Abstract
Background: Acute myeloid leukemia (AML) is the most common form of acute leukemias in adults which is clinically and molecularly heterogeneous. Several risk and genetic factors have been widely investigated to characterize AML. However, the concomitant epigenetic factors in controlling the gene expression lead to AML transformation was not fully understood. This study was aimed to identify epigenetically regulated genes in AML cell lines induced by epigenetic modulating agents, Trichostatin A (TSA) and 5-Azacytidine (5-Aza).
Materials and Methods: MV4-11 and Kasumi 1 were treated with TSA and/or 5-Aza at IC50 concentration. Gene expression profiling by microarray was utilized using SurePrint G3 Human Gene Expression v3. Gene ontology and KEGG pathway annotations were analyzed by DAVID bioinformatics software using EASE enrichment score. mRNA expression of the differentially expressed genes were verified by quantitative real time PCR.
Results: Gene expression analysis revealed a significant changes in the expression of 24,822, 15,720, 15,654 genes in MV4-11 and 12,598, 8828, 18,026 genes in Kasumi 1, in response to TSA, 5-Aza and combination treatments, respectively, compared to non-treated (p<0.05). 7 genes (SOCS3, TUBA1C, CCNA1, MAP3K6, PTPRC, STAT6 and RUNX1) and 4 genes (ANGPTL4, TUBB2A, ADAM12 and PTPN6) shown to be predominantly expressed in MV4-11 and Kasumi 1, respectively (EASE<0.1). The analysis also revealed phagosome pathway commonly activated in both cell lines.
Conclusion: Our data showed a distinct optimal biological characteristic and pathway in different types of leukemic cell lines. These finding may help in the identification of cell-specific epigenetic biomarker in the pathogenesis of AML.
Key Words: Acute myeloid leukemia, Epigenetics* Histone deacetylase inhibitors, 5-Azacytidine, Gene expression
Introduction
Acute myeloid leukemia (AML) is characterized by a block in early progenitor differentiation leading to accumulation of immature and highly proliferative leukemic stem cells (LSCs) in the bone marrow and peripheral blood 1 . The 2017 World Health Organization (WHO) has provided guidelines on the cut-off value of blast percentage of AML by; 200 and 500 cells-leukocytes differential counts in the peripheral blood and in the bone marrow, respectively 2 . For a diagnosis of AML, a marrow or blood blast count of 20% or more is required, except for AML with t(15;17), t(8;21), inv(16) or t(16;16), and some cases of erythroleukemia. AML is the most common form of acute leukemias in adults which affected 32% adults. Although the overall mortality rate has decreased by 1.0% each year from 2001 to 2010, the overall incidence rate was increased by 0.2% each year. In 2018, the American Cancer Society estimated that 19,520 of new cases and 10,670 deaths from AML. The 5-years overall survival rate was also poor with only 24% 3 .
For many years, gene expression profiling by microarray was used as a traditional method to search abnormalities in cancers, including in AML 4 . These presented data was invaluable and accessible to the identification of disease’s class discovery, class prediction, and class comparison. Class discovery refers to the identification of a new subgroup, that later was class predicted by gene expression data. The first and second class already had a diagnostic implication. While the third class, which is class comparison refer to the identification of genes that were deregulated in certain subgroups, that may address biological function 5 .
It has long established that AML is clinically heterogeneous disease characterized by an accumulation of continuous genetic abnormalities 6 and prior epigenetic lesions 7 resulting in clonal evolution and expansion. The considerable complexities disrupt the genetic and epigenetic landscapes by changes in gene expression 8 which profoundly affecting treatment response and patients’ survival. Earlier epigenetic alteration established cellular identities initiating tumorigenesis by inappropriate activation or inhibition of cellular signaling pathways 9 . For example, promoter hypermethylation of a tumor suppressor genes is commonly implicated in cancer 10 , involving genes controlling the cell cycle and DNA repair 11 . On the other hand, modification to histone protein in nucleosome modulates the transcriptional burst frequency specifically through histone acetylation 12 . Both epigenetic mechanisms endow the regulation in gene expression. Hence, targeting the epigenetically-regulated genes in the control of AML licensed a promising outcome.
In this study, high-throughput microarray technique was used to analyze epigenetic-derived molecular mechanism by modulating gene expression using a classical DNA methyltransferase (DNMT) inhibitor; 5-Azacytidine (5-Aza) and a histone deacetylase (HDAC) inhibitor, Trichostatin A (TSA). The aim of this study was to induce the epigenetic response via gene re-expression or down-expression in two types of AML cell lines; MV4-11 and Kasumi 1. It was hypothesized that the silencing of a tumor suppressor gene and the activation of oncogenes in AML were due to epigenetic mechanisms of DNA hypermethylation and histone deacetylation.
MATERIALS AND METHODS
MV4-11 and Kasumi 1 cell culture
MV4-11 is a human AML cell line established from blasts cells of 10 years old male with biphenotypic B-myelomonocytic leukemia (AML FAB M5) that carry translocation t(4;11) and a FLT3-ITD mutation. Kasumi 1 is a human AML cell line established from peripheral blast cells from 7 years old juvenile male Japanese that carry translocation t(8;21) and AML1-ETO (also known as RUNX1-CBF2T1) fusion genes. The AML cell lines were originally purchased from the American Type Culture Collection (ATCC, VA, USA). Both AML cell lines were cultured in RPMI-1640 (Gibco®, CA, USA) supplemented with 10% Fetal bovine serum (Sigma-Aldrich, MO, USA) and 0.1% penicillin/streptomycin (Invitrogen, CA, USA) in humidified temperature containing 5% carbon dioxide (CO2) at 37°C.
TSA and/or 5-Aza treatment
TSA (Sigma-Aldrich, MO, USA) and 5-Aza (Sigma-Aldrich, MO, USA) were dissolved in DMSO (Sigma-Aldrich, MO, USA) and RPMI-1640, respectively to a stock concentration of 500 µM, and further diluted to the desired working concentrations. MV4-11 and Kasumi 1 were seeded in 6-wells plate to 80-90% confluency at the initial cell number of 1 x 105 cells/mL prior to the drug treatment for 24 hours. The cell lines were treated with varying concentration of TSA (0, 1.25, 2.5, 5.0, 10.0 µM) and 5-Aza (0, 5.0, 10.0, 20.0, 50, 100 µM) and incubated for 24 hours under humidified temperature.
Cell Viability Assay
Percentage viability of non-treated and treated MV4-11 and Kasumi 1 after the 24 hours exposure to TSA and 5-Aza treatments were measured by Trypan Blue Exclusion Assay (Life Technologies, CA, USA). The half maximal inhibitory concentration (IC50) was determined by GraphPad Prism 6.0 (GraphPad, CA, USA).
Total RNA extraction and quality control
Total RNA was extracted from treated and untreated MV4-11 and Kasumi 1 using Total RNA Isolation Kit (Promega, SA, USA) according to the manufacturer’s protocol. The final elution step was performed using 30 µl of elution buffer for a highly concentrated RNAs. The isolated RNA concentration and purity were determined by Nanodrop ND-1000 spectrophotometer (Thermo-Fisher Scientific, WA, USA). Prior to the gene expression profiling, the RNA integrity was assessed by 1.5% agarose gel electrophoresis and their RIN (RNA integrity number) values were determine by Agilent 2100 Bioanalyzer (Agilent, CA, USA). The qualified RNAs (absorbance 280/260 1.8-2.1 ratio; highly intact 28S and 18S ribosomal RNA and RIN above 7) were stored at -80 ºC until further analysis.
Microarray analysis
Whole genome expression profiling was performed using One-Color SurePrint G3 Human Gene Expression v3, 8 x 60K slides contained array probe (Agilent Technologies, CA, USA). Prior to Cyanine 3 (Cy3) labeling, RNA spiked-In dilution was prepared using RNA spiked-In Kit (Agilent Technologies, CA, USA) to each sample using T7 RNA polymerase (RNA reference target) for normalization. Cy3-labeled cRNA was generated from 25 ng input total RNA using Low Input Quick Amp Labeling Kit (Agilent Technologies, CA, USA). The fluorescent-labeled cRNA was purified by RNAeasy Mini Kit and RNAase-free DNAase Set (Qiagen, CA, USA) and quantified by Nanodrop ND-1000 spectrophotometer. 25 ng of fluorescein-labeled and amplified cRNA was hybridized into array slides containing 60,000 probes (Agilent Technologies, CA, USA) at 65 degree Celsius for 17 hours. After hybridization and washing steps, the array slides were scanned using SureCan Microarray Scanner (Agilent Technologies, CA, USA) to measure the fluorescence intensity of Cy3 labeled RNA bound to the microarray slide. The resulted images were processed using the Feature Extraction (FE) software v.12 (Agilent Technologies, CA, USA) for data filtering. Raw data obtained was analyzed by Genespring GX v12.6 software (Agilent Technologies, CA, USA).
Database screening
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis annotations were utilized by the Database for Annotation, Visualization and Integrated Discovery (DAVID) Bioinformatics Resources v6.8 (https://david.ncifcrf.gov/) to characterize and predict epigenetically regulated genes in treated AML cell lines. The Enhanced AL Scoring Engine (EASE) scoring system (a modified Fisher Exact p-value, p<0.1) was implemented for statistical analysis to provide enriched GO terms and pathways annotation within gene lists. EASE analysis produces a consistent and similar functional annotation with numerous analytical methods 13 , and Venn diagram was constructed to analyze genes with differential expression pattern after TSA and 5-Aza treatment in MV4-11 and Kasumi 1. The analysis was conducted by the Venny 2.1 software (http://bioinfogp.cnb.csic.es/tools/venny/).
Quantitative Real-time PCR (qRT-PCR)
To validate microarray data, qRT-PCR analysis on selected up-regulated and down-regulated genes was performed by Taqman gene expression assays and analyzed using Applied Biosystem (ABI)® 7500 Real-Time PCR Machine (Applied Biosystem, CA, USA). Total RNAs from untreated and treated cell lines were reverse transcribed using High-Capacity cDNA Reverse Transcription Kit (Applied Biosystem, CA, USA). Pre-designed assays (PrimeTime® Pre-designed Assays) (IDT Inc., IA, USA) [ANGPTL4 (assay ID: Hs.PT58.25480012), TUBB2A (assay ID: Hs.PT58.40767003), PTPN6 (assay ID: Hs.PT58.23073507) and ADAM12 (assay ID: Hs.PT58.26423628)], and custom-designed primers and probes (SOCS3, TUBA1C, CCNA1, MAP3K6, STAT6, PTPRC and RUNX1 genes) were amplified by PrimeTime® Gene Expression Master Mix (IDT Inc., IA, USA). Assay sequences were confirmed using web Basic Local Alignment Search Tool (BLAST) by the National Center for Biotechnology Information (NCBI) (U.S. National Library of Medicine, MD, USA). The qRT-PCR amplification conditions were: 95°C for 3 min for enzyme activation, 40 cycles of denaturation at 95°C for 15 s and 60°C for 1 min for annealing and extension. B2M and GAPDH were used as endogenous control genes and expression levels were estimated using relative quantitation (RQ) of duplicated samples calculated by 2-∆∆CT method (∆∆CT=∆CTTreated–∆CTUntreated, ∆CT=CtSelected Genes –CtB2M/GAPDH).
