Abstract
The mode of binding of oligosaccharides to griffithsin, an antiviral lectin from the red alga Griffithsia sp., was investigated by a combination of X‐ray crystallography, isothermal titration calorimetry, and molecular modeling. The structures of complexes of griffithsin with 1→6α‐mannobiose and with maltose were solved and refined at the resolution of 2.0 and 1.5 Å, respectively. The thermodynamic parameters of binding of 1→6α‐mannobiose, maltose, and mannose to griffithsin were determined. Binding profiles of 1→6α‐mannobiose and mannose were similar with K d values of 83.3 μM and 102 μM, respectively. The binding of maltose to griffithsin was significantly weaker, with a fourfold lower affinity (K d = 394 μM). In all cases the binding at 30°C was entropically rather than enthalpically driven. On the basis of the experimental crystal structures, as well as on previously determined structures of complexes with monosaccharides, it was possible to create a model of a tridentate complex of griffithsin with Man9GlcNAc2, a high mannose oligosaccharide commonly found on the surface of viral glycoproteins. All shorter oligomannoses could be modeled only as bidentate or monodentate complexes with griffithsin. The ability to mediate tight multivalent and multisite interactions with high‐mannose oligosaccharides helps to explain the potent antiviral activity of griffithsin. Proteins 2007. © 2007 Wiley‐Liss, Inc.
Keywords: Lectin, anti‐HIV, anti‐viral, oligosaccharide binding, carbohydrate modeling
INTRODUCTION
Griffithsin is one of a number of lectins that have been shown to exhibit significant activity against human immunodeficiency virus (HIV), as well as against several other enveloped viruses. 1 Griffithsin was originally isolated from the red alga Griffithsia sp., collected from the waters off New Zealand, as part of a program at the National Cancer Institute to screen natural product extracts for their ability to inhibit the cytopathic effects of HIV.2 This lectin was shown to inhibit the cytopathic effects of different isolates of HIV‐1 at concentrations as low as 43 pM,2 as well as exhibited activity against viruses such as the coronavirus that causes severe acute respiratory syndrome (SARS).3 Crystal structures of two different recombinant forms of griffithsin,3 cloned with and without additional residues on the N terminus of the protein,4 have shown it to be a domain‐swapped dimer belonging to the β‐prism‐I (jacalin‐related) family,5 with each molecule exhibiting almost perfect internal three‐fold symmetry. Crystal structures of the complexes of griffithsin with monosaccharides such as mannose and N‐acetyl glucosamine, obtained by cocrystallization and/or soaking, have shown that each molecule contains three binding sites that form an almost perfect equilateral triangle, with each side ∼15 Å long. We have postulated that multivalency of binding might be the reason for the potency of this lectin,3 yet attempts to obtain experimental structures of complexes in the presence of oligosaccharides, such as Man9GlcNAc2, that are commonly found on viral envelope glycoproteins such as HIV1 gp120,6 Ebola virus gp17 and the SARS coronavirus spike proteins8 have so far been unsuccessful. The unique structure of griffithsin, which includes six independent sugar binding sites, provides opportunities to bind multiple individual sugars on large oligosaccharides such as Man9GlcNAc2, while viral envelope glycoproteins such as gp120 present an estimated 11 high mannose oligosaccharides to which griffithsin can bind.8 The importance of multivalency for antiviral lectin potency has previously been elaborated for the anti‐HIV protein cyanovirin‐N.9 The combination of multiple targets on one oligosaccharide together with an antiviral protein containing several lectin domains results in multivalent interactions, which are likely responsible for the unusual potency of griffithsin and also the difficulty in obtaining structural data on these large complexes. Thus a combination of crystallography, modeling, and biophysical measurements was found to be necessary in order to explain the antiviral properties of this potential future pharmaceutical agent.
MATERIALS AND METHODS
Crystallization and Structure Determination of Disaccharide Complexes
Recombinant griffithsin was expressed in Nicotiana benthamiana plants and purified according to methods described previously. 3 Crystals of the complexes of griffithsin with 1→6α‐mannobiose (Sigma, St. Louis, MO) and maltose (α‐D‐glucopyranosyl‐(1→4)‐α‐D‐glucopyranose) were obtained by the hanging drop, vapor diffusion method. The crystals of both complexes were grown in 1.8 M MgSO4, 0.1 M MES, pH 6.5, with 1:10 molar ratio of griffithsin monomers to disaccharide. Crystals grew in 5 days to the final size of ∼ 0.2 × 0.3 × 0.05 mm3. Before flash freezing, the crystals of the complex with maltose were transferred into a cryoprotectant solution containing 10% ethylene glycol, whereas no cryoprotectant was used for the mannobiose complex. Crystals of the complex with maltose belonged to space group P212121 (Table I), contained two molecules in the asymmetric unit, and diffracted to 1.5 Å resolution. The complex of griffithsin with 1→6α‐mannobiose crystallized in space group C2 (Table 1), also with two molecules in the asymmetric unit, and the crystals diffracted to 2 Å resolution.
Table I.
Statistics of Data Collection and Structure Refinement
| Crystal | ||
|---|---|---|
| Complex with maltose | Complex with 1 → 6α‐mannobiose | |
| Data collection | ||
| Space group | P212121 | C2 |
| Cell parameters | a = 34.82 Å, | a = 139.4 Å, |
| b = 53.26 Å, | b = 34.23 Å, | |
| c = 118.5 Å; | c = 55.11 Å; | |
| α = β = γ = 90° | α = 90°; β = 110.5°; γ = 90° | |
| Molecules/a.u. | 2 | 2 |
| Resolution (Å) | 30–1.5 | 30–2.0 |
| Total reflections | 584200 | 266987 |
| Unique reflections | 35264 | 15194 |
| Completeness (%)b | 99.9 (99.3) | 90.4 (45.4) |
| Avg. I/σ | 35.38 (3.3) | 15.03 (1.45) |
| R merge (%)c | 6.9 (44.6) | 10.1 (34.2) |
| Refinement statistics | ||
| R (%)d | 15.39 | 17.83 |
| R free (%)e | 18.54 | 24.25 |
| Rms deviations bond lengths (Å) | 0.021 | 0.021 |
| Rms deviations angles (degrees) | 1.96 | 1.97 |
| Pdb code | 2hyr | 2hyq |
aThe number of unique reflections is given first with Friedel pairs unmerged, and after merging in parentheses.
