Graphical abstract
Keywords: Breast cancer, Somatic mutations, Target sequencing, Ion torrent sequencing, Next Generation Sequencing
Highlights
-
•
Identifying somatic mutations associated with Egyptian breast cancer tumors.
-
•
Identifying breast cancer mutation driver genes in the studied Egyptian patients.
-
•
Identify genetic mutations in BC tumors help developing personalized treatment protocols or combination therapies.
-
•
Identifying novel variants that may be associated with Egyptian breast cancer patients.
-
•
Help in customization of Egyptian related breast cancer panels as a routine work
Abstract
Breast cancer (BC) incidence is progressively increasing in Egypt. However, there is insufficient knowledge of the acquired somatic mutations in Egyptian BC patients which limit our understanding of its progression. To the best of our knowledge, this is the first Egyptian cohort to sequence a multiple-gene panel of cancer related genes on BC patients. Four hundred and nine cancer related genes were sequenced in 46 fresh breast tumors of Egyptian BC patients to identify somatic mutations and their frequencies.
TP53 and PIK3CA were the most top two frequently mutated genes. We detected 15 different somatic mutations in TP53 and 8 different ones in PIK3CA, each in 27 samples (58.7%). According to Clinvar database; we found 19 pathogenic somatic mutations: 7 in Tp53, 5 in PIK3CA, and single variants of VHL, STK11, AKT1, KRAS, IDH2, PTEN and ERBB2. We also identified 5 variants with uncertain significance (4 in TP53 and 1 in CEBPA) and 4 variants with conflicting interpretations of pathogenicity (2 in TP53 and 1 in each of APC and JAK3). Moreover, one drug response variant (p.P72R) in TP53 was detected in 8 samples. Furthermore, four novel variants were identified in JAK2, MTOR, KIT and EPHB. Further analysis, by Ingenuity Variant Analysis software (IVA), showed that PI3K/AKT signaling is altered in greater than 50% of Egyptian BC patients which implicates PI3K/AKT signaling as a therapeutic target. In this cohort, we shed the light on the most frequently detected somatic mutations and the most altered pathway in Egyptian BC patients.
Introduction
Breast Cancer (BC) is the second most common lethal malignancy in women and the leading cause of cancer-related death in women worldwide [1]. It has been reported that over half (52%) of new BC cases and 62% of deaths occur in economically developing countries [2]. In Egypt, BC is the most common cancer among females accounting for 37.7% of about 12,000–13,000 new cases per year. These estimates have been confirmed in many regional Egyptian cancer registries [3], [4].
Recently, the Next Generation Sequencing (NGS) technology has provided a fast and cost effective means to characterize the mutations in the individual patient genome, which shed light on the mutated cancer genes involved in cancer progression [5]. NGS has been used to study the mutation pattern in BC patients from different ethnic groups and at different disease stages. In many studies, whole genome and whole exome sequencing have been used to study BC mutations on large number of patients [6], [7].
Further focused analysis using targeted sequencing was later conducted in many studies: Pereira et al. studied the somatic mutation profile conducted on 2433 patients using a custom gene panel of 173 genes and identified 40 mutation-driver genes [8]. Meric-Bernstam et al. used panel of 182 genes and determined the spectrum of genomic alterations in primary and metastatic BC [9]. Also, Wiesman et al. used a custom gene pane of 229 genes and identified the key somatic mutations previously reported in triple negative BC [10]. Moreover, Smith et al. could detect clinically actionable mutations in BC solid tumors using the MammaSeq [11]. On the other hand, Liu et al. and Bai et al. used Ion Torrent Ampliseq Cancer Panel to identify genetic mutations in BC tumors to help developing personalized treatment protocols or combination therapies [12], [13]. The previous studies covered mostly European and North American populations. Little is done to study the somatic profile for other ethnic groups; we could only locate the work on the Chinese [14], [15], Mexico and Vietnam [16] populations. To great extent, these studies were successful in understanding the molecular basis of the disease. Therefore, it was necessary to conduct such targeted sequencing studies on the Egyptian population to answer an important question: how the Egyptian patients are different in terms of mutations and affected genes from other populations? Answering this question helps understanding the Egyptian BC profiling which will help in the future evaluating drug efficacy and treatment protocols for that population. So, we developed this cohort study to explore the landscape of somatic mutations in Egyptian BC patients. We used the Ion Torrent sequencing technology (Ion AmpliSeq Comprehensive Cancer Panel) to sequence 409 tumor suppressor genes and oncogenes from 46 BC tumors of various subtypes.
Patients and methods
Ethics statement
All human subject protocols and procedures were approved by the Institutional Review Board (IRB number: IRB00004025) of National Cancer Institute (NCI), Cairo University, Egypt which conducted the study in accordance with ICH-GCP guidelines (approval number: MD2010014038.3). A written informed consent was obtained from each patient during the enrollment into this study.
Patient samples
Tissue samples used in this cohort were recruited from the Egyptian National Cancer Institute from October 2016 to March 2018. Forty-six fresh tissue samples from Egyptian BC female patients were collected at surgery. Included patients were naïve to treatment and those receiving neoadjuvant chemotherapy were excluded. Patients were classified according to age, histological type, histological grade, hormone receptor status (estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (Her2)) and molecular subtype (Luminal A, Luminal B, Her2- over-expressing and triple negative). All the clinico-pathological features of the studied patients were collected from the clinical records.
DNA preparation
Twenty-five mg of fresh tissues were collected from 46 BC female patients. DNA was isolated using QIAamp® DNA Mini Kit (Qiagen, Germany: Cat. No. 51304) following manufacturer’s instructions. For each sample, the isolated DNA was quantified using Nanodrop 2000 Spectrophotometer (Thermo Fisher Scientific) and Qubit 4 Fluorometer (Thermo Fisher Scientific). Moreover, DNA was checked by electrophoresis using 2% Ethidium-Bromide-stained agarose gel and visualized under UV trans-illuminator to confirm its integrity.
