Summary
To investigate the potential of antibody derivatives to provide passive protection against enteric infections when supplied orally in crude plant extracts, we have expressed a small immune protein (SIP) in plants using two different plant virus vectors based on potato virus X (PVX) and cowpea mosaic virus (CPMV). The ɛSIP molecule consisted of a single‐chain antibody (scFv) specific for the porcine coronavirus transmissible gastroenteritis virus (TGEV) linked to the ɛ‐CH4 domain from human immunoglobulin E (IgE). In some constructs, the sequence encoding the ɛSIP molecule was flanked by the leader peptide from the original murine antibody at its N‐terminus and an endoplasmic reticulum retention signal (HDEL) at its C‐terminus to allow the expressed protein to be directed to, and retained within, the endoplasmic reticulum. Western blot analysis of samples from Nicotiana clevelandii or cowpea tissue infected with constructs revealed the presence of SIP molecules which retained their ability to dimerize. The analysis of crude plant extracts revealed that the plant‐expressed ɛSIP molecules could bind to and neutralize TGEV in tissue culture, the levels of binding and neutralization reflecting the level of expression. Oral administration of crude extracts from SIP‐expressing plant tissue to 2‐day‐old piglets demonstrated that the extracts which showed the highest levels of in vitro neutralization could also provide in vivo protection against challenge with TGEV.
Keywords: cowpea mosaic virus, oral immunization, potato virus X, small immune protein, transmissible gastroenteritis virus
Introduction
Plants are attractive expression systems for the production of heterologous proteins, such as pharmaceuticals, as they produce large amounts of biomass relatively simply and cheaply without the need for fermentation apparatus and without the danger of contamination by animal pathogens. Furthermore, plants offer the prospect of supplying immunologically active material orally without the need for extensive downstream processing. Particular interest has focused on the production of antibodies (often termed ‘plantibodies’ when expressed in plants), and a significant number of antibody and antibody‐based derivatives have been produced in a variety of plant species (2002, 2005). Although plant‐expressed antibodies specific for proteins from Streptococcus mutans and herpes simplex virus have been shown to be capable of preventing disease when supplied topically (Ma et al., 1998; Zeitlin et al., 1998), there is no report of protection being afforded by the oral route.
Protection against enteric infections can be provided by the oral administration of neutralizing antibodies. This approach, termed ‘passive immunization’, is a particularly attractive method for protecting newborn animals against such infections. Transmissible gastroenteritis virus (TGEV) is a coronavirus which is an important pathogen that infects both the respiratory and enteric tissues of pigs. In newborn pigs, TGEV causes close to 100% mortality (Enjuanes and van der Zeijst, 1995), and it would be of great benefit if passive immunization could be used to protect such animals. The major antigenic sites of TGEV, involved in the induction of virus‐neutralizing antibodies, are located in the globular portion of the spike (S) protein (Gebauer et al., 1991). The monoclonal antibody (mAb) 6A.C3 has been shown to recognize a highly conserved epitope in the S protein and can neutralize all TGEV isolates tested, as well as TGEV‐related coronaviruses infecting pigs, dogs and cats (Suñe et al., 1990; Gebauer et al., 1991). The recombinant immunoglobulin A (IgA) form of mAb 6A.C3 has been shown to be highly efficient at neutralization when expressed in the milk of transgenic mice (1997, 1998; Sola et al., 1998), raising the possibility of basing passive immunization against TGEV on this molecule.
Recent work has shown that small immune proteins (SIPs) derived from mAb 6A.C3, expressed in mammalian cells, can neutralize TGEV infections in tissue culture and can confer protection against TGEV infection when supplied orally to newborn pigs (M. Bestagno et al., in preparation). SIPs are derivatives of single‐chain antibodies (scFvs), in which the scFv sequence is fused to the constant domain of a heavy chain that is responsible for dimerization (Li et al., 1997; Figure 1a, b). SIPs combine the advantages of the bivalency of full‐length antibodies with the small size of scFvs, and have been shown to have higher tissue penetration than complete antibodies and slower clearance than scFvs (Borsi et al., 2002). Thus, they are attractive candidates for passive immunotherapy. In the case of ɛSIPs, the scFv sequence is fused to the CH4 domain of the S2 subclass of IgE, which permits efficient and stable dimerization via a C‐terminal cysteine residue (Batista et al., 1996; Borsi et al., 2002).