Results
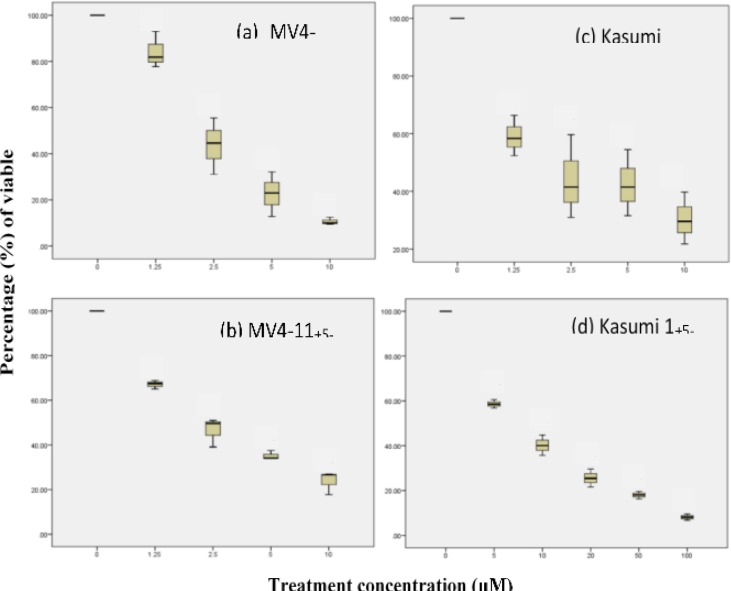
A significant decrease in cell viability was observed after the TSA and 5-Aza treatments (One-way ANOVA, p<0.05). The half maximal inhibitory concentration (IC50) was acquired at 2.2 µM and 2.3 µM for MV4-11 and; 6.25 µM and 6.95 µM for Kasumi 1 in TSA and 5-Aza, respectively. TSA and 5-Aza treatments have higher potency in MV4-11 due to their lower IC50 value compared to Kasumi 1 (Figure 1).
Figure 1.
Effect of TSA and 5-Aza treatment on cell viability by percentage (%) inhibition of MV4-11 and Kasumi 1 cell lines relative to non-treated cell lines. Significant inhibition of MV4-11 after (a) TSA and (b) 5-Aza treatment at increasing concentration (0.0, 1.25, 2.5, 5.0 and 10.0 µM) for 24 h. Significant inhibition of Kasumi 1 after (c) TSA treatment at increasing concentration (0.0, 1.25, 2.5, 5.0 and 10.0 µM) and (d) 5-Aza (0.0, 5.0, 10.0, 20.0, 50.0 and 100.0 µM) for 24 h calculated by Trypan Blue Exclusion Assay (TBEA) (One-Way ANOVA, LSD multiple comparison, p<0.05).
Gene expression profile of MV4-11 and Kasumi 1 in response to TSA and 5-Aza
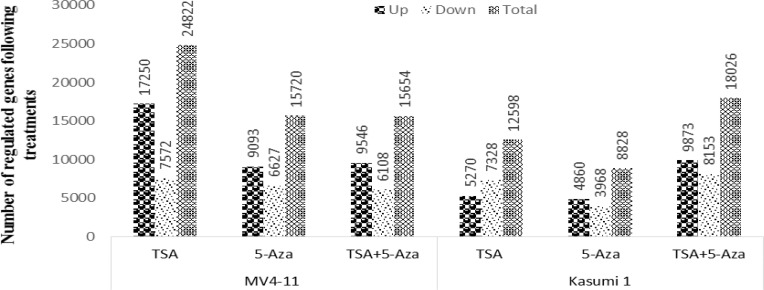
The gene expression profile of MV4-11 and Kasumi 1 after 24 hours of TSA, 5-Aza and combination (TSA+5-Aza) treatments at IC50 concentration. The exploratory microarray analysis was carried out to short-list the differentially expressed genes induced by the drug treatments analyzed by GeneSpring software 12.1 (the cut-off value; fold change ≥ 2.0, significance level, Pearson, P <0.05). 33,150 and 24,668 genes passed the FE filtering in MV4-11 and Kasumi 1, respectively. In MV4-11, 24,822 genes’ expressions were altered (either up or down-regulated) in TSA, 15,720 in 5-Aza and 15,654 in TSA+5-Aza. Whereas in Kasumi 1, 12,598 genes were altered in TSA, 8828 genes in 5-Aza and 18,026 genes in TSA+5-Aza treatments, normalized to non-treated cells (Figure 2). The most up-regulated and down-regulated genes in TSA, 5-Aza and TSA+5-Aza treatments and their folds change were listed in Tables 1 and 2. Genes were selected according to these three criteria: 1. Relevant genes with the highest fold-change different and commonly regulated across all treatments, 2. Relevant genes reported having an association with AML and other myeloid neoplasms from the previous study and/or Pubmed literature, 3. Genes with not otherwise classified under both criteria but could be interesting due to their implication in pathways in cancer.
Figure 2.
Microarray gene expression analysis for MV4-11 and Kasumi 1 treated with TSA, 5-Aza and TSA+5-Aza. Number of up-regulated and down-regulated genes was created by Genespring software analysis. Further analysis to obtain gene entities were performed using Moderated T-test with multiple correction (Benjamini Hochberg FDR) with p-value <0.05 and fold change of >2.0 as a significant.
Table 1(a).
Most up- and down-regulated genes in TSA treated MV4-11
|
Gene Bank
Accession |
Gene symbol | Gene description ( Homo sapiens) |
*Folds
Change |
|---|---|---|---|
| NM_001082 | CYP4F2 | Cytochrome P450, family 4, subfamily F, polypeptide 2 | 1094.05 |
| NM_014971 | EFR3B | EFR3 homolog B (S. cerevisiae) | 360.59 |
| NM_006569 | CGREF1 | Cell growth regulator with EF-hand domain 1 | 348.85 |
| NM_017702 | DEF8 | Differentially expressed in FDCP 8 | 325.92 |
| NM_003914 | CCNA1 | Cyclin A1 | 298.44 |
| NM_003255 | TIMP2 | TIMP metallopeptidase inhibitor 2 | 281.56 |
| NM_031313 | ALPPL2 | Alkaline phosphatase, placental-like 2 | 250.36 |
| NM_032704 | TUBA1C | Tubulin, alpha 1c | 234.14 |
| NM_003955 | SOCS3 | Suppressor of cytokine signaling 3 | 176.76 |
| NM_001204054 | NDUFC2 | NADH dehydrogenase (ubiquinone) 1, subcomplex unknown 2 | 166.94 |
| NR_027028 | GUSBP1 | Glucuronidase, beta pseudogene 1 | 153.18 |
| NM_004522 | KIF5C | Kinesin family member 5C | 153.59 |
| NM_003520 | HIST1H2BN | Histone cluster 1, H2bn | 150.13 |
| NM_006321 | ARIH2 | Ariadne RBR E3 ubiquitin protein ligase 2 | 133.61 |
| NM_000612 | IGF2 | Insulin-like growth factor 2 | 131.09 |
| NM_177424 | STX12 | Syntaxin 12 | 103.73 |
| NM_006086 | TUBB3 | Tubulin, beta 3 class III | 80.38 |
| NM_004672 | MAP3K6 | Mitogen-activated protein kinase kinase kinase 6 | 39.50 |
| NM_001025300 | RAB12 | member RAS oncogene family | 38.83 |
| NM_139314 | ANGPTL4 | Angiopoietin-like 4 | 26.79 |
| NM_018437 | HEMGN | Hemogen | -518.75 |
| NM_024913 | CPED1 | Cadherin-like and PC-esterase domain containing 1 | -243.96 |
| NM_003152 | STAT5A | Signal transducer and activator of transcription 5A | -159.83 |
| NM_002838 | PTPRC | Protein tyrosine phosphatase, receptor type C | -138.75 |
| NM_080612 | GAB3 | GRB2-associated binding protein 3 | -117.26 |
| NM_003126 | SPTA1 | Spectrin, alpha, erythrocytic 1 | -107.30 |
| NM_015401 | HDAC7 | Histone deacetylase 7 | -88.16 |
| NM_006563 | KLF1 | Kruppel-like factor 1 (erythroid) | -85.08 |
| NM_015660 | GIMAP2 | GTPase, IMAP family member 2 | -73.83 |
| NM_006163 | NFE2 | Nuclear factor, erythroid 2 | -69.24 |
| NM_213674 | TPM2 | Tropomyosin 2 (beta) | -57.76 |
| NM_006287 | TFPI | Tissue factor pathway inhibitor | -55.30 |
| NM_005021 | ENPP3 | pyrophosphatase/phosphodiesterase 3 | -49.49 |
| NM_004688 | NMI | N-myc (and STAT) interactor | -47.85 |
| NM_000037 | ANK1 | Ankyrin 1, erythrocytic, transcript variant 3 | -46.78 |
| NM_013427 | ARHGAP6 | Rho GTPase activating protein 6 | -42.54 |
| NM_006546 | IGF2BP1 | Insulin-like growth factor 2 mRNA binding protein 1 | -42.54 |
| NM_033306 | CASP4 | Caspase 4, apoptosis-related cysteine peptidase | -42.42 |
| NM_080588 | PTPN7 | Protein tyrosine phosphatase, non-receptor type 7 | -39.69 |
| NM_004753 | DHRS3 | dehydrogenase/reductase (SDR family) member 3 | -36.59 |
| NR_026812 | RUNX1-IT1 | RUNX1 intronic transcript 1 | -22.05 |
| NM_003153 | STAT6 | signal transducer and activator of transcription 6 | -10.04 |
*Folds-change of treatment group compared to control analyzed by Genespring software analysis, Moderated T-test, p<0.05)
Table 1(b).