Ion AmpliSeq™ DNA library preparation, template preparation and sequencing
Ion AmpliSeq™ DNA Library was constructed using the Ion AmpliSeq™ Library Kit 2.0 (Cat. No. 4480441) which is designed for preparation of amplicon libraries using Ion AmpliSeq Comprehensive Cancer panels (Ion AmpliSeq CCP, Life Technologies, Cat. no. 4477685). For this, four amplicon pools per sample covering the 409 genes were quantified by qPCR with the Ion Library Quantitation Kit (Life Technologies, Cat. no. 4468802). The concentration and size of the library were determined by Agilent 2100 BioAnalyzer and DNA High-Sensitivity Lab Chip (Agilent Technologies). The quality of the libraries was assessed by QIAxcel advanced (QIAGEN). Then, the quantified libraries were preceded to template preparation on the ion chef using the Ion PI Hi-Q Chef Kit (Life Technologies, Cat. No. A27198) and loaded into an Ion PI Chip (Life Technologies, Cat. No. A26770) to be sequenced on the Ion proton using the Ion Proton Sequencing 200 Kit v2 (Life Technologies, Cat. No. 4485149).
Variant calling and variant classification
The bioinformatics analysis pipeline started with checking the QC step where the reads of each NGS run were examined and low quality parts were trimmed out. We then ran the alignment of the reads to the human reference genome (version hg19). For that step, we used the Torrent Suite as recommended by the manufacturer. A run was accepted only if the total number of aligned reads covered at least 95% of the target regions with an average depth of coverage of 668X. After alignment, we ran the TSVC module of the Torrent Suite to call the variants, using the options for calling somatic variants with hotspots. The qualified variants were annotated using an in-house developed system composed of different databases: we used the ANNOVAR [17] package to annotate the variants with all available public population information. For cancer Few variants databases, we used the Catalogue of Somatic Mutations in Cancer COSMIC [18] and CIViC [19]. The possible impact of amino acid changes was assessed with PolyPhen-2 [20], Sift [21], and CADD [22] prediction tools to understand their potential role in carcinogenesis. The identified variants were classified as benign or pathogenic according to Clin Var database [23]. For identifying somatic mutations and filtering out germline variants in the absence of normal tissues, we used the in silico methods of [24], [25] using our database sets. The filtration algorithm has the following steps: the variants with accepted quality, depth of coverage, and existence in our hotspot regions are retained for further analysis. Variants that exist in COSMIC database or those that exist in population databases (including our in-house one) with minimum allele frequency (MAF) less than 1% were retained. Variants that do not exist in cancer CiViC or COSMIC were filtered out. Finally, the remaining variants were inspected manually on IGV to revise their alignments and neighboring sequences.
Results
In this cohort study, we sequenced 46 BC samples from Egyptian patients ranging from 29 to 73 years of age. Patients classification was based on their age, histological type, histological grade, receptor status (ER, PR, and Her-2 Neu), and molecular classification as shown in Table 1. In this study, we used Ion AmpliSeq™ Comprehensive Cancer Panel which was designed to target 409 tumor suppressor genes and oncogenes across multiple gene families to identify somatic mutations among Egyptian BC patients and their frequencies.
Table 1.
Clinical features of the studied 46 Egyptian patients.
| Characteristics | Number | ||
|---|---|---|---|
| Age (Years) | Mean: 54.75 Median: 55 Range: 29–77 |
<40 y | 5 |
| 40–50 y | 8 | ||
| 50–60 y | 18 | ||
| ≥60 y | 15 | ||
| HR status | ER+ | 33 | |
| ER- | 13 | ||
| PR+ | 27 | ||
| PR- | 19 | ||
| Grade | I | 3 | |
| II | 33 | ||
| III | 10 | ||
| Histological type | Invasive duct carcinoma | 41 | |
| Invasive tubular carcinoma | 1 | ||
| Invasive micropapillary carcinoma | 1 | ||
| Invasive tubulolobular carcinoma | 2 | ||
| Invasive duct & Invasive lobular carcinoma (mixed) | 1 | ||
| Molecular classification | Luminal A | 25 | |
| Luminal B | 9 | ||
| Her2- enriched | 8 | ||
| Basal like (triple negative) | 4 | ||
Our analysis showed that there were 44 out of 46 patients had somatic mutations. Initial filtering yielded 79 variants. By looking up these variants in the most recent version of the COSMIC database (version 90), we found that 28 of them have been reclassified as SNP. Therefore, they were excluded from further discussion. This reclassification was due to their frequencies in the ExAC and GnomAD databases. From the remaining 51 variants, there were 10 benign ones according to Clinvar database with frequency higher than 1% in ExAC and GnomAD databases except for one variant (p.E168D) in MET gene. The final remaining set of somatic variants includes 38 variants; out of them there were 4 novel variants, as they did not show up in any of the public databases. Three of these novel variants (p.F151V, p.H263Q & p.T600I) were predicted to be damaging by CADD-phred and PolyPhen2 prediction tools.
