. Rapid assembly dynamics of the Escherichia coli FtsZ-ring demonstrated by fluorescence recovery after photobleaching. Proc Natl Acad Sci U S A 99:3171–3175. doi: 10.1073/pnas.052595099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lyu Z, Coltharp C, Yang X, Xiao J. 2016. Influence of FtsZ GTPase activity and concentration on nanoscale Z-ring structure in vivo revealed by three-dimensional superresolution imaging. Biopolymers 105:725–734. doi: 10.1002/bip.22895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Du S, Lutkenhaus J. 2019. At the heart of bacterial cytokinesis: the Z ring. Trends Microbiol 27:781–791. doi: 10.1016/j.tim.2019.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pichoff S, Lutkenhaus J. 2005. Tethering the Z ring to the membrane through a conserved membrane targeting sequence in FtsA. Mol Microbiol 55:1722–1734. doi: 10.1111/j.1365-2958.2005.04522.x. [DOI] [PubMed] [Google Scholar]
- 15.Wang X, Huang J, Mukherjee A, Cao C, Lutkenhaus J. 1997. Analysis of the Interaction of FtsZ with Itself, GTP, and FtsA. J Bacteriol 179:5551–5559. doi: 10.1128/jb.179.17.5551-5559.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Din N, Quardokus EM, Sackett MJ, Brun YV. 1998. Dominant C-terminal deletions of FtsZ that affect its ability to localize in Caulobacter and its interaction with FtsA. Mol Microbiol 27:1051–1063. doi: 10.1046/j.1365-2958.1998.00752.x. [DOI] [PubMed] [Google Scholar]
- 17.Ma X, Margolin W. 1999. Genetic and functional analyses of the conserved C-terminal core domain of Escherichia coli FtsZ. J Bacteriol 181:7531–7544. doi: 10.1128/JB.181.24.7531-7544.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan K, Pearce KH, Payne DJ. 2000. A conserved residue at the extreme C-terminus of FtsZ is critical for the FtsA-FtsZ interaction in Staphylococcus aureus. Biochem Biophys Res Commun 270:387–392. doi: 10.1006/bbrc.2000.2439. [DOI] [PubMed] [Google Scholar]
- 19.Haney SA, Glasfeld E, Hale C, Keeney D, He Z, de Boer P. 2001. Genetic analysis of the Escherichia coli FtsZ.ZipA interaction in the yeast two-hybrid system. Characterization of FtsZ residues essential for the interactions with ZipA and with FtsA. J Biol Chem 276:11980–11987. doi: 10.1074/jbc.M009810200. [DOI] [PubMed] [Google Scholar]
- 20.Meier EL, Razavi S, Inoue T, Goley ED. 2016. A novel membrane anchor for FtsZ is linked to cell wall hydrolysis in Caulobacter crescentus. Mol Microbiol 101:265–280. doi: 10.1111/mmi.13388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buske PJ, Levin PA. 2013. A flexible C-terminal linker is required for proper FtsZ assembly in vitro and cytokinetic ring formation in vivo. Mol Microbiol 89:249–263. doi: 10.1111/mmi.12272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Howell M, Aliashkevich A, Sundararajan K, Daniel JJ, Lariviere PJ, Goley ED, Cava F, Brown PJB. 2018. Agrobacterium tumefaciens divisome proteins regulate the transition from polar growth to cell division. bioRxiv 10.1101/412759. [DOI] [PMC free article] [PubMed]
- 23.Huecas S, Ramírez-Aportela E, Vergoñós A, Núñez-Ramírez R, Llorca O, Díaz JF, Juan-Rodríguez D, Oliva MA, Castellen P, Andreu JM. 2017. Self-organization of FtsZ polymers in solution reveals spacer role of the disordered C-terminal tail. Biophys J 113:1831–1844. doi: 10.1016/j.bpj.2017.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gardner K, Moore DA, Erickson HP. 2013. The C-terminal linker of Escherichia coli FtsZ functions as an intrinsically disordered peptide. Mol Microbiol 89:264–275. doi: 10.1111/mmi.12279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sundararajan K, Vecchiarelli A, Mizuuchi K, Goley ED. 2018. Species- and C-terminal linker-dependent variations in the dynamic behavior of FtsZ on membranes in vitro. Mol Microbiol 110:47–63. doi: 10.1111/mmi.14081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuru E, Tekkam S, Hall E, Brun YV, Van Nieuwenhze MS. 2015. Synthesis of fluorescent d-amino acids and their use for probing peptidoglycan synthesis and bacterial growth in situ. Nat Protoc 10:33–52. doi: 10.1038/nprot.2014.