Most up- and down-regulated genes in 5-Aza treated MV4-11
| Gene Bank Accession | Gene symbol | Gene description ( Homo sapiens) |
*Folds
change |
|---|---|---|---|
| NM_001145191 | FAM200B | family with sequence similarity 200, member B | 461.79 |
| NM_032905 | RBM17 | RNA binding motif protein 17 | 336.98 |
| NM_017702 | DEF8 | differentially expressed in FDCP 8 homolog | 277.69 |
| NM_024097 | C1orf50 | chromosome 1 open reading frame 50 | 207.14 |
| NM_001204054 | NDUFC2 | NADH dehydrogenase | 185.92 |
| NM_006321 | ARIH2 | ariadne RBR E3 ubiquitin protein ligase 2 | 158.81 |
| NR_027028 | GUSBP1 | glucuronidase, beta pseudogene 1, non-coding RNA | 157.88 |
| NM_032704 | TUBA1C | tubulin, alpha 1c | 154.28 |
| NM_031925 | TMEM120A | transmembrane protein 120A | 135.01 |
| NM_003955 | SOCS3 | suppressor of cytokine signaling 3 | 120.31 |
| NM_015046 | SETX | Homo sapiens senataxin | 95.04 |
| NM_016256] | NAGPA | N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase | 93.98 |
| NM_001031713 | MCUR1 | mitochondrial calcium uniporter regulator 1 | 92.49 |
| NM_033028 | BBS4 | Bardet-Biedl syndrome 4 | 90.09 |
| NM_177424 | STX12 | syntaxin 12 | 89.59 |
| NM_003520 | HIST1H2BN | histone cluster 1, H2bn | 89.53 |
| NM_052936] | ATG4A | autophagy related 4A, cysteine peptidase | 85.61 |
| NM_014884 | SUGP2 | SURP and G patch domain containing 2 | 70.67 |
| NM_138501 | TECR | trans-2,3-enoyl-CoA reductase | 69.28 |
| NM_004672 | MAP3K6 | mitogen-activated protein kinase kinase kinase 6 | 48.45 |
| NM_005614 | RHEB | Homo sapiens Ras homolog enriched in brain | 45.97 |
| NM_013230 | CD24 | CD24 molecule | 45.50 |
| NM_001025300 | RAB12 | RAB12, member RAS oncogene family | 44.06 |
| NM_173698 | FAM133A | family with sequence similarity 133, member A | -101.93 |
| NM_014653 | WSCD2 | WSC domain containing 2 | -30.48 |
| NM_145290 | GPR125 | G protein-coupled receptor 125 | -29.51 |
| NM_020353 | PLSCR4 | phospholipid scramblase 4 | -28.02 |
| NM_001099921 | MAGEB16 | melanoma antigen family B, 16 | -27.19 |
| NM_033306 | CASP4 | caspase 4, apoptosis-related cysteine peptidase | -23.01 |
| NM_004126 | GNG11 | guanine nucleotide binding protein (G protein), gamma 11 | -22.73 |
| NM_144722 | SPEF2 | sperm flagellar 2 | -20.86 |
| NM_015660 | GIMAP2 | GTPase, IMAP family member 2 | -19.99 |
| NR_027755 | LINC00922 | long intergenic non-protein coding RNA 922, long non-coding RNA | -19.17 |
| NM_018437 | HEMGN | hemogen | -18.55 |
| NM_001005285 | OR2AT4 | olfactory receptor, family 2, subfamily AT, member 4 | -18.19 |
| NM_000537 | REN | renin | -17.26 |
| NM_000519 | HBD | hemoglobin, delta | -16.75 |
| NM_213674 | TPM2 | tropomyosin 2 (beta) | -16.59 |
| NM_002421 | MMP1 | matrix metallopeptidase 1 | -12.23 |
| NM_000361 | THBD | thrombomodulin | -11.98 |
| NM_005807 | PRG4 | proteoglycan 4 | -11.81 |
| NM_080429 | AQP10 | aquaporin 10 | -11.33 |
| NM_139022 | TSPAN32 | tetraspanin 32 | -10.78 |
| NM_024711 | GIMAP6 | GTPase, IMAP family member 6 | -10.55 |
| NM_002145 | HOXB2 | homeobox B2 | -10.22 |
| NM_019032 | ADAMTSL4 | ADAMTS-like 4 | -9.71 |
| NM_002838 | PTPRC | Protein tyrosine phosphatase, receptor type C | -7.81 |
| NR_026812 | RUNX1-IT1 | RUNX1 intronic transcript 1 | -5.91 |
| NM_003153 | STAT6 | signal transducer and activator of transcription 6 | -4.07 |
Table 1(c).
Most up- and down-regulated genes in TSA+5-Aza treated MV4-11
| Gene Bank Accession | Gene symbol | Gene description ( Homo sapiens) |
*Folds
change |
|---|---|---|---|
| NM_001145191 | FAM200B | Family with sequence similarity 200, member B | 521.92 |
| NM_197958 | LARP6 | La ribonucleoprotein domain family, member 6 | 506.68 |
| NM_017702 | DEF8 | differentially expressed in FDCP 8 homolog | 268.16 |
| NR_027028 | GUSBP1 | Homo sapiens glucuronidase, beta pseudogene 1 | 243.94 |
| NM_032905 | RBM17 | RNA binding motif protein 17 | 160.05 |
| NM_014773 | KIAA0141 | KIAA0141 (KIAA0141) | 157.47 |
| NM_001204054 | NDUFC2 | NADH dehydrogenase (ubiquinone) 1, subcomplex unknown 2 | 155.54 |
| NM_016256 | NAGPA | N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase | 141.82 |
| NM_032704 | TUBA1C | tubulin, alpha 1c | 139.42 |
| NM_013268 | LGALS13 | lectin, galactoside-binding, soluble 13 | 132.17 |
| NM_004187 | KDM5C | lysine (K)-specific demethylase 5C | 116.85 |
| NM_024097 | C1orf50 | chromosome 1 open reading frame 50 | 113.21 |
| NM_006321 | ARIH2 | ariadne RBR E3 ubiquitin protein ligase 2 | 97.43 |
| NM_014035 | SNX24 | sorting nexin 24 | 94.35 |
| NM_000600 | IL6 | interleukin 6 (interferon, beta 2) | 91.55 |
| NM_138433 | KLHDC7B | kelch domain containing 7B | 89.54 |
| NM_033028 | BBS4 | Bardet-Biedl syndrome 4 | 87.94 |
| NM_177424 | STX12 | syntaxin 12 | 87.27 |
| NM_015046 | SETX | senataxin | 87.24 |
| NM_001031713 | MCUR1 | mitochondrial calcium uniporter regulator 1 | 85.70 |
| NM_001010893 | SLC10A5 | solute carrier family 10, member 5 | 79.58 |
| NM_031925 | TMEM120A | transmembrane protein 120A | 78.16 |
| NM_006945 | SPRR2D | small proline-rich protein 2D | 71.36 |
| NM_052936 | ATG4A | Homo sapiens autophagy related 4A, cysteine peptidase | 70.34 |
| NM_014945 | ABLIM3 | actin binding LIM protein family, member 3 | 68.78 |
| NM_015701 | ERLEC1 | endoplasmic reticulum lectin 1 | 61.29 |
| NM_004672 | MAP3K6 | mitogen-activated protein kinase kinase kinase 6 | 59.79 |
| NM_006415 | SPTLC1 | serine palmitoyltransferase, long chain base subunit 1 | 59.76 |
| NM_001025300 | RAB12 | RAB12, member RAS oncogene family | 59.16 |
| NM_005988 | SPRR2A | small proline-rich protein 2A | 58.97 |
| NM_001080541 | MGA | Homo sapiens MGA, MAX dimerization protein | 56.75 |
| NM_144569 | SPOCD1 | Homo sapiens SPOC domain containing 1 | 54.22 |
| NM_018357 | LARP6 | Homo sapiens La ribonucleoprotein domain family, member 6 | 54.17 |
| NM_206818 | OSCAR | osteoclast associated, immunoglobulin-like receptor | 53.30 |
| NM_017956 | TRMT12 | tRNA methyltransferase 12 homolog (S. cerevisiae) | 52.10 |
| NM_005614 | RHEB | Ras homolog enriched in brain | 50.16 |
| NM_012337 | CCDC19 | coiled-coil domain containing 19 | 50.03 |
| NM_014884 | SUGP2 | SURP and G patch domain containing 2 | 47.37 |
| NM_015335 | MED13L | mediator complex subunit 13-like | 47.11 |
| NM_173698 | FAM133A | family with sequence similarity 133, member A | -153.62 |
| NM_145290 | GPR125 | G protein-coupled receptor 125 | -78.33 |
| NM_017521 | FEV | Homo sapiens FEV | -77.72 |
| NM_001541 | HSPB2 | Homo sapiens heat shock 27kDa protein 2 | -67.21 |
| NM_032501 | ACSS1 | Homo sapiens acyl-CoA synthetase short-chain family member 1 | -63.80 |
| NM_021992 | TMSB15A | thymosin beta 15a | -55.18 |
| NM_012449 | STEAP1 | six transmembrane epithelial antigen of the prostate 1 | -44.93 |
| NM_017414 | USP18 | ubiquitin specific peptidase 18 | -44.70 |
| NM_001803 | CD52 | CD52 molecule | -44.63 |
| NM_004126 | GNG11 | guanine nucleotide binding protein (G protein), gamma 11 | -42.81 |
| NM_000519 | HBD | hemoglobin, delta | -40.08 |
| NM_033258 | GNG8 | guanine nucleotide binding protein (G protein), gamma 8 | -38.65 |
| NM_138444 | KCTD12 | potassium channel tetramerization domain containing 12 | -35.88 |
| NM_002866 | RAB3A | RAB3A, member RAS oncogene family | -35.15 |
| NM_014697 | NOS1AP | nitric oxide synthase 1 (neuronal) adaptor protein | -35.11 |
| NM_018437 | HEMGN | hemogen | -34.39 |
| NM_207459] | TEX19 | testis expressed 19 | -33.52 |
| NM_004982 | KCNJ8 | potassium inwardly-rectifying channel, subfamily J, member 8 | -33.13 |
| NM_013251 | TAC3 | tachykinin 3 222335545766788WWSSF BBGTT | -30.44 |
| NM_032333 | FAM213A | family with sequence similarity 213, member A | -29.38 |
| NM_213599 | ANO5 | anoctamin 5 | -29.37 |
| NM_130776 | XAGE3 | X antigen family, member 3 | -28.64 |
| NM_002585 | PBX1 | pre-B-cell leukemia homeobox 1 | -28.42 |
| NM_001110199 | SRRM3 | Homo sapiens serine/arginine repetitive matrix 3 | -28.20 |
| NM_000537 | REN | renin | -27.47 |
*Folds-change of treatment group compared to control analyzed by Genespring software analysis, Moderated T-test, p<0.05)
Table 2(a).
Most up- and down-regulated genes in TSA treated Kasumi 1
| Gene Bank Accession | Gene symbol |
Gene description
( Homo sapiens) |
*Folds
change |
|---|---|---|---|
| NM_139314 | ANGPTL4 | angiopoietin-like 4 | 791.26 |
| NM_182908 | DHRS2 | dehydrogenase/reductase (SDR family) member 2 | 612.16 |
| NM_001069 | TUBB2A | tubulin, beta 2A class IIa | 574.87 |
| NM_001080434 | LMTK3 | lemur tyrosine kinase 3 | 356.19 |
| NM_138345 | VWA5B2 | von Willebrand factor A domain containing 5B2 | 331.00 |
| NM_030630 | HID1 | HID1 domain containing | 331.00 |
| NM_006928 | PMEL | premelanosome protein | 323.68 |
| NM_145056 | DACT3 | dishevelled-binding antagonist of beta-catenin 3 | 269.03 |
| NM_144698 | ANKRD35 | ankyrin repeat domain 35, | 258.42 |
| NM_014971 | EFR3B | EFR3 homolog B (S. cerevisiae) | 248.79 |
| NM_004933 | CDH15 | cadherin 15, type 1, M-cadherin (myotubule) | 221.35 |
| NM_006086 | TUBB3 | tubulin, beta 3 class III | 205.73 |
| NM_000088 | COL1A1 | collagen, type I, alpha 1 | 122.33 |
| NM_017577 | GRAMD1C | GRAM domain containing 1C | 109.67 |
| NM_080860 | RSPH1 | radial spoke head 1 homolog | 109.55 |
| NM_003835 | RGS9 | regulator of G-protein signaling 9 | 103.85 |
| NM_001098722 | GNG4 | guanine nucleotide binding protein (G protein), gamma 4 | 102.41 |
| NM_005325 | HIST1H1A | histone cluster 1, H1a | 99.67 |
| NM_018667 | SMPD3 | sphingomyelin phosphodiesterase 3, neutral membrane (neutral sphingomyelinase II) | 98.71 |
| NM_033103 | RHPN2 | rhophilin, Rho GTPase binding protein 2 | 91.75 |
| NM_007224 | NXPH4 | neurexophilin 4 | 88.57 |
| NM_014226 | MOK | MOK protein kinase | 73.56 |
| NM_001077621 | VPS37D | vacuolar protein sorting 37 homolog D | 69.03 |
| NM_001145028 | PALM3 | paralemmin 3 | 66.97 |
| NM_177403 | RAB7B | RAB7B, member RAS oncogene family | -264.07 |
| NM_005574 | LMO2 | Homo sapiens LIM domain only 2 (rhombotin-like 1) | -215.33 |
| NM_001004196 | CD200 | CD200 molecule | -162.39 |
| NM_001146 | ANGPT1 | angiopoietin 1 | -159.45 |
| NM_003474 | ADAM12 | ADAM metallopeptidase domain 12 | -137.13 |
| NM_003942 | RPS6KA4 | Homo sapiens ribosomal protein S6 kinase, 90kDa, polypeptide 4 | -136.39 |
| NM_080588 | PTPN7 | protein tyrosine phosphatase, non-receptor type 7 | -133.96 |
| NM_130782 | RGS18 | regulator of G-protein signaling 18 | -119.12 |
| NM_033101 | LGALS12 | lectin, galactoside-binding, soluble, 12 | -94.20 |
| NM_002005 | FES | FES proto-oncogene, tyrosine kinase | -93.71 |
| NM_080387 | CLEC4D | C-type lectin domain family 4, member D | -93.00 |
| NM_024888 | LPPR3 | lipid phosphate phosphatase-related protein type 3 | -80.70 |
| NM_012252 | TFEC | transcription factor EC | -77.90 |
| NM_001805 | CEBPE | CCAAT/enhancer binding protein (C/EBP), epsilon | -69.46 |
| NM_014682 | ST18 | suppression of tumorigenicity 18, zinc finger | -67.63 |
| NM_002467 | MYC | v-myc avian myelocytomatosis viral oncogene homolog | -65.46 |
| NM_005263 | GFI1 | growth factor independent 1 transcription repressor | -64.45 |
| NM_153615 | RGL4 | ral guanine nucleotide dissociation stimulator-like 4 | -63.06 |
| NM_002287 | LAIR1 | leukocyte-associated immunoglobulin-like receptor 1 | -59.78 |
| NM_002586 | PBX2 | pre-B-cell leukemia homeobox 2 | -58.11 |
| NM_005211 | CSF1R | colony stimulating factor 1 receptor | -55.40 |
| NM_002831 | PTPN6 | protein tyrosine phosphatase, non-receptor type 6 | -52.38 |
| NM_000442 | PECAM1 | platelet/endothelial cell adhesion molecule 1 | -52.24 |
*Folds-change of treatment group compared to control analyzed by Genespring software analysis, Moderated T-test, p<0.05)
Table 2(b).