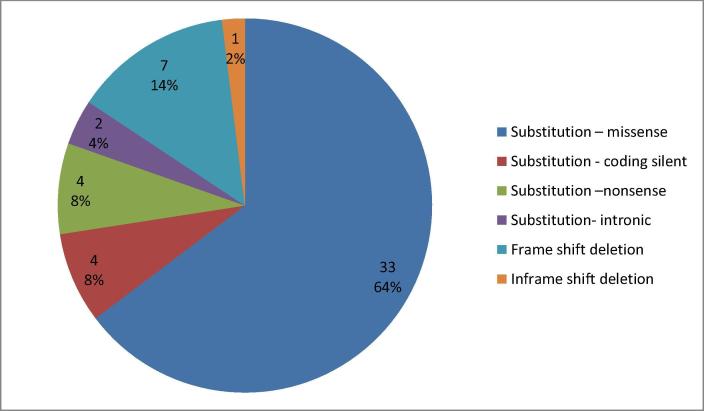
We detected different somatic mutations (Substitution – missense, Frame shift deletion, Substitution – coding silent, Substitution – nonsense, In Frame shift insertion, and Substitution- intronic). Summary of types and numbers of the detected somatic mutations is shown in Fig. 1a. Fifty one somatic mutations were detected in 22 genes, out of them there were: 19 pathogenic or likely pathogenic variants, 10 benign or likely benign variants, 5 variants of uncertain significance, 4 variants with conflicting interpretation of pathogenicity, 8 variants not reported in Clinvar database, 4 novel variants, an 1 drug response variant as shown in Fig. 1b. The distribution of somatic mutations in the studied BC patients was shown in the Oncoplot (Fig. 2). We also analyzed variants with Ingenuity Variant Analysis software (IVA; QIAGEN) for further variant annotation and interpretation. IVA showed that PI3K/AKT signaling was up- regulated in 54% of our patients as shown in Fig. 3a and 3b.
Fig. 1a.
Summary of types and numbers of the detected somatic mutations.
Fig. 1b.
Classification of the identified variants.
Fig. 2.
Oncoplot showing the distribution of somatic mutations in the studied breast cancer patients. The Oncoplot provided an overview of somatic mutations in particular genes (rows) affecting individual samples (columns). According to the logic of oncoplot, if a sample has more variants, it is counted once, and not with the total frequencies. The plot shows total positive 44 samples. The substitution mutations were shown in green, indels were shown in red.
Fig. 3.
a. PI3K/AKT signaling pathway identified using ingenuity variant analysis (IVA). Blue represents loss of function, orange represents gain of function, and grey inferred normal.
Fig. 3b.
PI3K/AKT signaling pathway identified using ingenuity variant analysis (IVA). Blue represents loss of function, orange represents gain of function, grey inferred normal, and entities outlined in red are potential drug targets.
Tumor protein TP53 (TP53), phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha (PIK3CA), proto-oncogene c-Kit (KIT), Phosphatase and Tensin Homolog (PTEN), proto-oncogene MET (MET), and Kirsten Rat Sarcoma (KRAS) were the most common mutated genes (≥2 variants per gene). TP53 was the most mutated one (15 different variants), followed by PIK3CA (8 different variants), KIT (3 variants), PTEN (2 variants), KRAS (2 variants), TSC1 (2 variants) and MET (2 variants). The detailed list of the affected genes, incurred somatic mutations, mutation type, frequency, zygosity and other data are listed in Table 2.
Table 2.
Somatic mutations detected in 46 Egyptian BC patients:
| Gene | Function | AA mutation | CDS mutation | Mutation type | Samples with mutation | Zygosity | CADD Phred prediction | Hot spot | Clinvar |
|---|---|---|---|---|---|---|---|---|---|
| KIT | Exonic; splicing | p.L862L | c.2586G > C | Substitution – coding silent | 10 | Het. | – | – | Benign |
| Exonic; splicing | p.M541L | c.1621A > C | Substitution – Missense | 6 | Het. | – | 4-55593464 | Benign | |
| Exonic; splicing | p.K546K | c.1638A > G | Substitution – coding silent | 5 | Het. | – | 4-55593481 | Benign | |
| TSC1 | Exonic | p.F608Lfs*21 | c.1824del | Frame shift deletion | 12 | Hom. | D | – | Not reported |
| Exonic | p.L203Cfs*7 | c.608del | Frame shift deletion | 2 | Hom. | D | – | Not reported | |
| TSC2 | Exonic | p.F298Lfs*65 | c.894del | Frame shift deletion | 3 | Hom. | D | – | Not reported |
| CEBPA | Exonic | p.A176V | c.527C > T | Substitution – Missense | 7 | Het. | D | – | Uncertain significance |
| KRAS | Splicing | – | c.575-9G > A | Substitution- Intronic | 6 | Het. | – | – | Benign |
| Exonic | p.G12V | c.35G > T | Substitution – Missense | 1 | Het. | D | 12-25398284 | Pathogenic | |
| PDGFRA | Exonic | – | c.2472C > T | Substitution – coding silent | 6 | Het. | – | 4-55152040 | Benign |
| PTEN | Exonic | p.E288fs | c.863delA | Frame shift deletion | 6 | Hom. | – | 10-89720712 | Not reported |
| 1 | Het. | – | |||||||
| Exonic | p.R130X | c.388C > T | Substitution – Nonense (stopgain) | 1 | Het. | D | 10-89692904 | Pathogenic | |
| VHL | Exonic | p.F148fs | c.440delT | Frame shift deletion | 5 | Hom. | – | 3-10188297 | Pathogenic |
| NOTCH4 | Exonic | p.K117Q | c. 349A > C | Substitution – Missense | 4 | Hom. | – | – | Not reported |
| STK11 | Exonic | p.G279fs | c.837delC | Frame shift deletion | 4 | Het. | – | 19-1221314 | Pathogenic |
| Gene | Function | AA mutation | CDS mutation | Mutation type | Samples with mutation | Zygosity | CADD Phred prediction | Hot spot | Clinvar |
|---|---|---|---|---|---|---|---|---|---|