Figure 1.
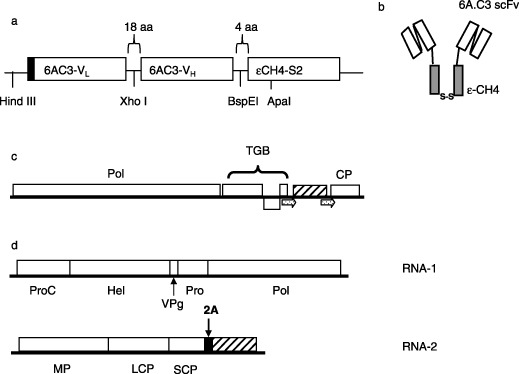
Constructs used to express ɛ‐small immune protein (ɛSIP) in plants. (a) Schematic representation of the ɛSIP portion of plasmid pcDNA‐6AC3‐huɛsip, with the regions derived from the variable domains of the light (VL) and heavy (VH) chains of monoclonal antibody 6A.C3 and the CH4 domain from the human immunoglobulin E (IgE) secretory isoform IgE‐S2 (ɛCH4‐S2) indicated. The leader peptide is indicated by the black box. (b) Secondary structure of dimerized ɛSIP with the regions derived from the 6A.C3 scFv and the CH4 domain shown as open and filled boxes, respectively. The disulphide bridge formed by the C‐terminal cysteine residue of each monomer is indicated. (c) Schematic representation of PVX‐hueSIP and PVX‐nakedhue‐SIP, which differ only in the presence or absence of the SIP leader peptide. The duplicated coat protein subgenomic promoter is indicated by arrows. (d) Structure of CPMV‐hueSIP. In (c) and (d), the sequence encoding ɛSIP is indicated by the hatched box. The various virus‐encoded proteins are indicated as follows: CP, coat protein; Hel, helicase; LCP, large coat protein; MP, movement protein; Pol, RNA‐dependent RNA polymerase; Pro, proteinase; ProC, proteinase cofactor; SCP, small coat protein; TGB, triple gene block proteins; VPg, genome‐linked viral protein. The position of the 2A catalytic peptide from foot‐and‐mouth disease virus (FMDV) is indicated.
If passive immunization against TGEV with mAb 6A.C3 or its derivatives is to become a practical reality, it is essential that large amounts of immunotherapeutic material be produced at low cost. Effective passive immunization through the administration of crude plant extracts requires that the original tissue contains high levels of the appropriate antibody. One way of achieving the necessary levels is through the use of virus‐based vectors rather than stable transformation (Porta and Lomonossoff, 2002). In this paper, we report the use of two different plant virus vectors based on potato virus X (PVX) and cowpea mosaic virus (CPMV) to express an anti‐TGEV ɛSIP molecule in two different plant species. The plant‐expressed SIP molecules retain their ability to dimerize, bind to TGEV particles and neutralize TGEV infections in vitro. Extracts from plants expressing high levels of ɛSIP were able to confer protective immunity in newborn piglets against TGEV infection when supplied orally, thus demonstrating the utility of plant‐derived antibodies in providing passive oral immunity.
Results
Construction of recombinant viruses
The sequence of the anti‐TGEV ɛSIP (Figure 1a,b) was inserted into the two plant virus‐based vectors in different ways to allow the release of a free protein in each case. For expression from PVX, the sequence of ɛSIP was inserted, with or without its leader peptide, behind a duplicated coat protein subgenomic promoter to give plasmids pGR106‐eSIP and pGR106‐eSIPnaked, respectively. To express ɛSIP using CPMV, the sequence was inserted downstream of a foot‐and‐mouth disease virus (FMDV) 2A catalytic peptide at the C‐terminus of the RNA‐2‐encoded polyprotein to give plasmid pBinP‐YP2. The 2A‐mediated cleavage reaction is at least 90% efficient and results in the release of a protein with an additional proline residue at its amino terminus. The sequence encoding ɛSIP was flanked by the leader peptide from the original 6A.C3 scFv at its N‐terminus and an endoplasmic reticulum (ER) retention signal (HDEL) at its C‐terminus to allow the expressed protein to be directed to, and retained in, the ER.