Most up- and down-regulated genes in 5-Aza treated Kasumi 1
| Gene Bank Accession | Gene symbol |
Gene description
( Homo sapiens) |
*Folds
change |
|---|---|---|---|
| NM_021120 | DLG3 | discs, large homolog 3 (Drosophila) | 14.12 |
| NM_033114 | ZCRB1 | zinc finger CCHC-type and RNA binding motif 1 | 12.82 |
| NM_001110514 | EBF4 | early B-cell factor 4 | 12.63 |
| NM_013271 | PCSK1N | proprotein convertase subtilisin/kexin type 1 inhibitor | 11.11 |
| NM_003278 | CLEC3B | C-type lectin domain family 3, member B | 9.44 |
| NM_003456 | ZNF205 | zinc finger protein 205 | 9.23 |
| NM_005252 | FOS | FBJ murine osteosarcoma viral oncogene homolog | 8.83 |
| NM_002840 | PTPRF | protein tyrosine phosphatase, receptor type F | 8.83 |
| NM_019058 | DDIT4 | DNA-damage-inducible transcript 4 | 8.17 |
| NM_002728 | PRG2 | proteoglycan 2, bone marrow | 7.82 |
| NM_001122962 | SIRPB2 | signal-regulatory protein beta 2 | 7.78 |
| NM_001039580 | MAP9 | microtubule-associated protein 9 | 7.46 |
| NM_080863 | ASB16 | ankyrin repeat and SOCS box containing 16 | 7.21 |
| NM_021158 | TRIB3 | tribbles pseudokinase 3 | 6.95 |
| NM_153334 | SCARF2 | scavenger receptor class F member 2 | 6.80 |
| NM_002390 | ADAM11 | ADAM metallopeptidase domain 11 | 5.63 |
| NM_032797 | AIFM2 | apoptosis-inducing factor, mitochondrion-associated 2 | 4.98 |
| NM_004626 | WNT11 | wingless-type MMTV integration site family, member 11 | 4.90 |
| NM_032271 | TRAF7 | TNF receptor-associated factor 7, E3 ubiquitin protein ligase | 3.67 |
| NM_001015053 | HDAC5 | histone deacetylase 5 | 3.67 |
| NM_001069 | TUBB2A | tubulin, beta 2A class IIa | 2.67 |
| NM_139314 | ANGPTL4 | angiopoietin-like 4 | 2.67 |
| NM_002831 | PTPN6 | protein tyrosine phosphatase, non-receptor type 6 | 2.27 |
| NM_001292030 | TTC39C | tetratricopeptide repeat domain 39C | -70.59 |
| NM_002844 | PTPRK | protein tyrosine phosphatase, receptor type K | -32.81 |
| NM_198481 | VSTM1 | V-set and transmembrane domain containing 1 | -32.49 |
| NM_000099 | CST3 | cystatin C | -26.47 |
| NM_001244008 | KIF1A | kinesin family member 1A | -22.49 |
| NM_001190467 | PRR36 | proline rich 36 | -21.97 |
| NM_024422 | DSC2 | desmocollin 2 | -20.96 |
| NM_001282735 | SPATS2L | spermatogenesis associated, serine-rich 2-like | -18.59 |
| NM_015238 | WWC1 | WW and C2 domain containing 1 | -16.52 |
| NM_021199 | SQRDL | sulfide quinone reductase-like (yeast) | -15.53 |
| NM_001838 | CCR7 | chemokine (C-C motif) receptor 7 | -13.97 |
| NM_000474 | TWIST1 | twist family bHLH transcription factor 1 | -13.27 |
| NM_012395 | CDK14 | cyclin-dependent kinase 14 | -13.19 |
| NM_000168 | GLI3 | GLI family zinc finger 3 | -12.65 |
| NM_024940 | DOCK5 | dedicator of cytokinesis 5 | -11.91 |
| NM_030906 | STK33 | serine/threonine kinase 33 | -11.90 |
| NM_001900 | CST5 | cystatin D | -11.86 |
| NM_006897 | HOXC9 | homeobox C9 | -11.74 |
| NM_005855 | RAMP1 | receptor (G protein-coupled) activity modifying protein 1 | -11.55 |
| NM_033292 | CASP1 | caspase 1, apoptosis-related cysteine peptidase | -11.50 |
| AK027605 | CYP2S1 | cytochrome P450, family 2, subfamily S, polypeptide 1 | -11.02 |
| NM_003474 | ADAM12 | ADAM metallopeptidase domain 12 | -7.681 |
| NM_172217 | IL16 | interleukin 16 | -4.46 |
| NM_001025300 | RAB12 | RAB12, member RAS oncogene f | -4.89 |
*Folds-change of treatment group compared to control analyzed by Genespring software analysis, Moderated T-test, p<0.05)
Table 2(c).
Most up- and down-regulated genes in TSA+5-Aza treated Kasumi 1
| Gene Bank Accession | Gene symbol | Gene description ( Homo sapiens) | *Folds change |
|---|---|---|---|
| NM_182908 | DHRS2 | dehydrogenase/reductase (SDR family) member 2 | 758.66 |
| NM_001080434 | LMTK3 | lemur tyrosine kinase 3 | 541.34 |
| NM_001069 | TUBB2A | tubulin, beta 2A class IIa | 435.79 |
| NM_139314 | ANGPTL4 | angiopoietin-like 4 | 429.60 |
| NM_138345 | VWA5B2 | von Willebrand factor A domain containing 5B2 | 398.46 |
| NM_030630 | HID1 | HID1 domain containing | 341.01 |
| NM_006928 | PMEL | premelanosome protein | 282.05 |
| NM_014971 | EFR3B | EFR3 homolog B (S. cerevisiae) | 263.45 |
| NM_144698 | ANKRD35 | ankyrin repeat domain 35 | 220.61 |
| NM_145056 | DACT3 | dishevelled-binding antagonist of beta-catenin | 219.77 |
| NM_004933 | CDH15 | cadherin 15, type 1, M-cadherin | 190.60 |
| NM_006086 | TUBB3 | tubulin, beta 3 class III | 173.87 |
| NM_001098722 | GNG4 | guanine nucleotide binding protein (G protein), gamma 4 | 167.50 |
| NM_080860 | RSPH1 | radial spoke head 1 homolog (Chlamydomonas) | 146.52 |
| NM_003835 | RGS9 | regulator of G-protein signaling 9 | 126.58 |
| NM_007224 | NXPH4 | neurexophilin 4 | 124.19 |
| NM_020770 | CGN | cingulin | 118.29 |
| NM_001145028 | PALM3 | paralemmin 3 | 114.39 |
| NM_000088 | COL1A1 | collagen, type I, alpha 1 | 111.63 |
| NM_003933 | BAIAP3 | BAI1-associated protein 3 | 107.26 |
| NM_017577 | GRAMD1C | GRAM domain containing 1C | 95.72 |
| NM_052899 | GPRIN1 | G protein regulated inducer of neurite outgrowth 1 | 95.72 |
| NM_005325 | HIST1H1A | histone cluster 1, H1a | 95.08 |
| NM_033141 | MAP3K9 | mitogen-activated protein kinase kinase kinase 9 | 92.48 |
| NM_198573 | ENHO | energy homeostasis associated | 92.06 |
| NM_001039570 | KREMEN1 | kringle containing transmembrane protein 1 | 91.54 |
| NM_018667 | SMPD3 | sphingomyelin phosphodiesterase 3 | 91.24 |
| NM_012253 | TKTL1 | transketolase-like 1 | 87.98 |
| NM_002599 | PDE2A | phosphodiesterase 2A, cGMP-stimulated | 84.11 |
| NM_033259 | CAMK2N2 | calcium/calmodulin-dependent protein kinase II inhibitor 2 | 80.49 |
| NM_014226 | MOK | MOK protein kinase | 79.66 |
| NM_001678 | ATP1B2 | ATPase, Na+/K+ transporting, beta 2 polypeptide | 78.33 |
| NM_006500 | MCAM | melanoma cell adhesion molecule | 75.94 |
| NM_001077621 | VPS37D | vacuolar protein sorting 37 homolog D | 74.87 |
| NM_052924 | RHPN1 | rhophilin, Rho GTPase binding protein 1 | 74.59 |
| NM_020127 | TUFT1 | tuftelin 1 | 73.36 |
| NM_001040709 | SYPL2 | synaptophysin-like 2 | 70.97 |
| NM_032432 | ABLIM2 | actin binding LIM protein family, member 2 | 70.76 |
| NM_001024401 | SBK1 | SH3 domain binding kinase 1 | 68.42 |
| NM_022742 | CCDC136 | coiled-coil domain containing 136 | 68.41 |
| NM_021979 | HSPA2 | heat shock 70kDa protein 2 | 67.51 |
| NM_000142 | FGFR3 | fibroblast growth factor receptor 3 | 65.65 |
| NM_033103 | RHPN2 | rhophilin, Rho GTPase binding protein 2 | 65.01 |
| NM_198196 | CD96 | CD96 molecule (CD96) | -228.86 |
| NM_001972 | ELANE | elastase, neutrophil expressed | -172.59 |
| NM_001244008 | KIF1A | kinesin family member 1A | -171.82 |
| NM_133374 | ZNF618 | zinc finger protein 618 | -169.32 |
| NM_020125 | SLAMF8 | SLAM family member 8 | -158.07 |
| NM_003974 | DOK2 | docking protein 2 | -153.14 |
| NM_080387 | CLEC4D | C-type lectin domain family 4, member D | -143.62 |
| NM_130782 | RGS18 | regulator of G-protein signaling 18 | -110.02 |
| NM_033101 | LGALS12 | lectin, galactoside-binding, soluble, 12 | -107.48 |
| NM_178443 | FERMT3 | fermitin family member 3 | -106.90 |
| NM_012072 | CD93 | CD93 molecule | -102.56 |
| NM_001946 | DUSP6 | dual specificity phosphatase 6 | -98.76 |
| NM_012252 | TFEC | transcription factor EC | -92.29 |
| NM_002467 | MYC | v-myc avian myelocytomatosis viral oncogene homolog | -91.05 |
| NM_001004196 | CD200 | CD200 molecule | -87.76 |
| NM_005814 | GPA33 | glycoprotein A33 (transmembrane) | -82.88 |
| NM_153615 | RGL4 | ral guanine nucleotide dissociation stimulator-like 4 | -81.77 |
| NM_080588] | PTPN7 | protein tyrosine phosphatase, non-receptor type 7 | -79.77 |
| NM_014795 | ZEB2 | zinc finger E-box binding homeobox 2 | -79.47 |
| NM_005211 | CSF1R | colony stimulating factor 1 receptor | -74.06 |
| NM_001146 | ANGPT1 | angiopoietin 1 | -70.80 |
| NM_006418 | OLFM4 | olfactomedin 4 | -70.64 |
| NM_014682 | ST18 | Homo sapiens suppression of tumorigenicity 18 | -68.89 |
| NM_177403 | RAB7B | RAB7B, member RAS oncogene family | -67.90 |
| NM_198481 | VSTM1 | V-set and transmembrane domain containing 1 | -66.89 |
| NM_005187 | CBFA2T3 | core-binding factor, runt domain, alpha subunit 2; translocated to, 3 | -61.51 |
| NM_003474 | ADAM12 | ADAM metallopeptidase domain 12 | -59.66 |
| NM_005574 | LMO2 | LIM domain only 2 | -58.27 |
| NM_080387 | CLEC4D | C-type lectin domain family 4, member D | -54.65 |
| NM_001805 | CEBPE | CCAAT/enhancer binding protein (C/EBP), epsilon | -48.73 |
*Folds-change of treatment group compared to control analyzed by Genespring software analysis, Moderated T-test, p<0.05)
Identification of an optimal Gene Ontology (GO) and KEGG pathway by DAVID software
GO analysis identified 13 optimal GO terms in MV4-11 after TSA, 5-Aza and TSA+5-Aza treatments constituted of 7 highly enriched biological processes (BP); Actin filament organization, Cytoskeleton organization, JAK-STAT, Blood coagulation, Positive regulation of activated T cell proliferation, Positive regulation of MAPK cascade and Cytoskeleton-dependent intracellular transport, related to 6 enriched molecular function (MF); GTPase activity, GTP binding, Structural constituent of cytoskeleton, Signal transducer activity, Polysaccharide binding, and Insulin-like growth factor receptor binding. The transduced GO terms were correspondent to 4 enriched KEGG pathway, which was Viral carcinogenesis, Hepatitis B, JAK-STAT and Phagosome (Table 3a).