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aaron M, Charbon G, Lam H, Schwarz H, Vollmer W, Jacobs-Wagner C. 2007. The tubulin homologue FtsZ contributes to cell elongation by guiding cell wall precursor synthesis in Caulobacter crescentus. Mol Microbiol 64:938–952. doi: 10.1111/j.1365-2958.2007.05720.x. [DOI] [PubMed] [Google Scholar]
- 28.Shaner NC, Lambert GG, Chammas A, Ni Y, Cranfill PJ, Baird MA, Sell BR, Allen JR, Day RN, Israelsson M, Davidson MW, Wang J. 2013. A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nat Methods 10:407–409. doi: 10.1038/nmeth.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohta N, Ninfa AJ, Allaire A, Kulick L, Newton A. 1997. Identification, characterization, and chromosomal organization of cell division cycle genes in Caulobacter crescentus. J Bacteriol 179:2169–2180. doi: 10.1128/jb.179.7.2169-2180.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Szwedziak P, Wang Q, Freund SMV, Löwe J. 2012. FtsA forms actin-like protofilaments. EMBO J 31:2249–2260. doi: 10.1038/emboj.2012.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Szwedziak P, Wang Q, Bharat TAM, Tsim M, Löwe J. 2014. Architecture of the ring formed by the tubulin homologue FtsZ in bacterial cell division. eLife 3:e04601. doi: 10.7554/eLife.04601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pichoff S, Shen B, Sullivan B, Lutkenhaus J. 2012. FtsA mutants impaired for self-interaction bypass ZipA suggesting a model in which FtsA's self-interaction competes with its ability to recruit downstream division proteins. Mol Microbiol 83:151–167. doi: 10.1111/j.1365-2958.2011.07923.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Geissler B, Shiomi D, Margolin W. 2007. The ftsA* gain-of-function allele of Escherichia coli and its effects on the stability and dynamics of the Z ring. Microbiology 153:814–825. doi: 10.1099/mic.0.2006/001834-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shiomi D, Margolin W. 2007. Dimerization or oligomerization of the actin-like FtsA protein enhances the integrity of the cytokinetic Z ring. Mol Microbiol 66:1396–1415. doi: 10.1111/j.1365-2958.2007.05998.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schoenemann KM, Krupka M, Rowlett VW, Distelhorst SL, Hu B, Margolin W. 2018. Gain-of-function variants of FtsA form diverse oligomeric structures on lipids and enhance FtsZ protofilament bundling. Mol Microbiol 109:676–693. doi: 10.1111/mmi.14069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Woldemeskel SA, McQuillen R, Hessel AM, Xiao J, Goley ED. 2017. A conserved coiled-coil protein pair focuses the cytokinetic Z-ring in Caulobacter crescentus. Mol Microbiol 105:721–740. doi: 10.1111/mmi.13731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meier EL, Daitch AK, Yao Q, Bhargava A, Jensen GJ, Goley ED. 2017. FtsEX-mediated regulation of the final stages of cell division reveals morphogenetic plasticity in Caulobacter crescentus. PLoS Genet 13:e1006999. doi: 10.1371/journal.pgen.1006999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang HC, Gayda RC. 1990. High-level expression of the FtsA protein inhibits cell septation in Escherichia coli K-12. J Bacteriol 172:4736–4740. doi: 10.1128/jb.172.8.4736-4740.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dai K, Lutkenhaus J. 1992. The proper ratio of FtsZ to FtsA is required for cell division to occur in Escherichia coli. J Bacteriol 174:6145–6151. doi: 10.1128/jb.174.19.6145-6151.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dewar SJ, Begg KJ, Donachie WD. 1992. Inhibition of cell division initiation by an imbalance in the ratio of FtsA to FtsZ. J Bacteriol 174:6314–6316. doi: 10.1128/jb.174.19.6314-6316.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martin ME, Trimble MJ, Brun YV. 2004. Cell cycle-dependent abundance, stability and localization of FtsA and FtsQ in Caulobacter crescentus. Mol Microbiol 54:60–74. doi: 10.1111/j.1365-2958.2004.04251.x. [DOI] [PubMed] [Google Scholar]