The values in parentheses relate to the highest resolution shell.
R merge = Σ|I − 〈I〉|/ΣI, where I is the observed intensity, and 〈I〉 is the average intensity obtained from multiple observations of symmetry‐related reflections after rejections.
R = Σ||Fo | − |Fc ||/Σ|Fo |, where F o and F c are the observed and calculated structure factors, respectively.
R free = defined by Brünger 22
X‐ray diffraction data were collected at the SER‐CAT beamline 22‐ID at the Advanced Photon Source (APS), Argonne, Illinois, on a MAR 300CCD detector. All data were processed and scaled using the HKL2000 package. 10 The structures of both complexes were solved by molecular replacement with PHASER,11 using as a model the coordinates of griffithsin dimer from the complex with mannose refined at 1.78 Å resolution (PDB code 2guc).3 For the complex with 1→6α‐mannobiose, the solution was found with the Z‐score of 20.7 after rotation function search and 39.0 after translation function search, and resulted in R = 0.238 and R free = 0.292 after a single run of restrained refinement with no rebuilding. The solution for the complex with maltose was found with the Z‐score of 14.3 after rotation function and 25.4 after translation function, and with R = 0.271 and R free = 0.288 after the first run of restrained refinement. The structures were further rebuilt with COOT12 and refined with REFMAC5.13 The results of the refinement are summarized in Table 1. Hydrogen bonding data were calculated with the program LIGPLOT.14
Isothermal Titration Calorimetry
All calorimetric titrations were carried out on a VP‐ITC titration calorimeter (Microcal Inc., Northampton, MA). In each experiment, 5 μL aliquots of a 12.6 mM solution of an oligosaccharide were titrated into a rapidly spinning (300 rpm) solution of 0.30 mM griffithsin (cell volume = 1.3472 mL) held at a constant temperature of 30.0°C. A total of 55 injections per experiment were conducted. All protein solutions were dialyzed and constituted in 10 mM PBS (3 mM NaN3), pH 7.4, and the exact concentration of griffithsin in each experiment was determined by amino acid analysis. The binding isotherms were corrected for the heats of dilution and were fitted to a one‐identical‐site model using the Origin ITC Analysis software according to manufacturer's protocols. From the binding curve, values for enthalpy, binding affinity and stoichiometry were extracted. Other thermodynamic parameters were calculated using ΔG = −RTlnK a (R = 1.986 (cal mol)/K) and ΔG = ΔH − TΔS. Protein content was quantified by amino acid analysis using a Hitachi model L‐8800 automated amino acid analyzer according to manufacturer protocols.
Modeling of Structures with High‐mannose Oligosaccharides
Molecular modeling was performed on a Silicon Graphics Fuel workstation using InsightII and Discover software (Accelrys Inc., San Diego, USA). Oligomannose structures (Man x GlcNAc2, x = 5–9) were generated using the database of glycosidic linkage conformations 15 and the results of conformational studies on Man9GlcNAc2 using NMR spectroscopy and molecular dynamics.16 A single domain of griffithsin was created from the coordinates of the 1.78 Å structure of the complex with mannose (PDB code 2guc)3 and included residues A1‐A18 and B19‐B121. The crystal structure of snowdrop agglutinin17 was obtained from the PDB database18 (accession code 1msa). Oligomannose glycans were docked to single domains or molecules of griffithsin and agglutinin, respectively, by overlaying the required terminal mannose residues of Man x GlcNAc2 on the experimentally determined mannose residues in the appropriate binding sites, and using simulated annealing to minimize the oligosaccharide structure while keeping the terminal oligosaccharide residues in the binding sites and the protein tertiary structure fixed. In all cases, minimal changes were needed to the protein side chains in order to accommodate the glycans. For bidentate binding of Man9GlcNAc2 to agglutinin, the side chain conformation of Lys38 had to be adjusted to remove steric clashes, and for tridentate binding of Man9GlcNAc2 to griffithsin, the side chain conformation of Tyr28 had to be adjusted very slightly in both the A and B molecules. The distances between the binding sites were taken as the distance between the C1 atoms of the monosaccharides in the binding sites. Similarly, the distance between an oligosaccharide residue and a binding site was taken as the distance between the C1 atom of the glycan residue and the C1 atom of the residue in the binding site. Oligomannose residues are numbered according to the standard nomenclature.19 The torsion angle nomenclature used for a 1‐x linkage is ϕ = O5—C1—O—C x′, ψ = C1—O—C x′—C x−1′ and ω = O—C6′—C5′—C4′.
Accession Numbers
Coordinates and structure factors have been deposited in the PDB with accession codes 2hyr and 2hyq for the experimentally determined complexes of griffithsin with maltose and 1→6α‐mannobiose, respectively, and 2i43 for the model of the griffithsin‐Man9GlcNAc2 complex.
RESULTS
Crystallographic Studies of the Mode of Binding of Disaccharides
Three forms of mannobiose (1→2α‐mannobiose, 1→3α‐ mannobiose, and 1→6α‐mannobiose) were used in crystallization trials. Extensive precipitation was observed after addition of 1→2α‐mannobiose and 1→3α‐mannobiose to the protein solution. and only crystallization of the complex with 1→6α‐mannobiose was successful. The resulting C‐centered monoclinic crystals (space group C2) contained a protein dimer in the asymmetric unit and were different from any previously observed crystal forms of griffithsin. The structure was solved by molecular replacement with data extending to 2.0 Å resolution, and was refined to the R factor of 0.178 (R free of 0.242). Superposition of these coordinates on the coordinates of the mannose complex of griffithsin solved at high resolution 3 yielded r.m.s. difference of 0.41 Å for the whole dimer. The largest differences were observed for Ser1A, Leu2A, Gly53B, and Ser54B. No significant movement of Tyr28, residue located in close proximity to the carbohydrate, was observed in either molecule.
Cocrystallization of griffithsin with another disaccharide, maltose, yielded an orthorhombic P212121 crystal form that was not isomorphous with any other previously observed crystals of this protein. The structure was solved by molecular replacement and refined at 1.5 Å resolution to an R of 0.154 and R free of 0.185. Despite the higher resolution of data, the temperature factors were still refined isotropically. The quality of the resulting electron density map was excellent, as exemplified by the almost perfect density of the hydrated Mg2+ ion that was found bound between two symmetry‐related molecules (Fig. 1A). A comparison of the structure of the maltose complex of griffithsin with the complex with mannose 3 yielded r.m.s. difference of 0.51 Å. The largest differences were observed for Ser1A, Ser19A, Gly41A, Gly55A, Gly41B, Ser42B, Gly43B, and Gly44B.