Table 3(a).
Gene ontology (GO) profile after TSA, 5-Aza and TSA+5-Aza treatments in MV4-11
|
GO IDs
|
GO term
|
Genes
|
p-value
|
|---|---|---|---|
| Biological processes | |||
| GO:0007015 | Actin filamen organization | ARHGAP6, SPTA1, TPM2, TMSB15A | 0.0084 |
| GO:0007010 | Cytoskeleton organization | ABLIM3, TUBA1C, ANK1, TSPAN32, TUBB3 | 0.014 |
| GO:0007259 | JAK-STAT cascade | NMI, STAT5A, SOCS3 | 0.015 |
| GO:0007596 | Blood coagulation | CYP4F2, HBD, NFE2, THBD, TFPI | 0.022 |
| GO:0042102 | Positive regulation of activated T cell proliferation | CD24, IGF2, IL6 | 0.047 |
| GO:0043410 | positive regulation of MAPK cascade | TIMP2, IGF2, IL6 | 0.080 |
| GO:0030705 | Cytoskeleton-dependent intracellular transport | KIF5C, TUBA1C | 0.099 |
| Molecular Functions | |||
| GO:0003924 | GTPase activity | GNG11, GNG8, RHEB, RAB3A, TUBA1C, TUBB3 | 0.010 |
| GO:0005525 | GTP binding | GIMAP2, GIMAP6, RAB12, RAB3A, RHEB, TUBA1C, TUBB3 | 0.021 |
| GO:0005200 | Structural constituent of cytoskeleton | ANK1, SPTA1, TUBA1C, TUBB3 | 0.024 |
| GO:0004871 | Signal transducer activity | CD24, GNG11, GNG8, STAT5A, STAT6 | 0.028 |
| GO:0030247 | Polysaccharide binding | ENPP3, PRG4 | 0.076 |
| GO:0005159 | Insulin-like growth factor receptor binding |
IGF2, REN
|
0.081 |
| Pathways | |||
| Viral carcinogenesis | CCNA1, HDAC7, HIST1H2BN, STAT5A | 0.069 | |
| Hepatitis B | CCNA1, IL6, STAT5A, STAT6 | 0.084 | |
| JAK-STAT | SOCS3, IL6, STAT5A, STAT6 | 0.084 | |
| Phagosome | STX12, TUBA1C, TUBB3 | 0.10 |
(DAVID software analysis, EASE score 0.1, Benjamini p<0.1)
In Kasumi 1, 16 optimal GO terms by BP were identified; Cell adhesion, Leukocyte migration, Bone mineralization, Regulation of G-protein coupled receptor protein signaling pathway, Positive regulation of cell motility, phagocytosis, Peptidyl-tyrosine dephosphorylation, Protein localization to cell surface, Negative regulation of apoptotic process, Protein phosphorylation, Negative regulation of cell death, Hematopoiesis, Negative regulation of cell proliferation, Response to drug, Angiogenesis and Microtubule-based process, related to 8 MF; Protein tyrosine phosphatase activity, Transmembrane receptor protein tyrosine phosphatase activity, Carbohydrate-binding, Protein kinase activity, Heparin-binding, Protein serine/threonine kinase activity, Beta-catenin binding and Transcription factor binding. The most optimal KEGG pathway induced in Kasumi 1 were; Transcriptional misregulation in cancer, MAPK signaling pathway, PI3K-Akt signaling pathway, Pathways in cancer, Hippo signaling pathway, Proteoglycans in cancer, Ras signaling and Phagosome (Table 3b).
Table 3(b).
Gene ontology (GO) profile after TSA, 5-Aza and TSA+5-Aza treatments in Kasumi 1
|
GO IDs
|
GO term
|
Genes
|
P-value
|
|---|---|---|---|
| Biological processes | |||
| GO:0007155 | Cell adhesion | ADAM12, CDH15, COL1A1, PTPRK, PTPRF, DSC2, ATP1B2, CD96, DSC2, COL1A1, MCAM | 0.00093 |
| GO:0050900 | Leukocyte migration | ANGPTL1, COL1A1, ATP1B2, PECAM1, PTPN6, DOK2 | 0.0013 |
| GO:0030282 | Bone mineralization | CLEC3B, WNT11, FGFR3, TUFT1 | 0.0014 |
| GO:0008277 | Regulation of G-protein coupled receptor protein signaling pathway |
GNG4, RGS18, RGS9,
RAMP1 |
0.0022 |
| GO:2000147 | Positive regulation of cell motility | CCR7, CSF1R, TWIST1 | 0.0037 |
| GO:0006909 | Phagocytosis | CEBPE, CD93, ELANE, PECAM1 | 0.0039 |
| GO:0035335 | Peptidyl-tyrosine dephosphorylation | PTPN6, PTPN7, PTPRK,PTPRF, DUSP6 | 0.0042 |
| GO:0034394 | Protein localization to cell surface | WNT11, ANGPTL1, PTPRK | 0.0051 |
| GO:0043066 | Negative regulation of apoptotic process | GLI3, WNT11, ANGPTL1, ANGPTL4, CSF1R, DHRS2, TWIST1, MYC | 0.0068 |
| GO:0006468 | Protein phosphorylation | FES, MOK, WNT11, CDK14, LMTK3, TRIB3, RPS6KA4 | 0.024 |
| GO:0060548 | Negative regulation of cell death | WNT11, CST3, MYC | 0.030 |
| GO:0030097 | Hematopoiesis | ANGPTL1, CSF1R, GFI1 | 0.034 |
| GO:0008285 | Negative regulation of cell proliferation | PTPN6, PTPRK, GL13, CSF1R, DHRS2, DLG3CBFA2T3 | 0.048 |
| GO:0042493 | Response to drug | FOS, COL1A1, CST3, HDAC5, MYC | 0.062 |
| GO:0001525 | Angiogenesis | ANGPTL1, ANGPTL4, PECAM1, RAMP1, MCAM | 0.096 |
| GO:0007017 | Microtubule-based process | TUBB2A, TUBB3 | 0.10 |
| Molecular Functions | |||
| GO:0004725 | Protein tyrosine phosphatase activity | PTPN6, PTPN7, PTPRF, PTPRK, DUSP6 | 0.0038 |
| GO:0005001 | Transmembrane receptor protein tyrosine phosphatase activity | PTPN6, PTPRF, PTPRK | 0.0051 |
| GO:0030246 | Carbohydrate binding | CLEC3B, CLEC4B, PRG2, LGALS12 | 0.036 |
| GO:0004672 | Protein kinase activity | MOK, TRIB3, CDK14, LMTK3, STK33, MAP3K9 | 0.078 |
| GO:0008201 | Heparin binding | CLEC3B, ELANE, PTPRF, PRG2 | 0.081 |
| GO:0004674 | protein serine/threonine kinase activity | MOK, SBK1, LMTK3, MAP3K9, RPS6KA4, STK33 | 0.091 |
| GO:0008013 | Beta-catenin binding | GLI3, DACT3, PTPRK | 0.095 |
| GO:0003700 | Transcription factor binding | FOS, PBX2, HDAC5, TWIST1, MYC | 0.100 |
| Pathways | |||
| Transcriptional misregulation in cancer | CEBPE, LMO2, CSF1R, CDK14, MYC, ELANE | 0.0014 | |
| MAPK signaling pathway | FOS, PTPN7, MYC, RPS6KA4 | 0.010 | |
| PI3K-Akt signaling pathway | DDIT4, GNG4, ANGPTL1, COL1A1, CSF1R, FGFR3, MYC | 0.041 | |
| Pathway in cancer | FOS, GNG4, GLI3, WNT11, CSF1R, FGFR3, MYC | 0.069 | |
| Hippo signaling pathway | WWCI, WNT11, MYC, DLG3 | 0.10 | |
| Proteoglycans in cancer | WNT11, PTPN6, TWIST1, MYC | 0.18 | |
| Ras signaling | GNG4, ANGPTL4, CSF1R, FGFR3 | 0.23 | |
| Phagosome | TUBB2A, TUBB3 | 0.10 |
(DAVID software analysis, EASE score, p< 0.1)
Identification of Differentially Expressed Genes by Venn Diagram Configuration
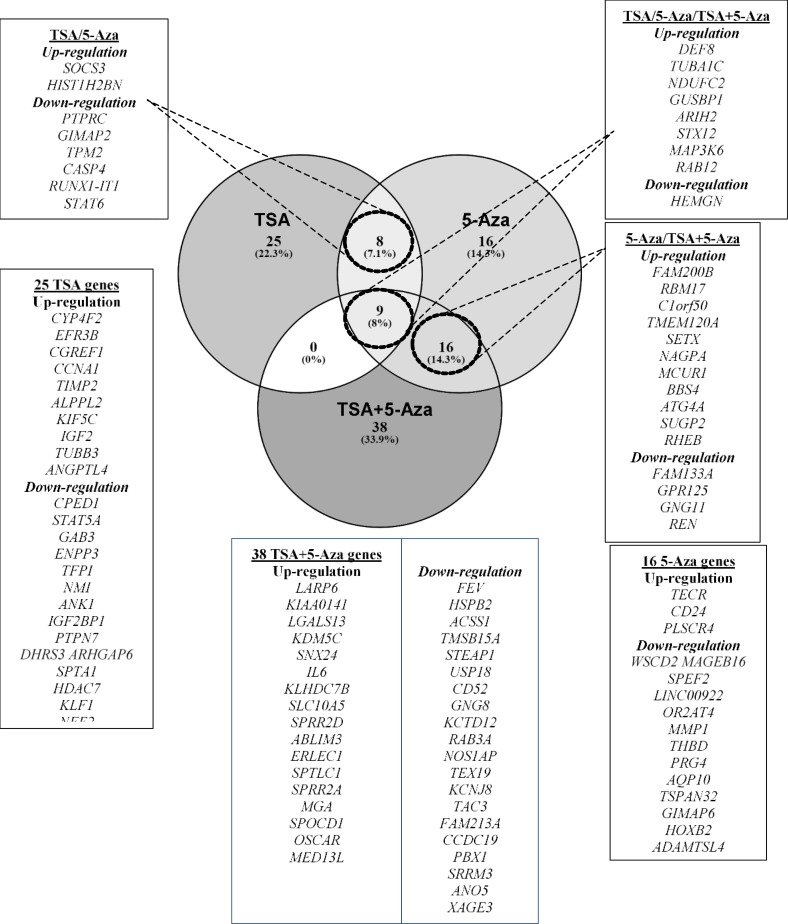
In MV4-11, out of 9 common differentially expressed genes between TSA, 5-Aza and TSA+5-Aza treatments, 8 genes (DEF8, GUSBP1, TUBA1C, NDUFC2, ARIH2, STX12, MAP3K6, and RAB12) were commonly up-regulated, while HEMGN was commonly down-regulated in all treatments. Between TSA and 5-Aza treatments, SOCS3 and HIST1H2BN were commonly up-regulated, but PTPRC, GIMAP2, TPM2, CASP4, RUNX1-IT1, and STAT6 were commonly down-regulated. 16 genes were commonly up-regulated in both 5-Aza and TSA+5-Aza treatments (FAM200B, RBM17, C1orf50, TMEM120A, SETX, NAGPA, MCUR1, BBS4, ATG4A, SUGP2, and RHEB). 5 down-regulated genes in 5-Aza (FAM133A, GPR125, GNG11, REN, and HBD) shared common down-regulation with TSA+5-Aza treatments. No gene in common was differentially expressed between TSA and TSA+5-Aza treatments. 25, 16 and 38 genes were exclusively expressed in TSA, 5-Aza and TSA+5-Aza, respectively as shown in Figure 3(a) (p<0.05).