| P53 | Exonic | p.R175H | c.524G>A | Substitution – Missense | 1 | Het. | D | 17-7578406 | Pathogenic |
| p.R282W | c.844C>T | Substitution – Missense | 1 | Het. | D | 17-7577094 | Pathogenic | ||
| p.C176W | c.528C>G | Substitution – Missense | 1 | Het. | D | 17-7578402 | Uncertain significance | ||
| p.Y234C | c.701A>G | Substitution – Missense | 1 | Het. | D | 17-7577580 | Pathogenic | ||
| p.R280G | c.838A>G | Substitution – Missense | 1 | Het. | D | 17-7577100 | conflicting interpretations of pathogenicity | ||
| P.G245D | c.734G>A | Substitution – Missense | 1 | Het. | D | 17-7577547 | Pathogenic | ||
| p.P72A | c.214C>G | Substitution – Missense | 4 | Het. | – | 17-7579473 | Uncertain significance | ||
| p.P72R | c.215C>G | Substitution – Missense | 8 | Hom. | – | 17-7579472 | Drug response | ||
| p.C135fs | c.403delT | Frame shift deletion | 2 | Hom. | – | 17-7578527 | Uncertain significance | ||
| 1 | Het. | ||||||||
| p.A276P | c.826G>C | Substitution – Missense | 1 | Het. | D | 17-7577112 | conflicting interpretations of pathogenicity | ||
| Exonic; splicing | p.Y220C | c.659A>G | Substitution – Missense | 1 | Het. | D | 17-7578190 | Pathogenic | |
| p.Q331* | c.991C>T | Substitution – Nonense (stopgain) | 1 | Het. | D | – | Not reported | ||
| p.K132R | c.395A>G | Substitution – Missense | 1 | Het. | D | 17-7578535 | Uncertain significance | ||
| p.R306* | c.916C>T | Substitution –Nonense (stopgain) | 1 | Het. | D | 17-7577022 | Pathogenic | ||
| Splicing | – | c.376-1G>A | Substitution- Intronic | 1 | Het. | D | 17-7578555 | Pathogenic |
| Gene | Function | AA mutation | CDS mutation | Mutation type | Samples with mutation | Zygosity | CADD Phred prediction | Hot spot | Clinvar |
| PIK3CA | Exonic | p.H1047R | c.3140A>G | Substitution – Missense | 10 | Het. | D | 3-178952085 | Pathogenic |
| p.I391M | c.1173A>G | Substitution – Missense | 7 | Het. | – | – | Benign | ||
| p.T1025T | c.3075C>T | Substitution – coding silent | 4 | Het. | – | 3-178952020 | Benign | ||
| p.E542K | c.1624G>A | Substitution – Missense | 1 | Hom. | D | 3-178936082 | Pathogenic | ||
| With | p.E80K | c.238G>A | Substitution – Missense | 1 | Het. | D | 3-178916851 | Not reported | |
| p.Q546R | c.1637A>G | Substitution – Missense | 1 | Het. | D | 3-178936095 | Pathogenic | ||
| p.E545K | c.1633G>A | Substitution – Missense | 2 | Het. | D | 3-178936091 | Pathogenic | ||
| p.E365K | c.1093G>A | Substitution – Missense | 1 | Het. | D | – | Pathogenic | ||
| JAK3 | Exonic | p.V722I | c.2164G>A | Substitution – Missense | 2 | Het. | D | 19-17945696 | conflicting interpretations of pathogenicity |
| APC | Exonic | p.G2502S | c.7504G>A | Substitution – Missense | 2 | Het. | D | – | Benign |
| p.E1317Q | c.3949G>C | Substitution – Missense | 1 | Het. | – | 5-112175240 | Conflicting interpretations of pathogenicity | ||
| MET | Exonic | p.N375S | c.1124A>G | Substitution – Missense | 1 | Het. | – | 7-116340262 | Benign |
| p.E168D | c.504G>T | Substitution – Missense | 1 | Het. | – | 7-116339642 | Benign | ||
| Gene | Function | AA mutation | CDS mutation | Mutation type | Samples with mutation | Zygosity | CADD Phred prediction | Hot spot | Clinvar |
| AKT1 | Exonic | p.E17K | c.49G>A | Substitution – Missense | 2 | Het. | D | 14-105246551 | Pathogenic |
| MAP2K4 | Exonic ; splicing | p.Q80* | c.238C>T | Substitution – Nonense (stopgain) | 1 | Het. | D | – | Not reported |
| IDH2 | Exonic | p.R140Q | c.419G>A | Substitution – Missense | 1 | Het. | D | 15-90631934 | Pathogenic |
| ERBB2 | Exonic | p.V777L | c.2329G>T | Substitution – Missense | 1 | Het. | D | 17-37881000 | Pathogenic |
| EPHB1 | Exonic | p.F151V | c.451T>G | Substitution – Missense | 14 | Het. | D | – | Novel |
| KIT | Exonic | p.H263Q | c.789T>G | Substitution – Missense | 10 | Het. | D | – | Novel |
| MTOR | Exonic; splicing | p.T600I | c.1799C>T | Substitution – Missense | 9 | Het. | D | – | Novel |
| JAK2 | Exonic | – | c.2041_2042delinsTA | inframeshift substitution | 6 | Het. | – | – | Novel |
Fifteen different somatic mutations were detected in TP53 gene; all, except only one, were within known hotspot regions and most of them were classified as pathogenic. Notably, one TP53 drug response variant (p.P72R) was detected in 8 samples.
Eight different somatic mutations were detected in PIK3CA gene. p.H1047R, p.E545K, p.E542K, p.E80K, and p.Q546R were found at known hotspot regions and classified as pathogenic. p.H1047R is the most frequently detected pathogenic somatic mutation in this study. Another PIK3CA variant (p.I391M) was detected in 7 samples and one more PIK3CA substitution coding silent variant (p.T1025T) was detected in 4 samples.