Agroinoculation was used to initiate infections for constructs based on the two viruses. Nicotiana clevelandii plants agroinoculated with the PVX constructs, with and without the leader peptide, developed systemic symptoms 7–9 days post‐inoculation (d.p.i.). The resulting viruses were termed PVX‐hueSIP and PVX‐nakedhueSIP, respectively (Figure 1c). In each case, the symptoms were milder than those obtained with the corresponding wild‐type construct. Reverse transcriptase‐polymerase chain reaction (RT‐PCR) analysis confirmed that the insert was retained until 10–14 d.p.i. After this time, additional, smaller PCR products, indicative of deletions within the insert, began to appear.
Cowpea plants agroinoculated with pBinP‐YP2 in the presence of RNA‐1 did not develop any detectable symptoms. However, when a sap extract enriched for virus particles (termed CPMV‐hueSIP; Figure 1d) was used to inoculate further healthy cowpea plants, these developed chlorotic local lesions at 10–14 d.p.i. The symptoms were less severe than those observed with wild‐type CPMV. RT‐PCR of RNA extracted from these first‐passage cowpea plants revealed that the SIP‐specific insert was retained in the RNA‐2 of CPMV‐hueSIP. Some leaves also showed the presence of variable amounts of smaller PCR products, indicating the presence of some deletion products (results not shown). To reduce the effect of deletion mutants, infected plant tissue was collected at 7–14 d.p.i. for all subsequent analyses.
Expression of ɛSIP in plant tissue
Western blot analysis using anti‐human ɛ‐chain antibodies of extracts from N. clevelandii leaves infected with PVX‐hueSIP revealed the presence of a protein of approximately 42 kDa in samples taken from either inoculated or systemically infected leaves at 7 d.p.i. (Figure 2a). This material corresponds to the monomeric form of ɛSIP, which was expected as the leaf material was analysed under reducing conditions. The material ran as a dimer when examined under non‐reducing conditions (data not shown). However, no material corresponding to monomeric ɛSIP was visible by 14 d.p.i., despite detection of the viral coat protein, when an anti‐coat protein serum was used to probe the Western blots (Figure 2a). Time‐course experiments showed that maximum expression in systemically infected leaves was achieved at 7–10 d.p.i., and expression decreased afterwards. The loss of ɛSIP expression correlated well with the detection of partially deleted sequences observed by PCR at late times post‐inoculation. Extracts of leaves infected with PVX‐nakedhueSIP, in which no leader peptide was present, did not contain detectable levels of ɛSIP at either time. The difference in ɛSIP accumulation in leaves infected with the PVX constructs with and without the leader peptide was not a result of differences in the replication of the viral constructs, as similar levels of coat protein could be detected in all the extracts (Figure 2a). Electrophoresis under non‐reducing conditions indicated that ɛSIP was capable of dimerization (data not shown).
Figure 2.