Figure 3(a).
Venn diagram illustrating the genes commonly and exclusively expressed after TSA, 5-Aza and TSA+5-Aza treatments in MV4-11 (adhered to gene selection criteria).
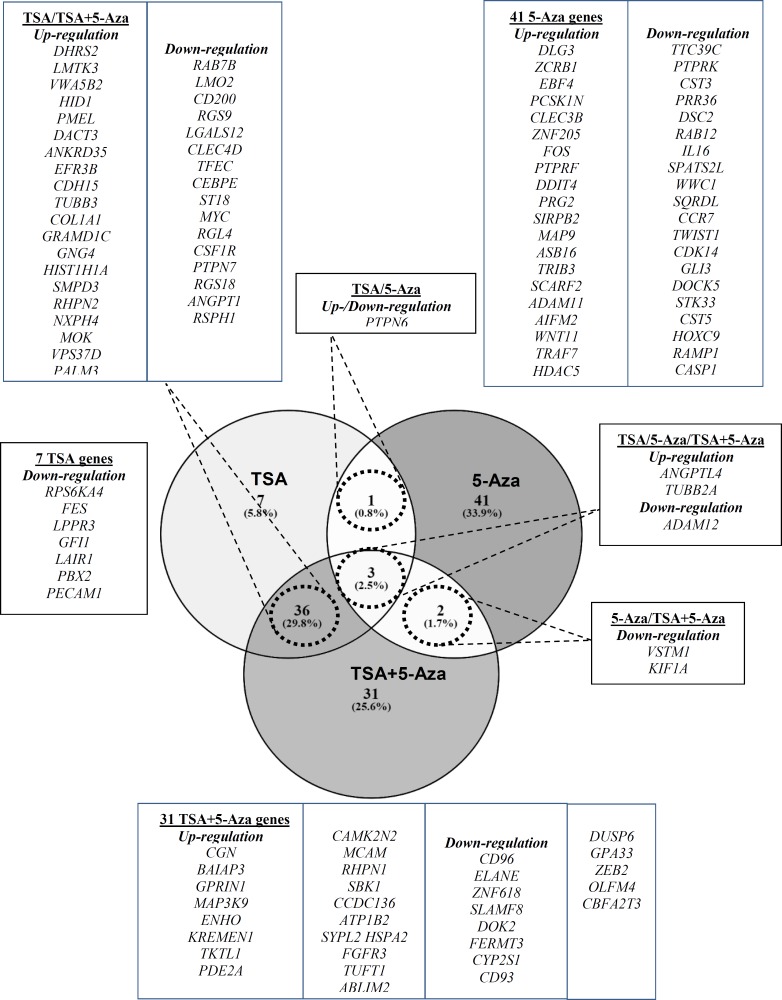
In Kasumi 1, there were 3 common differentially expressed genes across all treatments; 2 genes (ANGPTL4 and TUBB2A) and 1 gene (ADAM12) were commonly up-regulated and down-regulated, respectively. Whereas PTPN6 was either up-regulated in 5-Aza treatment or down-regulated in TSA. VSTM1 and KIF1A were commonly down-regulated in 5-Aza and TSA+5-Aza treatments. There were 36 genes commonly expressed in TSA and TSA+5-Aza treatments with 20 up-regulated and 16 down-regulated genes. 7, 41 and 31 genes were exclusively expressed in TSA, 5-Aza and TSA+5-Aza, respectively as shown in Figure 3(b) (p<0.05).
Figure 3(b).
Venn diagram illustrating the genes commonly and exclusively expressed after TSA, 5-Aza and TSA+5-Aza treatments in Kasumi 1(adhered to gene selection criteria).
Quantitative real-time PCR (qRT-PCR)
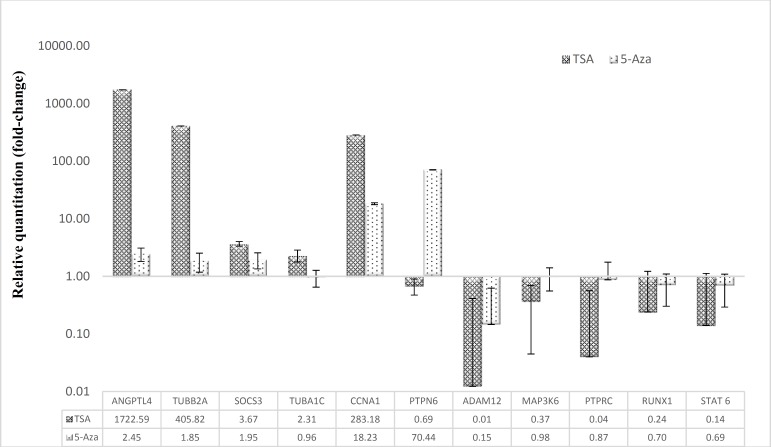
To verify the expression of genes, commonly up-regulated genes; SOCS3, TUBA1C, CCNA1, and MAP3K6 in MV4-11; ANGPTL4 and TUBB2A in Kasumi-1, and commonly down-regulated genes; STAT6, PTPRC and RUNX1 in MV4-11, ADAM12 and differentially expressed gene, PTPN6 in Kasumi 1 were selected for validation by qRT-PCR. The results were consistent with that of microarray in both MV4-11 and Kasumi 1 cell lines except for MAP3K6 in MV4-11 (Figure 4).
Figure 4.
Validation of expression levels of selected genes by qRT- PCR
The qRT-PCR results revealed a significant up- and down regulation of several genes in MV4-11 and Kasumi 1 treated with TSA and 5-Aza compared to non-treated cell lines. GAPDH and B2M were used as endogenous controls to which the expression was normalized. Shown in the bar graph is the standard error (SE) of duplicated samples.
Discussion
It was recognized that epigenetic changes serve as a mediator in cancer progression by the changes of gene expression. Epigenetic alterations are reported to concurrently disrupt the essential signaling pathway predisposed cell to uncontrolled growth, longer survival, and metastasis 14 . Histone modifications and DNA hypermethylation are two known epigenetic mechanisms that largely impact the regulation of gene transcription. Histone modification by acetylation has been found to be significantly deficient in acute leukemia patients, compared with the normal individual 15 . In this study, TSA acts by increasing the acetylation level by inhibiting HDAC activity in human leukemic cell lines. Histone acetylation is known to enhance the expression of specific genes that elicit extensive cellular morphology and metabolic changes, such as growth arrest, differentiation, and apoptosis 16 .
Aberrant DNA methylation was the most common epigenetic alteration in leukemia in which an increased level of DNA methylation was observed in AML at remission 17 . 5-Aza reverts DNA methylation to induce antineoplastic activity either by global hypomethylation and direct cytotoxicity on abnormal hematopoietic cells in the bone marrow 18 . 5-Aza inhibits DNMT thus to induce re-expression of the silenced genes to halt tumor growth, and to cause modest differentiation in transformed leukemic cell lines and primary AML 19 . The current study found that both TSA and 5-Aza inhibit the growth of MV4-11 and Kasumi 1 cell lines in a dose-dependent manner. The IC50 of both treatments at 24 hours were lower in MV4-11, compared to Kasumi 1 which could suggest the inhibitory effect of the drugs were less sensitive in Kasumi 1 harboring t(8;21) than in MV4-11 with FLT3-ITD mutation. The variation in the IC50 values would also represent different expression signature in response to TSA and 5-Aza treatments.
It is proposed that the genes which were commonly expressed within TSA, 5-Aza and TSA+5-Aza treatments were epigenetically regulated and involved in the pathogenesis of AML and may serve as candidates for potential biomarkers although they did not share similar GO profile and targeted different signaling pathways. DEF8, NDUFC2, GUSBP1, ARIH2, STX12 and HIST1H2BN were highly re-expressed (more than 100 folds) in either treatment of MV4-11, have not been previously discussed on their role in cancer except for HIST1H2BN. DEP8 is located at chromosome 16 encodes for an activator of intracellular signal transduction reported to carry single nucleotide polymorphism (SNP) rs4268748 at 16q24 with significantly associated with cell cycle regulator, CDK10 expression 20 . GUSBP1 which was located at chromosome 5 were involved in transcriptional regulation by putative alternative promoters (PAPs) 21 . ARIH2 primarily functions in neuronal differentiation was found to be tumor-specific in Glioblastoma multiforme (GBM) correlated with growth suppression in GBM cell lines 22 . Treatment with 5-aza-2′-deoxycytidine resulted in gene re-expression of HIST1H2BN in malignant ovarian cancer 23 . Differential down-regulation of HIST1H2BN was observed in meningiomas was associated with malignant progression 24 . RAB12 is a member of RAS oncogene family, function as small GTPase for intracellular protein transport, activated in stimulus-dependent pattern and promote microtubules-dependent of the cell secretary-granule in mast cell 25 and its up-regulation has been linked with colorectal cancer 26 .