Three different somatic mutations were detected in KIT gene; p.L862L, p.M541L, p.K546K in 9, 6, and 5 samples respectively. On the other hand, two different PTEN somatic mutations were detected; one variant (p.E288fs) in 6 samples as homozygous mutation and in one sample as heterozygous mutation and the other variant (p.R130X) in one sample. While KRAS gene had one substitution intronic splicing variant in 6 samples and 1 sample harbored one missense variant (p.G12V). Interestingly, we detected 2 frame shift deletions (p.F148fs and p.G279fs) in VHL and STK11 genes in 5 and 4 samples, respectively. These two variants were recorded as pathogenic in NCBI Clin Var database. In addition, 3 frame shift deletions were detected; 2 (p.F608Lfs*21 and p.L203Cfs*7) in TSC1 gene and 1 (p.F298Lfs*65) in TSC2 gene. Moreover, one substitution-intronic variant in platelet derived growth factor receptor alpha (PDGFRA) was detected in 6 samples and this variant is pathogenic according to FATHMM score.
Discussion
The identification of somatic mutations analyzed by Next- Generation Sequencing is increasingly used in clinical research as it allows deepening the knowledge of cancer progression. In this work we used Ion AmpliSeq Comprehensive caner panel to target most frequently cited and mutated cancer related genes and to report the frequency of the detected somatic mutations among 46 Egyptian BC patients. To the best of our knowledge, this is the first Egyptian cohort to sequence a multiple-gene panel of cancer related genes on BC patients.
In this cohort, we detected somatic mutations in genes previously reported to be frequently mutated in BC: TP53, PIK3CA, PTEN, AKT1, and MAP2K4. In addition, other somatic mutations were detected in genes recently reported to be mutated in BC; KRAS, PDGFRA, VHL, STK11, APC, MET, JAK3, SMARCB1, ERBB2, and IDH2 [26]. Matching with several previous studies, TP53 and PIK3CA were the most top two frequently mutated genes in BC proving their importance in carcinogenic process [14]. On the contrary, we detected variants at a relatively high frequency in STK11, KRAS, and ERBB2 genes, which were previously reported to occur infrequently in BC [7], [27].
TP53 is a key transcription factor that participates in repair of DNA damage, cell cycle check point control, and apoptosis induction. TP53 is mutated in BC in around 30–35% of cases and losing its normal functions causes tumorigenesis. In this cohort, 15 different TP53 somatic mutations were present in 58.7% (27 out of 46) of patients and they were all (except one) within known hotspot regions. Of interest, two polymorphic variants of TP53 gene were detected; the most frequent one was TP53 p.P72R (COSM250061) in 8 patients (17.4%) followed by TP53 p.P72A (COSM3738520) in 4 patients (8.7%). There are contradictory results about if TP53 codon 72 polymorphism is associated with BC risk or not. In meta-analysis by Zhang et al., it was reported that TP53 P72R contributes to BC susceptibility [28]. On the contrary, Ma et al. found that TP53 P72R showed no significant association with BC risk [29]. This null significant association was verified again in updated meta-analysis by Cheng et al. [30]. Therefore, an additional large study is required to validate this association in our Egyptian BC patients.
On the other hand, it was reported that TP53 polymorphism may influence individual responsiveness to cancer chemotherapy via modulation of TP73-dependent apoptosis [31]. The ability of mutant TP53 to bind to TP73, the TP53-family member, and inactivate it is influenced by TP53 codon 72 [32]. Xu et al., indicated that BC patients with the Pro/Pro genotype were less sensitive to a neoadjuvant chemotherapy regimen that included 5-fluorouracil, cyclophosphamide, and the anthracycline-based neoadjuvant chemotherapy than those with the Arg/Arg or Arg/Pro genotypes [33]. Similar results were reported in head and neck carcinoma [sullivan 2004]. It was also reported that Arg/Arg genotype induces apoptosis more effectively than Pro/Pro genotype, which may be due to enhanced mitochondrial localization of TP53 protein in cells with the Arg/Arg genotype [34]. Furthermore, it was reported that mutant TP53 with the R72 variant was significantly associated with poor prognosis in women with BC [35]. Thus, it is suggested that TP53 codon 72 might be a strong predictive marker for chemotherapy response in BC patients.
Genetic alteration causes abnormalities in PI3K/AKT/mTOR pathway and results in deregulated kinase activity and malignant transformation. Thus, target therapies are being actively investigated to inhibit this pathway including; PI3K inhibitors such as pictilisib, alpelisib, copanlisib, and taselisib; AKT inhibitors such as ipatasertib; and mTOR inhibitors such as everolimus [36]. PIK3CA gene is an important component of the phosphoinositide-3 kinase (PI3K) pathway which is frequently altered in human cancers. Deregulation of the PI3K pathway, through the acquired somatic mutations, contributes to tumors development and progression. In The Cancer Genome Atlas (TCGA) and COSMIC databases, it was reported that PIK3CA gene is mutated in ~36% of BC [37]. In patients with PIK3CA mutations, recent study showed promising results in progression‐free survival when using buparlisib, PI3K inhibitor; in combination with endocrine therapy [38]. Many other studies revealed that patients with PIK3CA mutations might benefit from PI3K‐selective inhibitor treatment [39], [40]. On the other hand, AKT gene is the PI3K effector and AKT signalling leads to increased cellular growth and survival. A somatic mutation (E17K) in AKT1 gene was discovered in 8% of BC [41]. At least one component of PI3K/AKT/mTOR pathway is altered in more than 50% of ER/PR positive tumors, and these alterations cause tumor growth and develop resistance to antihormone therapies. Thus, hormone receptor positive tumors will benefit from using PI3K/AKT inhibitors in combination with endocrine therapy [42]. We detected eight different PIK3CA mutations in 27 patients (58.7%). H1047R, E545K, E542K, Q546R, p.E80K hotspots accounted for 55.6% of all PIK3CA mutations in our cohort. We also detected PIK3CA I391M polymorphism; which is far from the binding site but can affect the protein function and change its dynamic action. It was suggested by Ahmadi et al. that this polymorphism may be involved in BC invasion [43]. This variant was present in 7 patients (15.2%) and may be used as marker for BC tumorigenesis. Another remaining PIK3CA (p.T1025T) polymorphism in our cohort is thought to be Arab specific variant. This SNP rs17849079 (p.T1025T) was reported at high prevalence among Arab BC patients and suggested to be used as a molecular marker for early diagnosis in this population [44]. So, further studies are needed to validate the use of such structural variants as SNP marker for BC early detection and invasion. Moreover, we found one hotspot mutation in AKT1 gene (exon3:c.49G > A: p.E17K) in two patients of luminal A and Triple negative subtypes. It was reported that BC patients with AKT p.E17K mutation are sensitive to AKT inhibitors [45]. Thus Egyptian BC patients carrying this mutation may represent good candidates for AKT inhibitors treatment. In this study and according to IVA, we identified many mutated genes that commonly up-regulate PI3K/AKT signaling pathway and promote carcinogenesis. Thus, we propose that Egyptian BC patients might benefit from PI3K/AKT inhibitors in combination with endocrine therapy.