- 42.Ducret A, Quardokus EM, Brun YV. 2016. MicrobeJ, a tool for high throughput bacterial cell detection and quantitative analysis. Nat Microbiol 1:16077. doi: 10.1038/nmicrobiol.2016.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paintdakhi A, Parry B, Campos M, Irnov I, Elf J, Surovtsev I, Jacobs-Wagner C. 2016. Oufti: an integrated software package for high-accuracy, high-throughput quantitative microscopy analysis. Mol Microbiol 99:767–777. doi: 10.1111/mmi.13264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Holden SJ, Pengo T, Meibom KL, Fernandez C, Collier J, Manley S. 2014. High throughput 3D super-resolution microscopy reveals Caulobacter crescentus in vivo Z-ring organization. Proc Natl Acad Sci U S A 111:4566–4571. doi: 10.1073/pnas.1313368111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lariviere PJ, Szwedziak P, Mahone CR, Löwe J, Goley ED. 2018. FzlA, an essential regulator of FtsZ filament curvature, controls constriction rate during Caulobacter division. Mol Microbiol 107:180–197. doi: 10.1111/mmi.13876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Modi K, Misra HS. 2014. Dr-FtsA, an actin homologue in Deinococcus radiodurans differentially affects Dr-FtsZ and Ec-FtsZ functions in vitro. PLoS One 9:e115918. doi: 10.1371/journal.pone.0115918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fujita J, Maeda Y, Nagao C, Tsuchiya Y, Miyazaki Y, Hirose M, Mizohata E, Matsumoto Y, Inoue T, Mizuguchi K, Matsumura H. 2014. Crystal structure of FtsA from Staphylococcus aureus. FEBS Lett 588:1879–1885. doi: 10.1016/j.febslet.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 48.Conti J, Viola MG, Camberg JL. 2018. FtsA reshapes membrane architecture and remodels the Z-ring in Escherichia coli. Mol Microbiol 107:558–576. doi: 10.1111/mmi.13902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Loose M, Mitchison TJ. 2014. The bacterial cell division proteins FtsA and FtsZ self-organize into dynamic cytoskeletal patterns. Nat Cell Biol 16:38–46. doi: 10.1038/ncb2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen Y, Huang H, Osawa M, Erickson HP. 2017. ZipA and FtsA* stabilize FtsZ-GDP miniring structures. Sci Rep 7:3650. doi: 10.1038/s41598-017-03983-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Woldemeskel SA, Alvarez L, Daitch AK, Zeinert R, Bhargava A, Gonzalez D, Collier J, Chien P, Cava F, Goley ED. 2019. The conserved transcriptional regulator CdnL is required for metabolic homeostasis and morphogenesis in Caulobacter. bioRxiv 10.1101/557637. [DOI] [PMC free article] [PubMed]
- 52.Radhakrishnan SK, Thanbichler M, Viollier PH. 2008. The dynamic interplay between a cell fate determinant and a lysozyme homolog drives the asymmetric division cycle of Caulobacter crescentus. Genes Dev 22:212–225. doi: 10.1101/gad.1601808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bowman GR, Comolli LR, Gaietta GM, Fero M, Hong S-H, Jones Y, Lee JH, Downing KH, Ellisman MH, McAdams HH, Shapiro L. 2010. Caulobacter PopZ forms a polar subdomain dictating sequential changes in pole composition and function. Mol Microbiol 76:173–189. doi: 10.1111/j.1365-2958.2010.07088.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.