The most optimal GO in MV4-11 were Cytoskeleton organization involving TUBA1C, JAK-STAT cascade involving SOCS3 and STAT6 and the cell cycle involving CCNA1, associated with Phagosome, JAK-STAT pathway and Viral carcinogenesis, respectively, CCNA1 was expressed after TSA treatment with high fold-change (298.44) in MV4-11, but was slightly re-expressed at a low level in 5-Aza and combination treatment (fold-change: 5.67 and 2.81, respectively) (results not shown). CCNA1, located at chromosome 13, encodes for activating regulatory subunit which binds to cyclin-dependent kinases 2 (CDK2) and cell division cycle 2 (CDC2) for the cell cycle machinery to progress into S phase 27 . In normal cells, CCNA1 was prominently expressed in testes, hematopoietic cells, and brain 28 . CCNA1 acts as tumor suppressor gene (TSG) which is epigenetically silenced by hypermethylation in cervical cancer 29 , ovarian, renal and lung carcinoma 30 . In AML, CCNA1 was found to be overexpressed especially in M3 and M2 AML with significant worse overall survival 31 . In addition, upregulation of CCNA1 was observed in leukemic cells in response to DNA damaging agents by increasing DNA repair process 32 . SOCS3, located at chromosome 17 is the known mediators in the JAK-STAT pathway which is strongly related to AML pathogenesis due to its function in blood lineage differentiation, apoptosis, and proliferation 33 . SOCS1, SOCS2 and SOCS3 negatively regulate JAK-STAT signaling in AML patients carrying a FLT3-ITD mutation 34 . SOCS3 has been extensively studied for over 20 years for their role in various diseases, especially in cancer. The most widely reported in SOCS3 was aberrant methylation affecting gene expression and protein function. Hypermethylation of promoter region of SOCS3 resulted in gene silencing implicated in cancer pathogenesis including hematological malignancies 35 , prostate cancer 36 , pancreatic cancer 37 , endometrial carcinoma 38 , hepatocellular carcinoma 39 and breast cancer 40 . Other candidate genes convoluted in the JAK-STAT pathway associated with hematological malignancies are STAT6 and RUNX1. TUBA1C, located at chromosome 12 is a member of tubulin family of microtubules ubiquitously expressed in the esophagus, bone marrow, appendix, brain, colon, bladder and placenta 41 . TUBA1C expression was significantly increased in hepatocellular carcinoma (HCC) on both mRNA and protein level, which predict a poor prognosis 42 , reduced expression in breast cancer associated invasive stage 43 and their expression was susceptible to colorectal cancer risk 44 . Cytochrome P450 (CYP4F2) was the highest re-expressed gene in TSA treatment with more than 1000 fold-change in MV4-11. CYP4F2 is a drug-metabolizing enzyme gene reported to have an epigenetic regulatory role with clinical implication 45 . Inhibition of DNMT and histone deacetylase (HDAC) by 5-Aza and TSA induced the demethylation of CYP1A1 and CYP1A2 leading to their up-regulation 46 .
In Kasumi 1, three common differentially expressed genes in either treatments were ANGPTL4, TUBB2A, and ADAM12 associated with angiogenesis, microtubule-based process, and cell-adhesion, respectively. ANGPTL4, located at chromosome 19 encodes a glycosylated, secreted protein containing a fibrinogen-like C-terminal domain, mainly induced by a nuclear receptor protein, peroxisome-proliferator-activated receptor (PPAR) 47 . It is the most studied among ANGPLT family, functions primarily in the regulation of lipid metabolism, glucose homeostasis, and insulin sensitivity 48 . ANGPTL4 has not been previously discussed in the context of AML. However previous studies have reported ANGPTL4 in various cancer types, including breast cancer, colorectal cancer, prostate cancer, hepatocarcinoma, and renal cell carcinoma, suggesting its important roles in cancer cell growth and progression 49 . In the current study, ANGPTL4 was mutually up-regulated in TSA treatment in both MV4-11 and Kasumi 1 cell lines, thus has wide potential for gene-specific therapy in AML. TUBB2A, located at chromosome 6 is another putative gene in AML with cell-specific expression. It forms a class ll beta-tubulin from six families of tubulins, including, alpha, gamma, delta, epsilon and zeta, and their protein may localize in extracellular exosome, cytoplasm and nucleus, involved in small GTPase activity, GTP binding, nucleotide binding acetylation and methylation 50 . Alpha and beta tubulin sub-families were studied for mutational analysis in human brain tumor and malformations was found in TUBB2A affecting the spectrum of "tubulinopathy" phenotypes51, 52. Mutations in TUBB2A were also explored in epilepsy 51 , gastric carcinoma and lung cancer 53 but not hematological malignancies. ADAM12, located at chromosome 10 was over-expression in non-Hodgkin’s lymphoma that lead to accelerate of proliferation and cell-adhesion 54 and was commonly methylated in chronic lymphocytic leukemia 55 . The roles of ADAM12 in leukemia pathogenesis is still obscure and need further study since the expression of this gene was similarly down-regulated in both treatments. PTPN6 (or SHP1) located at chromosome 12 was differentially regulated in TSA and 5-Aza treatments (re-expressed only in 5-Aza but not TSA). Our previous study showed a positive correlation of PTPN6 re-activation due to hypomethylation in MV4-11 that carry a FLT3-ITD mutation after the 5-Aza treatment 56 . PTPN6 expression has been studied in lymphoma, leukemia and other cancers such as breast cancer, ovarian cancer, prostate cancer, and pancreatic cancer 57 , and in hepatocellular carcinoma 58 . PTPN6 is a downstream mediator in the JAK-STAT pathway, and together with SOCS3 they potentially serve as molecular indicators for pathway-targeted therapy in AML. Another example of the methylation-related gene is PRG2. In the Venn diagram, PRG2 was exclusively expressed in 5-Aza treatment, but not in TSA treatment. The differentially expressed PRG2 was reported in three human leukemic cell lines (K562, THP1, and HL-60) 59 . We also previously reported that the expression of PRG2 was restored after 5-Aza treatment in PKC-412 (Midostaurin) resistant leukemic cell line 56 . DHRS2 and LMTK3 were another highly up-regulated genes in TSA treatment in Kasumi 1 with up to 500 fold change. Their up-regulation was due to histone acetylation.
Finally, despite thousands of genes generated by microarray expression profiling, the highly re-expressed and down-expressed genes perceived in this study were thought to be convoluted with epigenetic regulation of gene transcription in AML. Although only several genes were selected for validation by qRT-PCR, there were many other genes as discussed earlier that may have important roles in cancer pathogenesis.
CONCLUSION
In conclusion, we have identified common differently expressed genes that are importants in epigenetic regulation of AML. Our finding also revealed that Phagosome pathway was the most optimal and common in both MV4-11 and Kasumi 1 AML cell lines. Although MV4-11 and Kasumi 1 transduced different optimal signaling pathways in response to drug treatment, it was shown that MV4-11 mainly targeted the genes in the JAK-STAT signaling, while Kasumi 1 targeted the genes in transcriptional misregulation in cancer, PI3K-Akt and MAPK signaling, which are all critical pathways in oncogenesis. These were due to their different molecular characteristics (FLT3-ITD vs t(8;21) AML1-ETO). The data presented here may serve as a preliminary finding and are useful for further study to explore epigenetic involvement in the pathogenesis of AML.
ACKNOWLEDGEMENTS
This study was financially assisted by Research University grant (1001/PPSP/813050) and Bridging grant (304/PPSP/6316146) from Universiti Sains Malaysia.
CONFLICT OF INTEREST
The authors have no conflict of interest.
References
- 1.Babon J, Nicola NA. The biology and mechanism of action of suppressor of cytokine signaling 3 (SOCS3) Growth Factors. 2012;30(4):207–19. doi: 10.3109/08977194.2012.687375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arber DA, Orazi A, Hasserji RP, et al. Introduction and overview of the classification of myeloid neoplasms. WHO classification of tumors of haematopoietic and lymphoid tissues. Revised 4th Edition ed. Geneva: World Health Organization (WHO) Press; 2017. pp. 172–75. [Google Scholar]
- 3.American Cancer Society:Cancer Facts & Figures. Atlanta: American Cancer Society; c1913-2019 [updated 20 November 2018] [Accessed 24 October 2018]. American Cancer Society. Available from: https://www.cancer.org/research/cancer-facts-statistics/all-cancer-facts-figures/cancer-facts-figures-2018.html.