Figure 1.
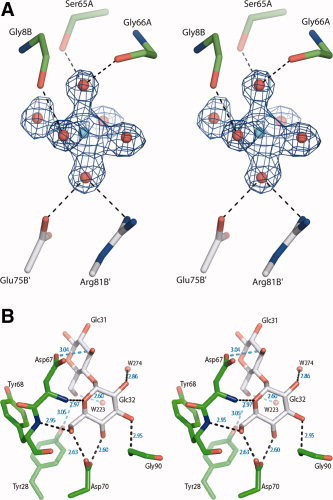
A complex of griffithsin with maltose. (A) Electron density (2F o‐F c, contour level 1σ) and the superimposed atomic model of the hydrated Mg2+ ion. The metal, found between two griffithsin molecules, is surrounded by six well‐determined water molecules, but makes no direct contacts with the protein. (B) Interactions of the maltose molecule with site 3 of molecule A of griffithsin, with hydrogen bonds indicated by dashed lines. Man32 is seen face on and Man31 edge on.
The interactions at the first sugar unit of 1→6α‐mannobiose or maltose (Fig. 1B) were very similar to interactions present in the complex with mannose. 3 The interactions between Man12, Man22, and Man32 with molecule A were almost identical to the interactions of Man42, Man52, and Man62 with molecule B at the complex with 1→6α‐mannobiose (in this nomenclature, the first digit after sugar name refers to its binding site on a griffithsin dimer, and the second to the sequence of the sugar in the oligosaccharide). Similarly, in the complex with maltose the interactions between Glc12, Glc22, and Glc32 and molecule A were very close to the interactions of Glc42, Glc52, and Glc62 with molecule B. A detailed comparison of the lengths of hydrogen bonds in the sugar binding site for the complexes is presented in Table II.
Table II.
Hydrogen Bonding (Å) in the Sugar Binding Sites for Complexes of Griffithsin With Different Carbohydrates
| Donor | Acceptor | Distance (Å) | ||||
|---|---|---|---|---|---|---|
| Complex with mannose | ||||||
| MAN | 1 | O4 | ASP A | 112 | OD2 | 2.55 |
| MAN | 1 | O6 | ASP A | 112 | OD1 | 2.79 |
| MAN | 1 | O6 | TYR A | 110 | O | 3.12 |
| TYR A | 110 | N | MAN | 1 | O6 | 2.79 |
| ASP A | 109 | N | MAN | 1 | O5 | 2.99 |
| ASP A | 109 | N | MAN | 1 | O6 | 2.99 |
| GLY A | 108 | N | MAN | 1 | O6 | 3.19 |
| GLY B | 12 | N | MAN | 1 | O3 | 2.84 |
| MAN | 2 | O4 | ASP A | 30 | OD2 | 2.46 |
| MAN | 2 | O6 | ASP A | 30 | OD1 | 2.70 |
| TYR A | 28 | N | MAN | 2 | O6 | 2.80 |
| SER A | 27 | OG | MAN | 2 | O5 | 3.70 |
| SER A | 27 | N | MAN | 2 | O5 | 2.94 |
| GLY A | 44 | N | MAN | 2 | O3 | 2.85 |
| MAN | 3 | O4 | ASP A | 70 | OD2 | 2.58 |
| MAN | 3 | O6 | ASP A | 70 | OD1 | 2.63 |
| TYR A | 68 | N | MAN | 3 | O6 | 2.87 |
| ASP A | 67 | N | MAN | 3 | O5 | 2.89 |
| GLY A | 66 | N | MAN | 3 | O6 | 3.06 |
| GLY A | 90 | N | MAN | 3 | O3 | 2.90 |
| GLY A | 12 | N | MAN | 4 | O3 | 2.87 |
| MAN | 4 | O4 | ASP B | 112 | OD2 | 2.53 |
| MAN | 4 | O6 | ASP B | 112 | OD1 | 2.73 |
| MAN | 4 | O6 | TYR B | 110 | O | 3.11 |
| TYR B | 110 | N | MAN | 4 | O6 | 2.76 |
| ASP B | 109 | N | MAN | 4 | O5 | 3.02 |
| ASP B | 109 | N | MAN | 4 | O6 | 2.98 |
| GLY B | 108 | N | MAN | 4 | O6 | 3.16 |
| GLY B | 44 | N | MAN | 5 | O3 | 2.95 |
| MAN | 5 | O4 | ASP B | 30 | OD2 | 2.46 |
| MAN | 5 | O6 | ASP B | 30 | OD1 | 2.61 |
| TYR B | 28 | N | MAN | 5 | O6 | 2.83 |
| SER B | 27 | N | MAN | 5 | O5 | 2.94 |
| GLY B | 90 | N | MAN | 6 | O3 | 2.81 |
| MAN | 6 | O4 | ASP B | 70 | OD2 | 2.59 |
| MAN | 6 | O6 | ASP B | 70 | OD1 | 2.69 |
| MAN | 6 | O6 | TYR B | 68 | O | 3.16 |
| TYR B | 68 | N | MAN | 6 | O6 | 2.93 |
| ASP B | 67 | N | MAN | 6 | O5 | 3.02 |
| Complex with 1→6α‐mannobiose | ||||||
| GLY B | 12 | N | MAN | 12 | O3 | 2.64 |
| MAN | 12 | O4 | ASP A | 112 | OD2 | 2.87 |
| MAN | 12 | O6 | ASP A | 112 | OD1 | 2.81 |
| MAN | 12 | O6 | TYR A | 110 | O | 3.21 |
| TYR A | 110 | N | MAN | 12 | O6 | 2.98 |
| ASP A | 109 | N | MAN | 12 | O5 | 3.16 |
| ASP A | 109 | N | MAN | 12 | O6 | 3.08 |
| GLY A | 108 | N | MAN | 12 | O6 | 3.12 |
| GLY A | 44 | N | MAN | 22 | O3 | 2.86 |
| MAN | 22 | O4 | ASP A | 30 | OD2 | 2.63 |
| MAN | 22 | O6 | ASP A | 30 | OD1 | 2.74 |
| MAN | 22 | O6 | TYR A | 28 | O | 2.96 |
| TYR A | 28 | N | MAN | 22 | O6 | 3.00 |
| SER A | 27 | N | MAN | 22 | O5 | 2.86 |
| GLY A | 90 | N | MAN | 32 | O3 | 2.82 |
| MAN | 32 | O4 | ASP A | 70 | OD2 | 2.63 |
| MAN | 32 | O6 | ASP A | 70 | OD1 | 2.78 |
| MAN | 32 | O6 | TYR A | 68 | O | 2.99 |
| TYR A | 68 | N | MAN | 32 | O6 | 2.65 |
| ASP A | 67 | N | MAN | 32 | O5 | 2.71 |
| ASP A | 67 | N | MAN | 32 | O6 | 2.90 |
| GLY A | 66 | N | MAN | 32 | O6 | 3.05 |
| GLY B | 90 | N | MAN | 42 | O3 | 3.01 |
| MAN | 42 | O4 | ASP B | 70 | OD2 | 2.60 |