Two pathogenic frame shift deletion variants in VHL and STK11 were detected in 5 and 4 samples, respectively. Other two frame shift deletion variants in TP53 and PTEN were identified. Interestingly, 2 patients were found to concomitantly harbor these four frame shift variants. This combination of mutation may contribute the BC development in those patients.
Loss of PTEN function, on basis of somatic mutations, mostly affects tumor development across tissues. In the nucleus, PTEN promotes chromosome stability and DNA repair. Consequently, loss of PTEN function increases genomic instability [46]. Also, improper PTEN function leads to uncontrolled activation of its downstream signals. One frame shift deletion and one stop gain variants in PTEN gene were identified. A deletion in codon 288, exon 8 of PTEN, resulting in a frame shift mutation (p.E288fs) was detected in 7 samples of Luminal A subtype. The stop gain variant (p.R130X) was detected in one sample which was stage I. A combination mutation in PIK3CA (p.H1047R) and PTEN (p.R130X) was also identified.
In addition, we identified rare hotspot point mutation in KRAS (exon2:c.35G > T: p.G12V) that have been previously reported in ductal carcinomas [47]. In our sample set, this mutation was found in one case co-occurred with another PIK3CA point mutation (p.T1025T) in luminal B (Her2+) subtype. This might explain the contribution of this co-occurrence in BC susceptibility as a driver mutation in tumor development. Furthermore, we identified an important pathogenic ERBB2 variant (p.V777L). In a study by Cocco et al, it was proposed that Neratinib is effective in breast tumors bearing both amplification and mutation of ERBB2 [48].
In conclusion NGS is a very useful tool to evaluate the mutational status of oncogenes and tumor suppressor genes to help identify the mutation drivers of BC [49]. In this cohort we shed the light on the most frequently detected somatic mutations and most altered pathways in Egyptian BC patients.
Recommendation
We recommend following up the patients until diagnosis of recurrence or metastasis and following up their response to treatment to give more focus on the association between survival data and the identified somatic mutations which may have important clinical implications for personalized medicine, target therapy, therapeutic guidance, and monitoring of recurrence or metastasis. Moreover, we recommend sequencing the most frequently detected genes from this preliminary study to confirm our findings on large number of BC patients. In addition, giving more focus on triple negative BC patients as it is the most aggressive and has a poor prognosis.
Author contributions
Abdel-Rahman N Zekri designed the study. M. Gomaa, Osman Mansour, Amany Abd-Elhameed Abou-Bakr and Samah A. Loutfy recruited patients and collected clinical data. Auhood Nassar, Mai M. Lotfy and Amira Salah El-Din Youssef performed the library preparation and NGS workflow. Ola s. ahmed, helped in NGS. Mohamed Abouelhoda and Auhood Nassar performed the bioinformatic analysis. Auhood Nassar drafted the manuscript. Mohamed M. Hafez, Hoda Ismail, and Abeer Bahnassy revised the manuscript. All authors read and approved the final version.
Declaration of Competing Interest
The authors declared that there is no conflict of interest.
Footnotes
Peer review under responsibility of Cairo University.
Contributor Information
Auhood Nassar, Email: auhood.nassar@nci.cu.edu.eg.
Osman Mansour, Email: osman.mansour@nci.cu.edu.eg.
Amira Salah El-Din Youssef, Email: amira.salah@nci.cu.edu.eg.
Abdel-Rahman N. Zekri, Email: abdelrhman.zekri@nci.cu.edu.eg.
References
- 1.Ferlay J., Soerjomataram I., Dikshit R., Eser S., Mathers C., Rebelo M. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Torre L.A., Bray F., Siegel R.L., Ferlay J., Lortet-Tieulent J., Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 3.The National Cancer Registry Program of Egypt (NCRPE). Reports and Statistics: Aswan, Damietta & El-Minia; 2013 http://www.cancerregistry.gov.eg/reports.aspx [accessed June 2013].
- 4.The Gharbiah Population-based Cancer Registry (GPCR). Cancer in Egypt, Gharbiah; 2007 http://www.emro.who.int/ncd/pdf/cancer_registry_Egypt.pdf [accessed June 2013].