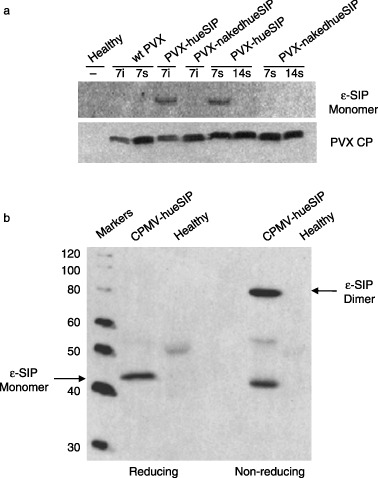
Western blot analysis of ɛ‐small immune protein (ɛSIP) expression in plants. (a) Extracts from healthy Nicotiana clevelandii plants or plants infected with the indicated potato virus X (PVX) construct were probed with antibodies specific for ɛSIP (top panel) or the PVX coat protein (bottom panel). Samples were taken from inoculated (i) or systemically infected (s) leaves at 7 and 14 days post‐inoculation. In each case, 10 µg of protein per sample was electrophoresed under reducing conditions. CP, coat protein; wt, wild‐type. (b) Extracts from either healthy cowpea plants or plants infected with CPMV‐hueSIP were probed with antibodies specific for ɛSIP. The extracts were analysed under reducing and non‐reducing conditions. The positions of the monomeric and dimeric forms of ɛSIP are indicated and the sizes of the marker proteins are indicated on the left‐hand side.
Western blot analysis using anti‐human ɛ‐chain antibodies of total protein extracted from CPMV‐hueSIP‐infected cowpea leaves at 22 d.p.i. revealed the presence of a band of the expected size (42 kDa) for the ɛSIP monomer when the sample was examined under reducing conditions (Figure 2b). Under non‐reducing conditions, approximately 70% of the ɛSIP‐specific material ran at approximately 84 kDa, consistent with efficient dimerization being promoted by the C‐terminal cysteine residue present in the CH4 domain of the SIP sequence. Enzyme‐linked immunosorbent assay (ELISA) with anti‐human ɛ‐chain antibodies of leaf extracts indicated that they contained levels of ɛSIP representing up to 2% (w/w) total soluble protein. Comparison, by Western blot analysis, indicated that the levels of ɛSIP obtained in N. clevelandii using PVX were only about 5%–6% of this level.
Plant‐expressed ɛSIP molecules can bind to and neutralize TGEV
To investigate whether plant‐expressed ɛSIP molecules retained their ability to bind to TGEV particles, ELISA was carried out using plates coated with partially purified TGEV. The level of binding obtained with extracts from leaves infected with each human ɛSIP‐expressing PVX and CPMV construct was compared with that obtained from equivalent extracts from leaves infected with the parental virus (negative controls) and with that observed with the same human ɛSIP expressed in mammalian cells (hueSIP; positive control). In the case of the PVX‐based constructs, an enhanced level of binding compared with the control extracts could be observed when the leader sequence was present (PVX‐hueSIP), but not in its absence (PVX‐nakedhueSIP; Figure 3a). However, the binding activity with the PVX‐hueSIP extracts was less than that obtained with supernatants from mammalian cells expressing the equivalent human ɛSIP. When extracts of cowpea leaves infected with CPMV‐hueSIP were analysed, levels of binding equivalent to those achieved with the positive control were observed (Figure 3a). However, in this case, the background signal obtained with extracts from wild‐type CPMV‐infected leaves was also quite high, making detailed comparisons difficult.
Figure 3.
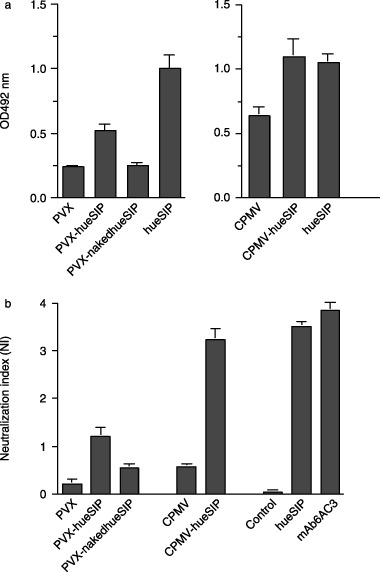
Activity of plant‐expressed ɛ‐small immune protein (ɛ‐SIP). (a) Enzyme‐linked immunosorbent assay (ELISA) of the binding of plant‐expressed ɛSIPs to partially purified transmissible gastroenteritis virus (TGEV) particles. Extracts from Nicotiana clevelandii plants infected with PVX‐hueSIP or PVX‐nakedhueSIP and cowpea plants infected with CPMV‐hueSIP were analysed. In each case, extracts from plants infected with the corresponding wild‐type virus [potato virus X (PVX) or cowpea mosaic virus (CPMV)] were analysed as a negative control and the binding activity of the same ɛSIP molecule expressed in mammalian cells (hueSIP) was used as a positive control. (b) TGEV neutralization assay of ɛSIP produced in plants. Plants infected with the wild‐type virus were used as negative controls for each plant sample. The neutralization index was calculated as the logarithm of the ratio of virus plaques in the absence of antibody to virus plaques after incubation with the antibody. The results obtained with the plant extracts were compared with those obtained with the parental antibody (monoclonal antibody 6A.C3) and ɛSIP (hueSIP) expressed in mammalian cells. The control contained a SIP of irrelevant specificity. Both the neutralization and ELISA data are the mean of at least two independent experiments.