| MAN | 42 | O6 | ASP B | 70 | OD1 | 2.61 |
| TYR B | 68 | N | MAN | 42 | O6 | 2.98 |
| ASP B | 67 | N | MAN | 42 | O5 | 2.89 |
| GLY B | 66 | N | MAN | 42 | O6 | 3.12 |
| GLY B | 44 | N | MAN | 52 | O3 | 3.10 |
| MAN | 52 | O4 | ASP B | 30 | OD2 | 2.70 |
| MAN | 52 | O6 | ASP B | 30 | OD1 | 2.71 |
| MAN | 52 | O6 | TYR B | 28 | O | 3.05 |
| TYR B | 28 | N | MAN | 52 | O6 | 2.92 |
| SER B | 27 | N | MAN | 52 | O5 | 2.81 |
| SER B | 27 | N | MAN | 52 | O6 | 2.94 |
| GLY B | 26 | N | MAN | 52 | O6 | 3.10 |
| MAN | 62 | O4 | ASP B | 112 | OD2 | 2.68 |
| MAN | 62 | O6 | ASP B | 112 | OD1 | 2.87 |
| MAN | 62 | O6 | TYR B | 110 | O | 3.08 |
| TYR B | 110 | N | MAN | 62 | O6 | 2.75 |
| ASP B | 109 | N | MAN | 62 | O5 | 2.86 |
| GLY B | 108 | N | MAN | 62 | O6 | 3.32 |
| GLY A | 12 | N | MAN | 62 | O3 | 3.03 |
| Second sugar unit | ||||||
| MAN | 31 | O4 | ASP A | 67 | OD2 | 2.99 |
| TYR B | 28 | OH | MAN | 41 | O2 | 2.97 |
| SER B | 27 | OG | MAN | 51 | O4 | 3.31 |
| Complex with maltose | ||||||
| GLC | 12 | O4 | ASP A | 112 | OD2 | 2.67 |
| GLC | 12 | O6 | ASP A | 112 | OD1 | 2.68 |
| GLC | 12 | O6 | TYR A | 110 | O | 3.09 |
| GLY B | 12 | N | GLC | 12 | O3 | 2.81 |
| TYR A | 110 | N | GLC | 12 | O6 | 2.97 |
| ASP A | 109 | N | GLC | 12 | O5 | 3.10 |
| GLY A | 108 | N | GLC | 12 | O6 | 3.29 |
| GLC | 22 | O4 | ASP A | 30 | OD2 | 2.58 |
| GLC | 22 | O6 | ASP A | 30 | OD1 | 2.81 |
| GLC | 22 | O6 | TYR A | 28 | O | 3.19 |
| TYR A | 28 | N | GLC | 22 | O6 | 2.92 |
| SER A | 27 | N | GLC | 22 | O5 | 2.95 |
| SER A | 27 | N | GLC | 22 | O6 | 3.03 |
| GLY A | 26 | N | GLC | 22 | O6 | 3.14 |
| GLY A | 44 | N | GLC | 22 | O3 | 2.95 |
| GLY A | 90 | N | GLC | 32 | O3 | 2.95 |
| GLC | 32 | O4 | ASP A | 70 | OD2 | 2.60 |
| GLC | 32 | O6 | ASP A | 70 | OD1 | 2.63 |
| GLC | 32 | O6 | TYR A | 68 | O | 3.19 |
| TYR A | 68 | N | GLC | 32 | O6 | 2.95 |
| ASP A | 67 | N | GLC | 32 | O5 | 2.97 |
| GLY A | 66 | N | GLC | 32 | O6 | 3.16 |
| GLC | 42 | O4 | ASP B | 112 | OD2 | 2.68 |
| GLC | 42 | O6 | ASP B | 112 | OD1 | 2.77 |
| TYR B | 110 | N | GLC | 42 | O6 | 2.88 |
| ASP B | 109 | N | GLC | 42 | O5 | 3.04 |
| ASP B | 109 | N | GLC | 42 | O6 | 3.07 |
| GLY A | 12 | N | GLC | 42 | O3 | 2.95 |
| GLY B | 44 | N | GLC | 52 | O3 | 2.83 |
| GLC | 52 | O4 | ASP B | 30 | OD2 | 2.58 |
| GLC | 52 | O6 | ASP B | 30 | OD1 | 2.64 |
| GLC | 52 | O6 | TYR B | 28 | O | 3.17 |
| TYR B | 28 | N | GLC | 52 | O6 | 2.97 |
| SER B | 27 | N | GLC | 52 | O5 | 2.98 |
| GLY B | 90 | N | GLC | 62 | O3 | 3.03 |
| GLC | 62 | O4 | ASP B | 70 | OD2 | 2.58 |
| GLC | 62 | O6 | ASP B | 70 | OD1 | 2.68 |
| GLC | 62 | O6 | TYR B | 68 | O | 3.12 |
| TYR B | 68 | N | GLC | 62 | O6 | 2.89 |
| ASP B | 67 | N | GLC | 62 | O5 | 3.05 |
| ASP B | 67 | N | GLC | 62 | O6 | 3.04 |
| GLY B | 66 | N | GLC | 62 | O6 | 3.08 |
| Second sugar unit | ||||||
| SER A | 27 | OG | GLC | 221 | O6 | 3.21 |
| GLC | 31 | O6 | TYR A | 28 | OH | 3.05 |
| GLC | 31 | O3 | ASP A | 67 | OD2 | 3.04 |
| TYR B | 28 | OH | GLC | 61 | O6 | 2.97 |
| GLC–GLC interactions | ||||||
| GLC | 21 | O3 | GLC | 22 | O2 | 2.94 |
| GLC | 62 | O2 | GLC | 61 | O4 | 2.75 |
| GLC | 52 | O2 | GLC | 51 | O4 | 2.80 |
| GLC | 51 | O3 | GLC | 52 | O2 | 3.28 |
| GLC | 42 | O2 | GLC | 41 | O4 | 2.67 |
| GLC | 41 | O3 | GLC | 42 | O2 | 3.29 |
| GLC | 32 | O2 | GLC | 31 | O4 | 2.78 |
| GLC | 221 | O3 | GLC | 22 | O2 | 2.98 |
| GLC | 22 | O2 | GLC | 21 | O4 | 2.77 |
| GLC | 11 | O3 | GLC | 12 | O2 | 3.04 |
| GLC | 22 | O2 | GLC | 221 | O4 | 2.77 |
The structures of all six maltose molecules bound to a griffithsin dimer could be superimposed almost exactly (Fig. 2A), whereas the deviations were much larger for 1→6α‐mannobiose (Fig. 2B). The quality of the electron density maps for the latter complex was significantly worse than for the former, as reflected by the temperature factors of the sugar residues. The average B factors were 32.9 Å2 for Glc11‐Glc61, and 19.6 Å2 for Glc12‐Glc62 of maltose, whereas the corresponding values were 83.7 Å2 for Man11‐Man61 and 52.4 Å2 for Man12‐Man62 of 1→6α‐mannobiose, indicating considerable flexibility of the carbohydrates in the latter complex, and resulting in the difficulties of establishing the exact conformation of the carbohydrate molecules.