- 5.Nik-Zainal S., Davies H., Staaf J., Ramakrishna M., Glodzik D., Zou X. Landscape of somatic mutations in 560 BC whole genome sequences. Nature. 2016;534(7605):47–54. doi: 10.1038/nature17676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stephens P.J., Tarpey P.S., Davies H., Van Loo P., Greenman C., Wedge D.C. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486(7403):400–404. doi: 10.1038/nature11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pereira B., Chin S.F., Rueda O.M., Vollan H.-K.M., Provenzano E., Bardwell H.A. The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat Commun. 2016;7:11479. doi: 10.1038/ncomms11479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meric-Bernstam F., Frampton G.M., Ferrer-Lozano J., Yelensky R., Pérez-Fidalgo J.A., Wang Y. Concordance of genomic alterations between primary and recurrent breast cancer. Mol Cancer Ther. 2014;13(5):1382–1389. doi: 10.1158/1535-7163.MCT-13-0482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weisman P.S., Ng C.K., Brogi E., Eisenberg R.E., Won H.H., Piscuoglio S. Genetic alterations of triple negative breast cancer by targeted next-generation sequencing and correlation with tumor morphology. Mod Pathol. 2016;29(5):476–488. doi: 10.1038/modpathol.2016.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith N.G., Gyanchandani R., Shah O.S., Gurda G.T., Lucas P.C., Hartmaier R.J. Targeted mutation detection in breast cancer using MammaSeq™. Breast Cancer Res. 2019;21(1):22. doi: 10.1186/s13058-019-1102-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bai X., Zhang E., Ye H., Nandakumar V., Wang Z. PIK3CA and TP53 gene mutations in human breast cancer tumors frequently detected by ion torrent DNA sequencing. PLoS ONE. 2014;9(6):e99306. doi: 10.1371/journal.pone.0099306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu S., Wang H., Zhang L., Tang C., Jones L., Ye H. Rapid detection of genetic mutations in individual breast cancer patients by next-generation DNA sequencing. Hum Genom. 2015;9(1):2. doi: 10.1186/s40246-015-0024-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li G., Guo X., Chen M., Tang L., Jiang H., Day J.X. Prevalence and spectrumof AKT1, PIK3CA, PTEN and TP53 somatic mutations in Chinese breast cancer patients. PLoS ONE. 2018;13(9):e0203495. doi: 10.1371/journal.pone.0203495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y., Cai Q., Shu X.-O., Gao Y.-T., Li C., Zheng W. Whole-exome sequencing identifies novel somatic mutations in Chinese breast cancer patients. J Mol Genet Med: Int J Biomed Res. 2015;9(4) doi: 10.4172/1747-0862.1000183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Banerji S., Cibul Skis K., Rangel-Escareno C., Brown K.K., Carter S.L., Frederick A.M. Sequence analysis of mutations and translocations across breast cancer subtypes. Nature. 2012;486(7403):405–409. doi: 10.1038/nature11154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang H., Wang K. Genomic variant annotation and prioritization with ANNOVAR and wANNOVAR. Nat Protoc. 2015;10:1556±1566. doi: 10.1038/nprot.2015.105. PMID: 26379229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tate J.G., Bamford S., Jubb H.C., Sondka Z. COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Res. 2019;47(D1):D941–D947. doi: 10.1093/nar/gky1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Griffith M., Spies N.C., Krysiak K., McMichael J.F., Coffman A.C., Danos A.M. CIViC is a community knowledgebase for expert crowdsourcing the clinical interpretation of variants in cancer. Nat Genet. 2017;49(2):170–174. doi: 10.1038/ng.3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Landrum M.J., Lee J.M., Benson M., Brown G.R., Chao C., Chitipiralla S. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018 doi: 10.1093/nar/gkx1153. PubMed PMID: 29165669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vaser R., Adusumalli S., Leng S.N., Sikic M., Ng P.C. SIFT missense predictions for genomes. Nat Protocols. 2016;11:1–9. doi: 10.1038/nprot.2015.123. [DOI] [PubMed] [Google Scholar]
- 22.Rentzsch P., Witten D., Cooper G.M., Shendure J., Kircher M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2018 doi: 10.1093/nar/gky1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hiltemann S., Jenster G., Trapman J., van der Spek P., Stubbs A. Discriminating somatic and germline mutations in tumor DNA samples without matching normals. Genome Res. 2015;25(9):1382–1390. doi: 10.1101/gr.183053.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sukhai M.A., Misyura M., Thomas M., Garg S., Zhang T., Stickle N. Somatic tumor variant filtration strategies to optimize tumor-only molecular profiling using targeted next-generation sequencing panels. J Mol Diagn. 2019;21(2):261–273. doi: 10.1016/j.jmoldx.2018.09.008. [DOI] [PubMed] [Google Scholar]
- 26.Ye Q., Qi F., Bian L., Zhang S.H., Wang T., Jiang Z.F. Circulating-free DNA mutation associated with response of targeted therapy in human epidermal growth factor Receptor 2-positive metastatic breast cancer. Chin Med J. 2017;130:522–529. doi: 10.4103/0366-6999.200542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Loi S., Michiels S., Lambrechts D., Fumagalli D. Somatic mutation profiling and associations with prognosis and trastuzumab benefit in early breast cancer. J Natl Cancer Inst. 2013;105:960–967. doi: 10.1093/jnci/djt121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Z., Wang M., Wu D., Tong N., Tian Y. P53 codon 72 polymorphism contributes to breast cancer risk: a meta-analysis based on 39 case–control studies. Breast Cancer Res Treat. 2010;120 doi: 10.1007/s10549-009-0480-4. 509–517 12. [DOI] [PubMed] [Google Scholar]