To confirm the functionality of the ɛSIP molecules, TGEV neutralization assays were performed. The results obtained with the plant extracts were compared with the neutralization obtained with mammalian cell‐expressed human ɛSIP (hueSIP) and ascitic fluid containing the original 6A.C3 mAb (positive controls), as well as with supernatants from mammalian cells expressing a SIP molecule of irrelevant specificity (negative control). The results obtained with the PVX constructs mirrored those of the binding assay, with the highest level of neutralization being obtained with extracts from N. clevelandii leaves infected with PVX‐hueSIP (Figure 3b). Substantially higher levels of neutralization, similar to those found with both the original 6A.C3 mAb and ɛSIP derived from it, were found in extracts of cowpea leaves infected with CPMV‐hueSIP (Figure 3b). In contrast with the binding assay, only low levels of neutralization were observed with extracts from wild‐type CPMV‐infected leaves. The lower levels of neutralization obtained with the PVX samples relative to the CPMV samples are consistent with the significantly lower levels of ɛSIP expression found in the former. As expected, negligible neutralization was observed when a SIP molecule of irrelevant specificity was analysed. The results confirmed that the ɛSIP molecules expressed in plants retained both their ability to bind to and neutralize TGEV.
In vivo protection of pigs
To assess the ability of the plant‐expressed ɛSIP molecules to protect pigs against challenge with TGEV, extracts were made from N. clevelandii or cowpea leaves infected with the constructs showing the highest levels of neutralization (PVX‐hueSIP and CPMV‐hueSIP, respectively) and from leaves infected with parental vectors. The binding and neutralization activities of the lyophilized extracts were reconfirmed before supplying them to the animals. Lyophilized samples retained 60%−70% of the specific neutralizing activity (relative to total protein content) relative to the fresh plant extracts. The loss of neutralizing activity, probably a result of the repeated freeze–thaw treatments, was similar for all samples within one experiment. The lyophilized plant extracts were reconstituted with water, mixed with milk and supplied orally to the piglets, which were simultaneously challenged with TGEV. Previous experiments (M. Bestagno et al., in preparation) have shown that supplying supernatants from mammalian cells containing ɛSIP molecules simultaneously with a TGEV challenge yields results similar to those achieved when the ɛSIP‐containing extract is supplied prior to challenge. Thus, the observed decrease in virus titre is not a result of virus neutralization prior to administration. As controls piglets were supplied with either ɛSIP expressed in mammalian cells or the 6A.C3 antibody from which it was derived, or with the corresponding null supernatants. Following further administration of the extracts over a 2‐day period, TGEV levels in the lung and gut were determined and expressed as a ratio of the virus titre obtained with antibody‐containing extract relative to the equivalent extract without antibody (Figure 4). The results showed that supplying pigs orally with milk containing plant extracts from N. clevelandii or cowpea plants expressing ɛSIP caused a clear decrease in virus titre in both the gut and lung compared with extracts from leaves infected with the parental virus (Figure 4). This decrease correlated with a decrease in the number of pulmonary lesions observed. The decrease in virus titre obtained with extracts from cowpea leaves infected with CPMV‐hueSIP (approximately 104‐ and 103‐fold in lung and gut, respectively) was significantly greater than that obtained with N. clevelandii leaves infected with PVX‐hueSIP (approximately 10 and 102‐fold in lung and gut, respectively), and, in gut tissue, was similar to that obtained with the mammalian cell‐expressed ɛSIP (Figure 4). In all cases, the decrease in virus