Figure 2.

Superposition of the coordinates of the disaccharide molecules bound to griffithsin. Molecules 21–26 are shown face down and molecules 11–16 face up. (A) Superposition of the six molecules of maltose. Atom names are identified. (B) Superposition of the six molecules of 1→6α‐mannobiose.
In both the 1→6α‐mannobiose and maltose complexes, the second sugar unit was more flexible and the pattern of interactions was not as regular as for the first one. In the complex with 1→6α‐mannobiose, the following contacts were made: O4 of Man31 with Asp67A (2.99 Å), O2 of Man41 with Tyr28B (2.97 Å), and O4 of Man51 with Ser27B (3.31 Å). In the complex with maltose, O6 of Glc21 was in contact with Ser27A OG (3.21 Å), O6 of Glc31 interacted with the hydroxyl group of Tyr28A (3.05 Å), O3 of Glc31 was in contact with Asp67A (3.04 Å), and O6 of Glc61 interacted with OH group of Tyr28B (2.97 Å).
Measurements of the Thermodynamic Properties of Carbohydrate Binding
Calorimetric titrations of mannose, 1→6α‐mannobiose and maltose with griffithsin revealed that the energetics of binding were similar for these sugars. As demonstrated by the isotherms (Fig. 3, Table III), all sugars exhibited weakly exothermic (negative ΔH values) binding with griffithsin. The binding isotherms were fitted to a one‐identical‐site model consistent with previous (and present) structural data, showing that glycans bind identically at all sites on griffithsin. The resulting equilibrium dissociation constants of the binding of mannose and 1→6α‐mannobiose to griffithsin were comparable (K d of 102 μM and 83 μM, respectively), with maltose binding much more weakly (K d of 394 μM).
Figure 3.

Calorimetric titrations of griffithsin with mannose (red), 1(6α‐mannobiose (blue) and maltose (green). The solid lines represent fits to the data. The overall ΔH (kcal/mol) of binding are compared by superposition of all three isotherms.
Table III.
Thermodynamic Parameters Determined for the Binding of Selected Oligosaccharides to Griffithsin
| Affinity (μM) | ΔH (cal/mol) | ΔG (kcal/mol) | −TΔS (kcal/mol) | Stoichiometry | |
|---|---|---|---|---|---|
| Mannose | 102.0 ± 13.0 | −81.1 ± 1.6 | −5.53 ± 0.08 | −5.45 ± 0.08 | 6.30 ± 0.18 |
| 1→6α‐mannobiose | 83.3 ± 17.5 | −71.6 ± 2.0 | −5.65 ± 0.12 | −5.58 ± 0.12 | 6.4 ± 0.13 |
| Maltose | 394.0 ± 192.6 | −47.6 ± 5.3 | −4.72 ± 0.26 | −4.67 ± 0.26 | 5.94± 0.35 |
Although the bindings of the glycans at 30°C were mildly exothermic, in all three cases the interactions were entropically favored (TΔS were two orders of magnitude larger than ΔH). Such a large positive change in the entropy could only be due to that of the water molecules being excluded from the binding interface, and this then providing the driving force for association, outweighing any loss of translational and configurational entropy of the ligand or the side chains of the protein at the binding site. The higher affinity of griffithsin for 1→6α‐mannobiose compared to mannose can then be rationalized by the larger glycan displacing more water from the binding site. The weaker binding of maltose compared to mannose or 1→6α‐mannobiose suggested that either less water was excluded or that conformational entropy of the ligand was lost upon binding (the latter being consistent with the much greater degree of order seen for the bound maltose glycan compared to 1→6α‐mannobiose, Fig. 2). The negative enthalpies of binding indicated that favorable protein‐ligand interactions (polar, van der Waals, and hydrogen bonds) compensated for any loss of solvation. The similarities in values of ΔH for the glycans were consistent with the glycans making similar numbers of contacts to the protein (an extra hydrogen bond, for example, would be worth an additional 2 kcal/mol).