- 29.Ma Y., Yang J., Liu Z., Zhang P., Yang Z., Wang Y. No significant association between the TP53 codon 72 polymorphism and breast cancer risk: a meta-analysis of 21 studies involving 24,063 subjects. Breast Cancer Res Treat. 2011;125:201–205. doi: 10.1007/s10549-010-0920-1. [DOI] [PubMed] [Google Scholar]
- 30.Cheng H., Biao Ma B., Jiang R., Wang W., Guo H. Individual and combined effects of MDM2 SNP309 and TP53 Arg72Pro on breast cancer risk: an updated meta-analysis. Mol Biol Rep. 2012;39:9265–9274. doi: 10.1007/s11033-012-1800-z. [DOI] [PubMed] [Google Scholar]
- 31.Bergamaschi D., Gasco M., Hiller L., Sullivan A., Syed N., Trigiante G. p53 polymorphism influences response in cancer chemotherapy via modulation of p73-dependent apoptosis. Cancer Cell. 2003;3(4):387–402. doi: 10.1016/s1535-6108(03)00079-5. [DOI] [PubMed] [Google Scholar]
- 32.Marin M.C., Jost C.A., Brooks L.A., Irwin M.S., O'Nions J., Tidy J.A. A common polymorphism acts as an intragenic modifier of mutant p53 behaviour. Nat Genet. 2000;25(1):47–54. doi: 10.1038/75586. [DOI] [PubMed] [Google Scholar]
- 33.Xu Y., Yao L., Ouyang T., Li J., Wang T., Fan Z. p53 Codon 72 polymorphism predicts the pathologic response to neoadjuvant chemotherapy in patients with breast cancer. Clin Cancer Res. 2005;11(20):7328–7333. doi: 10.1158/1078-0432.CCR-05-0507. [DOI] [PubMed] [Google Scholar]
- 34.Dumont P., Leu J.I., Della Pietra A.C., 3rd, George D.L., Murphy M. The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nat Genet. 2003;33(3):357–365. doi: 10.1038/ng1093. [DOI] [PubMed] [Google Scholar]
- 35.Basu S., Gnanapradeepan K., Barnoud T., Kung C.P., Tavecchio M., Scott J. Mutant p53 controls tumor metabolism and metastasis by regulating PGC-1α. Genes Dev. 2018;32(3–4):230–243. doi: 10.1101/gad.309062.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Araki K., Miyoshi Y. Mechanism of resistance to endocrine therapy in breast cancer: the important role of PI3K/Akt/mTOR in estrogen receptor-positive, HER2-negative breast cancer. Breast Cancer. 2018;25(4):392–401. doi: 10.1007/s12282-017-0812-x. [DOI] [PubMed] [Google Scholar]
- 37.Chen L., Yang L., Yao L., Kuang X.Y., Zuo W.J., Li S. Characterization of PIK3CA and PIK3R1 somatic mutations in Chinese breast cancer patients. Nat Commun. 2018;9(1):1357. doi: 10.1038/s41467-018-03867-9. PMID: 29636477; PMCID: PMC5893593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baselga J., Im S.A., Iwata H., Cortes J., De Laurentiis M., Jiang Z. Buparlisib plus fulvestrant versus placebo plus fulvestrant in postmenopausal, hormone receptor-positive, HER2-negative, advanced breast cancer (BELLE-2): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2017;18:904–916. doi: 10.1016/S1470-2045(17)30376-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kodahl A.R., Ehmsen S., Pallisgaard N., Jylling A.M.B., Jensen J.D., Laenkholm A.V. Correlation between circulating cell-free PIK3CA tumor DNA levels and treatment response in patients with PIK3CA-mutated metastatic breast cancer. Mol Oncol. 2018;12(6):925–935. doi: 10.1002/1878-0261.12305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berns K., Horlings H.M., Hennessy B.T., Madiredjo M., Hijmans E.M., Beelen K. A functional genetic approach identifies the PI3K pathway as a major determinant of trastuzumab resistance in breast cancer. Cancer Cell. 2007;12(4):395–402. doi: 10.1016/j.ccr.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 41.Engelman J.A. Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nat Rev Cancer. 2009;9:550–562. doi: 10.1038/nrc2664. [DOI] [PubMed] [Google Scholar]
- 42.Polo M.L., Riggio M., May M. Activation of PI3K/Akt/mTOR signaling in the tumor stroma drives endocrine therapy-dependent breast tumor regression. Oncotarget. 2015;6(26):22081–22097. doi: 10.18632/oncotarget.4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ahmadi S., Ramezani S., Ghafouri H., Hosseini S.M., Najafi A. Association between the PIK3CA Ile391Met polymorphism and the risk of breast cancer in an Iranian population. J Appl Biotechnol Rep. 2018;5(1):8–12. [Google Scholar]
- 44.Karakas B., Colak D., Kaya N., Ghebeh H., Al-Qasem A., Hendrayani F. Prevalence of PIK3CA mutations and the SNPrs17849079 in Arab breast cancer patients. Cancer Biol Ther. 2013;14(10):888–896. doi: 10.4161/cbt.25945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hyman D.M., Smyth L.M., Donoghue M.T.A., Westin S.N., Bedard P.L., Dean E.J. AKT inhibition in solid tumors with AKT1 mutations. J Clin Oncol: Off J Am Soc Clin Oncol. 2017;35(20):2251–2259. doi: 10.1200/JCO.2017.73.0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dillon L.M., Miller T.W. Therapeutic targeting of cancers with loss of PTEN function. Curr Drug Targets. 2014;15(1):65–79. doi: 10.2174/1389450114666140106100909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Myers M.B., Banda M., McKim K.L., Wang Y. Breast cancer heterogeneity examined by high-sensitivity quantification of PIK3CA, KRAS, HRAS, and BRAF mutations in normal breast and ductal carcinomas. Neoplasia. 2016;18:253–263. doi: 10.1016/j.neo.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cocco E., Javier Carmona F., Razavi P., Won H.H., Cai Y., Rossi V. Neratinib is effective in breast tumors bearing both amplification and mutation of ERBB2 (HER2) Sci Signal. 2018;11(551):eaat9773. doi: 10.1126/scisignal.aat9773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Uscanga-Perales G.I., Santuario-Facio S.K., Sanchez-Dominguez C.N., Cardona-Huerta S., Muñoz-Maldonado G.E., Ruiz-Flores P. Genetic alterations of triple negative breast cancer (TNBC) in women from Northeastern Mexico. Oncol Lett. 2019;17(3):3581–3588. doi: 10.3892/ol.2019.9984. [DOI] [PMC free article] [PubMed] [Google Scholar]