- 4.Pollack JR. A perspective on DNA microarrays in pathology research and practice. Am J Pathol. 2007;171(2):375–85. doi: 10.2353/ajpath.2007.070342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Golub TR, Slonim DK, Tamayo P, et al. Molecular Classification of Cancer: Class Discovery and Class Prediction by Gene Expression Monitoring. Science. 1999;286(5439):531–37. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 6.Kumar CC. Genetic abnormalities and challenges in the treatment of acute myeloid leukemia. Genes Cancer. 2011;2(2):95–107. doi: 10.1177/1947601911408076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li S, Mason CE, Melnick A. Genetic and epigenetic heterogeneity in acute myeloid leukemia. Curr Opin Genet Dev. 2016;36:100–06. doi: 10.1016/j.gde.2016.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.You JS, Jones PA. Cancer genetics and epigenetics: two sides of the same coin? Cancer Cell. 2012;22(1):9–20. doi: 10.1016/j.ccr.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis. 2010;31(1):27–36. doi: 10.1093/carcin/bgp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer. 2011;11(10):726–34. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hatziapostolou M, Iliopoulos D. Epigenetic aberrations during oncogenesis. Cell Mol Life Sci. 2011;68(10):1681–702. doi: 10.1007/s00018-010-0624-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nicolas D, Zoller B, Suter DM, et al. Modulation of transcriptional burst frequency by histone acetylation. Proc Natl Acad Sci USA. 2018;115(27):7153–58. doi: 10.1073/pnas.1722330115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hosack DA, Dennis G Jr, Sherman BT, et al. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4(10):R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kagohara LT, Stein-O'Brien GL, Kelley D, et al. Epigenetic regulation of gene expression in cancer: techniques, resources and analysis. Brief Funct Genomics. 2018 ;17(1):49–63. doi: 10.1093/bfgp/elx018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiao L, Huang Y, Zhen R, et al. Deficient Histone Acetylation in Acute Leukemia and the Correction by an Isothiocyanate. Acta Haematol. 2010;123(2):71–76. doi: 10.1159/000264628. [DOI] [PubMed] [Google Scholar]
- 16.Shankar S, Srivastava RK. Histone deacetylase inhibitors: mechanisms and clinical significance in cancer: HDAC inhibitor-induced apoptosis. Adv Exp Med Biol. 2008;615:261–98. doi: 10.1007/978-1-4020-6554-5_13. [DOI] [PubMed] [Google Scholar]
- 17.Agrawal S, Unterberg M, Koschmieder S, et al. DNA methylation of tumor suppressor genes in clinical remission predicts the relapse risk in acute myeloid leukemia. Cancer Res. 2007;67(3):1370–7. doi: 10.1158/0008-5472.CAN-06-1681. [DOI] [PubMed] [Google Scholar]
- 18.NCI Drug Dictionary: Azacitidine. Bethesda: US National Cancer Institute; [updated 1 August 2018]; [Accessed 25 October 2018]. Available from: https://www.cancer.gov/publications/dictionaries/cancer-drug/def/azacitidine. [Google Scholar]
- 19.Leone G, D'Alo F, Zardo G, et al. Epigenetic treatment of myelodysplastic syndromes and acute myeloid leukemias. Curr Med Chem. 2008;15(13):1274–87. doi: 10.2174/092986708784534947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Asgari MM, Wang W, Ioannidis NM, et al. Identification of Susceptibility Loci for Cutaneous Squamous Cell Carcinoma. J Invest Dermatol. 2016;136(5):930–37. doi: 10.1016/j.jid.2016.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kimura K, Wakamatsu A, Suzuki Y, et al. Diversification of transcriptional modulation: large-scale identification and characterization of putative alternative promoters of human genes. Genome Res. 2006;16(1):55–65. doi: 10.1101/gr.4039406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harisankar A. Identification of novel genes with important functions in glioblastoma multiforme and acute myeloid leukemia. Huddinge. Huddinge: Institute för medicine; 2018. [Google Scholar]
- 23.Liao YP, Chen LY, Huang RL, et al. Hypomethylation signature of tumor-initiating cells predicts poor prognosis of ovarian cancer patients. Hum Mol Genet. 2014;23(7):1894–906. doi: 10.1093/hmg/ddt583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pérez ME, Rodríguez de LÁ, Ribalta T, et al. Differential expression profiling analyses identifies downregulation of 1p, 6q, and 14q genes and overexpression of 6p histone cluster 1 genes as markers of recurrence in meningiomas. Neuro Oncol. 2010;12(12):1278–90. doi: 10.1093/neuonc/noq081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Efergan A, Azouz NP, Klein O, et al. Rab12 Regulates Retrograde Transport of Mast Cell Secretory Granules by Interacting with the RILP–Dynein Complex. J Immunol. 2016;196(3):1091–101. doi: 10.4049/jimmunol.1500731. [DOI] [PubMed] [Google Scholar]
- 26.Yoshida T, Kobayashi T, Itoda M, et al. Clinical omics analysis of colorectal cancer incorporating copy number aberrations and gene expression data. Cancer Inform. 2010;9:147–61. doi: 10.4137/cin.s3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lapenna S, Giordano A. Cell cycle kinases as therapeutic targets for cancer. Nat Rev Drug Discov. 2009;8(7):547–66. doi: 10.1038/nrd2907. [DOI] [PubMed] [Google Scholar]
- 28.National Cancer for Biotechnology Information (NCBI) Gene ID: 8900. CCNA1 cyclin A1 [Homo sapiens (human)] Bethesda: U.S. National Library of Medicine; c1988-2019 [updated updated 7 September 2018]; [Accessed 7 October 2018]. Available from: https://www.ncbi.nlm.nih.gov/gene/84790. [Google Scholar]
- 29.Yang N, Eijsink JJH, Lendvai Á, et al. Methylation Markers for CCNA1 & C13ORF18 Are Strongly Associated with High-Grade Cervical Intraepithelial Neoplasia and Cervical Cancer in Cervical Scrapings. Cancer Epidemiol Biomarkers Prev. 2009;18(11):3000. doi: 10.1158/1055-9965.EPI-09-0405. [DOI] [PubMed] [Google Scholar]
- 30.Rivera A, Mavila A, Bayless KJ, et al. Cyclin A1 is a p53-induced gene that mediates apoptosis, G2/M arrest, and mitotic catastrophe in renal, ovarian, and lung carcinoma cells. Cell Mol Life Sci. 2006;63(12):1425–39. doi: 10.1007/s00018-006-5521-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ekberg J, Holm C, Jalili S, et al. Expression of cyclin A1 and cell cycle proteins in hematopoietic cells and acute myeloid leukemia and links to patient outcome. Eur J Haematol. 2005;75(2):106–15. doi: 10.1111/j.1600-0609.2005.00473.x. [DOI] [PubMed] [Google Scholar]
- 32.Federico M, Symonds CE, Bagella L, et al. R-Roscovitine (Seliciclib) prevents DNA damage-induced cyclin A1 upregulation and hinders non-homologous end-joining (NHEJ) DNA repair. Mol Cancer. 2010;9:208. doi: 10.1186/1476-4598-9-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vainchenker W, Constantinescu SN. JAK/STAT signaling in hematological malignancies. Oncogene. 2013;32(21):2601–13. doi: 10.1038/onc.2012.347. [DOI] [PubMed] [Google Scholar]
- 34.Kazi JU, Ronnstrand L. Suppressor of cytokine signaling 2 (SOCS2) associates with FLT3 and negatively regulates downstream signaling. Mol Oncol. 2013;7(3):693–703. doi: 10.1016/j.molonc.2013.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fourouclas N, Li J, Gilby DC, et al. Methylation of the suppressor of cytokine signaling 3 gene in myeloproliferative disorders. Haematologica. 2008;93(11):1635. doi: 10.3324/haematol.13043. [DOI] [PubMed] [Google Scholar]
- 36.Pierconti F, Martini M, Pinto F, et al. Epigenetic silencing of SOCS3 identifies a subset of prostate cancer with an aggressive behavior. The Prostate. 2010;71(3):318–25. doi: 10.1002/pros.21245. [DOI] [PubMed] [Google Scholar]
- 37.Wang J, Zhou H, Han Y. SOCS3 methylation in synergy with Reg3A overexpression promotes cell growth in pancreatic cancer. J Mol Med (Berl) 2014;92(12):1257–69. doi: 10.1007/s00109-014-1184-8. [DOI] [PubMed] [Google Scholar]
- 38.Chen H, Zhang C, Sheng Y, et al. Frequent SOCS3 and 3OST2 promoter methylation and their epigenetic regulation in endometrial carcinoma. Am J Cancer Res. 2014 Dec 15;5(1):180–90. [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang X, You Q, Zhang X, et al. SOCS3 Methylation Predicts a Poor Prognosis in HBV Infection-Related Hepatocellular Carcinoma. Int J Mol Sci. 2015;16(9) doi: 10.3390/ijms160922662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barclay JL, Anderson ST, Waters MJ, et al. SOCS3 as a tumor suppressor in breast cancer cells, and its regulation by PRL. Int J Cancer. 2009;124(8):1756–66. doi: 10.1002/ijc.24172. [DOI] [PubMed] [Google Scholar]
- 41.National Cancer for Biotechnology Information (NCBI) Gene ID: 84790. TUBA1C tubulin alpha 1c [Homo sapiens (human)] Bethesda: U.S. National Library of Medicine; c1988-2019 [updated 7 September 2018]; [Accessed 7 October 2018]. Available from: https://www.ncbi.nlm.nih.gov/gene/84790. [Google Scholar]
- 42.Wang J, Chen W, Wei W, et al. Oncogene TUBA1C promotes migration and proliferation in hepatocellular carcinoma and predicts a poor prognosis. Oncotarget. 2017;8(56):96215–24. doi: 10.18632/oncotarget.21894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen D, Li Y, Wang L, et al. SEMA6D Expression and Patient Survival in Breast Invasive Carcinoma. Int J Breast Cancer. 2015;2015:539721. doi: 10.1155/2015/539721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Closa A, Cordero D, Sanz-Pamplona R, et al. Identification of candidate susceptibility genes for colorectal cancer through eQTL analysis. Carcinogenesis. 2014;35(9):2039–46. doi: 10.1093/carcin/bgu092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tang X, Chen S. Epigenetic Regulation of Cytochrome P450 Enzymes and Clinical Implication. Curr Drug Metab. 2015;16(2):86–96. doi: 10.2174/138920021602150713114159. [DOI] [PubMed] [Google Scholar]
- 46.Park HJ, Choi YJ, Kim JW, et al. Differences in the Epigenetic Regulation of Cytochrome P450 Genes between Human Embryonic Stem Cell-Derived Hepatocytes and Primary Hepatocytes. PLoS One. 2015;10(7):e0132992–e92. doi: 10.1371/journal.pone.0132992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.La Paglia L, Listi A, Caruso S, et al. Potential Role of ANGPTL4 in the Cross Talk between Metabolism and Cancer through PPAR Signaling Pathway. PPAR Res. 2017;2017:8187235. doi: 10.1155/2017/8187235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Genecards Human gene Database (GCID:GC19P008363) ANGPTL4 Gene (Protein Coding) Israel: Weizmann Institute of Science; c1996-2019 [updated 10 September 2018]; [Accessed 4 October 2018]. Available from: https://www.genecards.org/cgi-bin/carddisp.pl?gene=ANGPTL4. [Google Scholar]
- 49.Tan MJ, Teo Z, Sng MK, et al. Emerging roles of angiopoietin-like 4 in human cancer. Mol Cancer Res. 2012;10(6):677–88. doi: 10.1158/1541-7786.MCR-11-0519. [DOI] [PubMed] [Google Scholar]
- 50.UniProtKB - Q13885 (TBB2A_HUMAN) Protein knowledgebase (UniProtKB) Bethesda: National Institute of Health; c2002-2019 [updated 16 March 2018] [Accessed 21 August 2018]. Available from: https:// www.uniprot.org/uniprot/Q13885.
- 51.Cushion Thomas D, Paciorkowski Alex R, Pilz Daniela T, et al. De Novo Mutations in the Beta-Tubulin Gene TUBB2A Cause Simplified Gyral Patterning and Infantile-Onset Epilepsy. Am J Hum Genet. 2014;94(4):634–41. doi: 10.1016/j.ajhg.2014.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Romaniello R, Arrigoni F, Bassi MT, et al. Mutations in α- and β-tubulin encoding genes: Implications in brain malformations. Brain Dev. 2015;37(3):273–80. doi: 10.1016/j.braindev.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 53.The Human Protein Atlas: TUBB2A. Knut & Alice Wallenberg foundation 2018. [Accessed 24 june 2018]. Available from: https://v18.proteinatlas.org/ENSG00000137267-TUBB2A/tissue.
- 54.Zhong F, Ouyang Y, Wang Q, et al. Upregulation of ADAM12 contributes to accelerated cell proliferation and cell adhesion-mediated drug resistance (CAM-DR) in Non-Hodgkin’s Lymphoma AU - Yin, Haibing. Hematology. 2017;22(9):527–535. doi: 10.1080/10245332.2017.1312205. [DOI] [PubMed] [Google Scholar]
- 55.Rahmatpanah FB, Carstens S, Hooshmand SI, et al. Large-scale analysis of DNA methylation in chronic lymphocytic leukemia. Epigenomics. 2009;1(1):39–61. doi: 10.2217/epi.09.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Al-jamal H, Asmaa MJ, Sidek M, et al. Restoration of PRG2 Expression by 5-Azacytidine Involves in Sensitivity of PKC-412 (Midostaurin) Resistant FLT3-ITD Positive Acute Myeloid Leukaemia Cells. J Hematol Thrombo Dis. 2015;3(1):1–7. [Google Scholar]
- 57.Wu C, Sun M, Liu L, et al. The function of the protein tyrosine phosphatase SHP-1 in cancer. Gene. 2003;306:1–12. doi: 10.1016/s0378-1119(03)00400-1. [DOI] [PubMed] [Google Scholar]
- 58.Wen LZ, Ding K, Wang ZR, et al. SHP-1 acts as a Tumor Suppressor in Hepatocarcinogenesis and HCC Progression. Cancer Res. 2018;78(16):4680–4691. doi: 10.1158/0008-5472.CAN-17-3896. [DOI] [PubMed] [Google Scholar]
- 59.Wang H, Hu H, Zhang Q, et al. Dynamic transcriptomes of human myeloid leukemia cells. Genomics. 2013;102(4):250–6. doi: 10.1016/j.ygeno.2013.06.004. [DOI] [PubMed] [Google Scholar]