Modeling of ManxGlcNAc2 Binding to Griffithsin
Only one monomer of griffithsin was used for the purpose of modeling, with the second molecule of the domain‐swapped dimer assumed to form a complex identical to that of the first one. The separation of the three adjacent mannose binding sites on griffithsin was relatively small, each approximately 15 Å apart from the others. Man9GlcNAc2 (Fig. 4A) was modeled to interact in a tridentate fashion to these three binding sites, in all three possible configurations (i.e. independent of which terminal mannose residue was bound to which binding site). In the model discussed here, Man11 corresponds to the terminal mannose of the D1 arm, Man8 to the terminal mannose of the D2 arm, and Man6 to the terminal mannose of the D3 arm of Man9GlcNAc2 (Fig. 4B). The resulting structure had all of the glycosidic linkages within their observed experimental ranges (Table IV), corresponding to one of the minimum energy conformations of the oligosaccharide. 16 None of the smaller oligomannose glycans appeared to be able to bind to griffithsin in a tridentate fashion. Each of the three Man8GlcNAc2 isomers (missing the terminal mannose on either of the D1, D2, or D3 arms) could form a bidentate complex using the two remaining terminal mannose residues, as could the three Man7GlcNAc2 isomers missing either residues D2 and A, residues D3 and B or residues D1 and C Man9GlcNAc2. The most common Man5GlcNAc2 isomer (missing terminal mannose residues on the D1, D2, D3 arms as well as the removal of a second mannose residue from the D1 arm) could only form a mondentate complex with griffithsin, binding of residue B giving a minimum distance of 6 Å between residue C and the nearest binding site. Thus, Man9GlcNAc2 could bind to griffithsin with high affinity by interacting with up to three binding sites whereas Man5GlcNAc2 could only bind in a mondentate fashion.
Figure 4.
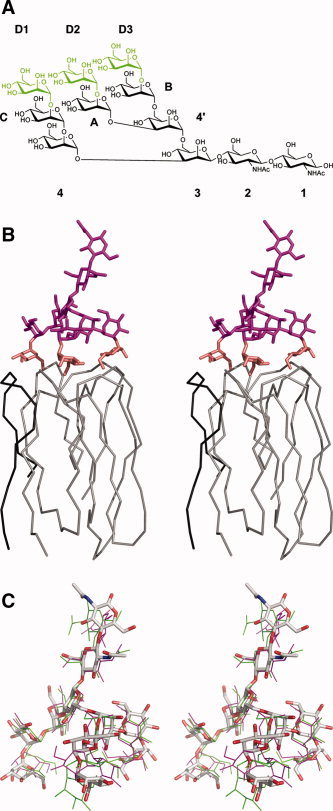
Modeling of the interactions of Man9GlcNAc2 with griffithsin. (A) Chemical structure of Man9GlcNAc2, with the three terminal mannose residues that make direct contact with griffithsin colored green. In the resulting model described here, mannose D1 is equivalent to Man11, D2 to Man8, and D3 to Man6. (B) Chain tracing of a single domain of griffithsin, consisting of residues A1–A18 (black) and B19–B121 (gray), together with the model of bound Man9GlcNAc2. The latter is colored in magenta, except of the three terminal residues that are colored in gold. (C) Superposition of the coordinates of the model of Man9GlcNAc2 complexed with griffithsin (colored sticks) with the experimentally determined structures of Man9GlcNAc2 in complex with the specific antibody 2G12 25 (thin green and magenta lines).
Table IV.
Glycosidic Linkage Conformations of Man9GlcNAc2 Modeled as a Tridentate Ligand to Griffithsin and a Bidentate Ligand to Snowdrop Agglutinin, Compared with the Database Values 11
| Linkage | Database | Torsion angles (ϕ,Ψ,ω) | ||
|---|---|---|---|---|
| Griffithsin | Agglutinin | |||
| 2‐1 | GlcNAcβ1‐4GlcNAc | −76, 119 | −75, 119 | −74, 120 |
| 3‐2 | Manβ1‐4GlcNAc | −87, 111 | −86, 112 | −87, 118 |
| 4‐3 | Manα1‐3Man | 72, −121 | 72, −118 | 69, −116 |
| C‐4 | Manα1‐2Man | 63, −180 | 74, −103 | 75, −99 |
| 71, −106 | ||||
| D1‐C | Manα1‐2Man | 63, −180 | 80, −107 | 43, −135 |
| 71, −106 | ||||
| 4′‐3 | Manα1‐6Man | 59, 94, 189 | 60,173,60 | 76, 161, 66 |
| 67, 179, 186 | ||||
| 65, 182, 60 | ||||
| A‐4′ | Manα1‐3Man | 72, −121 | 63, −126 | 71, −121 |
| D2‐A | Manα1‐2Man | 63, −180 | 68, −121 | 77,‐98 |
| 71, −106 | ||||
| B‐4′ | Manα1‐6Man | 59, 94, 189 | 64,187,174 | 75, −167,185 |
| 67, 179, 186 | ||||
| 65, 182, 60 | ||||
| D3‐B | Manα1‐2Man | 63, −180 | 64, −114 | −88, −79 |
| 71, −106 | ||||
Modeling of the Binding of ManxGlcNAc2 to Snowdrop Agglutinin
Snowdrop agglutinin resembles griffithsin in having three carbohydrate‐binding sites arranged as an equilateral triangle, although these proteins do not share any other structural resemblance. 17 Since the anti‐retroviral activity of this agglutinin was found to be at least three orders of magnitude weaker than that of griffithsin,2, 20 its structure could serve as a negative control for the attempts to find the structural basis of the activity of griffithsin. It was not possible to model tridentate binding of Man9GlcNAc2 to snowdrop agglutinin (i.e. with all three terminal mannose residues binding to the protein) because of the relatively large separation of the mannose binding sites on the protein (binding sites are separated by 20 Å or more). It was possible to dock Man9GlcNAc2 to snowdrop agglutinin with two of the terminal mannose residues (on the D1 and D3 arms) binding to the protein (either sites A112 and B112, or sites C112 and D112 labeled as in the PDB file). However, this required significant distortions to the usual torsion angles for at least two of the glycosidic linkages in the oligosaccharide (Table IV). For instance, the Manα1→2Man linkage of the D3 residue had to be in a conformation with ϕ, ψ values in the region of −90°, −80°. This linkage is normally only observed with ϕ, ψ values in the region +70°, −105°.15 It was not possible to model bidentate binding of any smaller oligomannose oligosaccharides to snowdrop agglutinin. It therefore seemed plausible that snowdrop agglutinin normally only binds in a monodentate fashion to oligomannose glycans, achieving its relatively high affinity20 through binding to multiply presented oligosaccharides.
DISCUSSION
The combination of crystallographic and calorimetric data for the complexes of griffithsin with carbohydrates was useful in elucidating a number of aspects of sugar binding to this protein, including its specificity for mannose. The only stereochemical difference between the glucose and mannose residues of maltose and 1→6α‐mannobiose was in the conformation at the C2 carbon of the sugar ring. Oxygen O2 pointed in the opposite direction in the structures of the two complexes, but in neither of them was it found to be involved in hydrogen bonds other than to solvent molecules. Therefore, it would appear that the principal binding site of griffithsin should not be specific for either mannose or glucose. However, in examining the calorimetric titration data, we observed clear distinction between the bindings of maltose and that of 1→6α‐mannobiose (or mannose). The mannose‐containing sugars bound griffithsin with more that fourfold greater affinity than did maltose. In addition, precipitation was observed only after addition of 1→2α‐mannobiose and 1→3α‐mannobiose to the protein solution but not 1→6α‐mannobiose. If this precipitation is due to crosslinking then the lack of precipitation with the 1→6α‐mannobiose could be explained by limited accessibility of the second mannose to additional griffithsin molecules. Additional studies are planned to further clarify this result.
Crystal structures and modeling results clearly indicated that in order to bind larger carbohydrates with high affinity, griffithsin utilizes multiple, monosaccharide‐specific binding sites in a multivalent binding mode. In this respect this algal lectin is very different from other antiviral proteins of cyanobacterial origin, such as cyanovirin, 21 scytovirin,22 or MVL,23 which achieved high affinity through an extended binding site. The latter lectins specifically recognized larger oligosaccharide structures on the terminal branch (cyanovirin), D3 arm (scytovirin), or four‐five units of the oligomannoside core (MVL). Griffithsin is also significantly different from other multivalent lectins such as mannose‐binding protein (MBP). MBP consists of trimeric units of carbohydrate recognition domains (CRDs), each CRD monomer of the trimer having one low‐affinity mannose binding site (K d ∼ 3 mM). Each CRD trimer is attached to a long collagen‐like domain and these oligomerize to form tetramers to hexamers of trimers, each MBP oligomer having twelve to fifteen mannose binding sites. However, the arrangement of these binding sites is such that a single oligomannose molecule cannot interact with more than one CRD. Thus, high affinity is only achieved by multiple binding to multiply presented oligomannose molecules on a cell surface,24 as discussed above for agglutinin. Conversely, such high affinity for individual oligomannose glycans is likely to be responsible for the potent antiviral activity displayed by griffithsin against enveloped viruses such as HIV and the SARS coronavirus.
We have shown in our previous studies of the interactions of griffithsin with monosaccharides, as well as in this study that involved disaccharides, that direct and specific interactions between the carbohydrates and the protein were limited principally to the terminal sugar. The second carbohydrate unit, whether mannose of the 1→6α‐mannobiose or glucose of maltose, made at most two direct hydrogen bonds with the protein. Partial stacking of mannose molecules 21 and 22, 41 and 42, and 51 and 52 is observed in the complex with 1→6α‐mannobiose. Nevertheless, the conformation of the second carbohydrate agreed very well with the model of binding of Man9GlcNAc2, even though only the structures of the three terminal mannose residues were used to create the model discussed here. In that model the core of the carbohydrate, including its reducing end, pointed away from the protein surface, thus providing easy access for binding to similar carbohydrates located on the surface of the viral envelope proteins.
It was instructive to compare the model of Man9GlcNAc2 bound to griffithsin with the experimental structures of that oligosaccharide complexed to a specific human antibody, 2G12 (Fig. 4C). That structure, refined at 3.0 Å resolution, 25 included two complete molecules (A and B) of Man9GlcNAc2. Each of them made extensive contacts with the protein and carbohydrate molecules related by crystallographic symmetry and they exhibited some significant differences, although their overall structures were generally similar (Fig. 4C). The largest differences, including a significant departure from ideal geometry, were present in the link between the D3 and B mannoses. The distances between the terminal sugars, defined as distances between their C1 atoms, were 13.2, 14.1, and 17.9 Å for molecule A, and 11.2, 13.2, and 13.7 Å for molecule B. The extreme distances (11.2 and 17.9 Å) both involved the D3 mannoses. By comparison, the distances measured in the model of the Man9GlcNAc2‐griffithsin complex, which were strictly based on the experimentally determined positions of the mannose residues observed in the monosaccharide complexes of griffithsin, were 13.7, 14.7, and 15.2 Å. Although all modeling results must be interpreted with care, we are confident that the proposed mode of interactions is at least feasible and it explains the known properties of griffithsin. The multiple binding sites of this lectin also explain why this protein was usually precipitated upon addition of multivalent carbohydrates. It must be stressed, however, that this property should not necessarily interfere with the antiviral activity of griffithsin, because obstruction of the carbohydrates on the viral envelope by crosslinking might be as efficient in preventing fusion as the proper, multivalent interactions with a single molecule. This does, however, explain our inability to obtain crystals of the complexes of griffithsin with carbohydrates that contain more than two sugar units. Interestingly, the terminal D1 mannose in the 2G12 complex contacts a different antibody molecule than the terminal mannoses on the D2 and D3 arms, proving that, even in that case, Man9GlcNAc2 is involved in making intermolecular interactions with molecules related by crystallographic symmetry.
Acknowledgements
We thank Drs. Nicole LaRonde‐LeBlanc and Sergei Pletnev for collecting some of the diffraction data used in this study and Cheryl Thomas for technical assistance in the ITC studies. We acknowledge the use of beamline 22‐ID of the Southeast Regional Collaborative Access Team (SER‐CAT), located at the Advanced Photon Source, Argonne National Laboratory.
This article is a US Government work and, as such, is in the public domain in the United States of America.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
REFERENCES
- 1. Botos I, Wlodawer A. Proteins that bind high‐mannose sugars of the HIV envelope. Prog Biophys Mol Biol 2005; 88: 233–282. [DOI] [PubMed] [Google Scholar]
- 2. Mori T, O'Keefe BR, Sowder RC, Bringans S, Gardella R, Berg S, Cochran P, Turpin JA, Buckheit RW,Jr. , McMahon JB, Boyd MR. Isolation and characterization of griffithsin, a novel HIV‐inactivating protein, from the red alga Griffithsia sp . J Biol Chem 2005; 280: 9345–9353. [DOI] [PubMed] [Google Scholar]
- 3. Zió𝔩̷kowska NE, O'Keefe BR, Mori T, Zhu C, Giomarelli B, Vojdani F, Palmer KE, McMahon JB, Wlodawer. Domain‐swapped structure of the potent antiviral protein griffithsin and its mode of carbohydrate binding. Structure 2006; 7: 1127–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Giomarelli B, Schumacher KM, Taylor TE, Sowder RC, Hartley JL, McMahon JB, Mori T. Recombinant production of anti‐HIV protein, griffithsin, by auto‐induction in a fermentor culture. Protein Expr Purif 2006; 47: 194–202. [DOI] [PubMed] [Google Scholar]
- 5. Raval S, Gowda SB, Singh DD, Chandra NR. A database analysis of jacalin‐like lectins: sequence‐structure‐function relationships. Glycobiology 2004; 14: 1247–1263. [DOI] [PubMed] [Google Scholar]
- 6. Geyer H, Holschbach C, Hunsmann G, Schneider J. Carbohydrates of human immunodeficiency virus. Structures of oligosaccharides linked to the envelope glycoprotein 120. J Biol Chem 1988; 263: 11760–11767. [PubMed] [Google Scholar]
- 7. Feldmann H, Nichol ST, Klenk HD, Peters CJ, Sanchez A. Characterization of filoviruses based on differences in structure and antigenicity of the virion glycoprotein. Virol 1994; 199: 469–473. [DOI] [PubMed] [Google Scholar]
- 8. Han DP, Kim HG, Kim YB, Poon LL, Cho MW. Development of a safe neutralization assay for SARS‐CoV and characterization of S‐glycoprotein. Virol 2004; 326: 140–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shenoy SR, Barrientos LG, Ratner DM, O'Keefe BR, Seeberger PH, Gronenborn AM, Boyde MR. NMR and calorimetric titration studies of the anti‐HIV protein cyanovirin‐N with branched oligomannosides: characterization of multivalent and multi‐site binding. Chem & Biol 2002; 10: 1109–1118. [DOI] [PubMed] [Google Scholar]
- 10. Otwinowski Z, Minor W. Processing of X‐ray diffraction data collected in oscillation mode. Methods Enzymol 1997; 276: 307–326. [DOI] [PubMed] [Google Scholar]
- 11. Storoni LC, McCoy AJ, Read RJ. Likelihood‐enhanced fast rotation functions. Acta Crystallogr 2004; D60: 432–438. [DOI] [PubMed] [Google Scholar]
- 12. Emsley P, Cowtan K. Coot: model‐building tools for molecular graphics. Acta Crystallogr 2004; D60: 2126–2132. [DOI] [PubMed] [Google Scholar]
- 13. Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum‐likelihood method. Acta Crystallogr 1997; D53: 240–255. [DOI] [PubMed] [Google Scholar]
- 14. Wallace AC, Laskowski RA, Thornton JM. LIGPLOT: a program to generate schematic diagrams of protein‐ligand interactions. Protein Eng 1995; 8: 127–134. [DOI] [PubMed] [Google Scholar]
- 15. Wormald MR, Petrescu AJ, Pao YL, Glithero A, Elliott T, Dwek RA. Conformational studies of oligosaccharides and glycopeptides: complementarity of NMR, X‐ray crystallography, and molecular modelling. Chem Rev 2002; 102: 371–386. [DOI] [PubMed] [Google Scholar]
- 16. Woods RJ, Pathiaseril A, Wormald MR, Edge CJ, Dwek RA. The high degree of internal flexibility observed for an oligomannose oligosaccharide does not alter the overall topology of the molecule. Eur J Biochem 1998; 258: 372–386. [DOI] [PubMed] [Google Scholar]
- 17. Hester G, Kaku H, Goldstein IJ, Wright CS. Structure of mannose‐specific snowdrop (Galanthus nivalis) lectin is representative of a new plant lectin family. Nat Struct Biol 1995; 2: 472–479. [DOI] [PubMed] [Google Scholar]
- 18. Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res 2000; 28: 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vliegenhart JFG, Dorland L, van Halbeek H. High‐resolution 1H nuclear magnetic resonance spectroscopy as a tool in the structural analysis of carbohydrates related to glycoproteins. In: Tipson RS, editors. Advances in carbohydrate chemistry and biochemistry. Elsevier; 1983. pp 209–374 [Google Scholar]
- 20. Balzarini J, Schols D, Neyts J, Van Damme E, Peumans W, De Clercq E. Alpha‐(1‐3)‐ and alpha‐(1‐6)‐d‐mannose‐specific plant lectins are markedly inhibitory to human immunodeficiency virus and cytomegalovirus infections in vitro. Antimicrob Agents Chemother 1991; 35: 410–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Botos I, O'Keefe BR, Shenoy SR, Cartner LK, Ratner DM, Seeberger PH, Boyd MR, Wlodawer A. Structures of the complexes of a potent anti‐HIV protein cyanovirin‐N and high mannose oligosaccharides. J Biol Chem 2002; 277: 34336–34342. [DOI] [PubMed] [Google Scholar]
- 22. Bokesch HR, O'Keefe BR, McKee TC, Pannell LK, Patterson GM, Gardella RS, Sowder RC, Turpin J, Watson K, Buckheit RW,Jr. , Boyd MR. A potent novel anti‐HIV protein from the cultured cyanobacterium Scytonema varium . Biochemistry 2003; 42: 2578–2584. [DOI] [PubMed] [Google Scholar]
- 23. Williams DC,Jr. , Lee JY, Cai M, Bewley CA, Clore GM. Crystal structures of the HIV‐1 inhibitory cyanobacterial protein MVL free and bound to Man3GlcNAc2: structural basis for specificity and high‐affinity binding to the core pentasaccharide from n‐linked oligomannoside. J Biol Chem 2005; 280: 29269–29276. [DOI] [PubMed] [Google Scholar]
- 24. Weis WI, Drickamer K. Structural basis of lectin‐carbohydrate recognition. Annu Rev Biochem 1996; 65: 441–473. [DOI] [PubMed] [Google Scholar]
- 25. Calarese DA, Scanlan CN, Zwick MB, Deechongkit S, Mimura Y, Kunert R, Zhu P, Wormald MR, Stanfield RL, Roux KH, Kelly JW, Rudd PM, Dwek RA, Katinger H, Burton DR, Wilson IA. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science 2003; 300: 2065–2071. [DOI] [PubMed] [Google Scholar]
- 26. Brünger AT. The free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature 1992; 355: 472–474. [DOI] [PubMed] [Google Scholar]