titre with the ɛSIP‐containing extracts was less than that found with the full‐length parental mAb 6A.C3. This may reflect the higher level of neutralization found with extracts containing the full‐length mAb compared with ɛSIP (Figure 3), or the fact that the full‐length mAb is able to mediate additional mechanisms of protection, such as antibody‐dependent cell‐dependent cytotoxicity (ADCC) and complement‐dependent cytotoxicity (CDC), which will not be invoked by SIP molecules as they lack the relevant portions of the constant region of the full‐length antibody. The difference in in vivo efficacy between the PVX‐ and CPMV‐expressed ɛSIP parallels the difference in binding and neutralization activity found in the respective extracts, and is consistent with the lower level of ɛSIP accumulation in the former case. Thus, the in vitro data on the expression levels provide a good indication of the ability of leaf extracts to confer protection in vivo.
Figure 4.
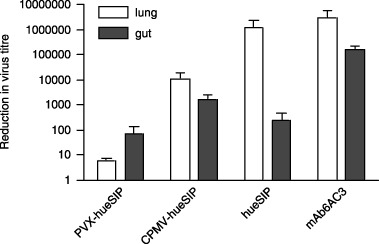
In vivo protection of newborn pigs against transmissible gastroenteritis virus (TGEV). The virus titres in the lung and gut were determined after the administration of extracts from infected plants to pigs which had been challenged with TGEV. Positive controls consisted of parental antibody 6A.C3 and ɛSIP expressed in Sp2/0 mammalian cells. The decrease in virus titre was determined as described in ‘Experimental procedures’. Mean values and standard deviations correspond to the decrease in virus titres in three to four piglets tested for each plantibody.
Discussion
The results presented here show that plant‐expressed SIP molecules retain their ability to dimerize and can neutralize TGEV infections of cells in culture. Furthermore, we have shown that it is possible to protect newborn farm animals against TGEV by providing them with milk containing crude extracts of plant tissue expressing SIP molecules. We chose to use the decrease in TGEV titre in gut and lung, the two main target organs for the virus, rather than the clinical symptoms of infection, such as diarrhoea, as a marker for protection, as this is a much more specific assay. Gastroenteritis, although a clinical manifestation of TGEV infections, is a common episode in newborn animals and can result from unrelated infections or even from the artificial feeding protocols necessary for the in vivo experiments. Although virus accumulation was not totally abolished in the in vivo experiments, the virus titres recovered after administration of plant‐expressed ɛSIP (< 104.5) were below the threshold previously shown to be necessary to induce diarrhoea and death in newborn animals. In contrast with diarrhoea, histopathological lesions, such as interstitial pneumonia and atrophy of intestinal villi, are specific signs associated with TGEV infection. Histopathological examination of tissues from piglets treated with the mAb 6A.C3 or cowpea extracts containing ɛSIP revealed that these signs were absent. In contrast, both signs were evident in animals who received no specific antibody (data not shown). These results indicate that both the parental mAb 6A.C3 and plant‐expressed ɛSIP prevent the appearance of lesions associated with disease. Therefore, the administration of plant‐expressed ɛSIP can be anticipated to afford protection against TGEV. We are therefore convinced that we have established the proof of principle that the protection of mammals against virus infection can be afforded by the oral administration of crude plant extracts containing virus‐neutralizing antibody derivatives.
Although the in vivo experiments were on a relatively small scale, they are important because they demonstrate that orally administered SIP molecules are stable during the preparation of plant extracts and are resistant to the enzymatic and pH conditions of the enteric tract. This is especially relevant as it validates the general approach of using the oral administration of SIP molecules to achieve passive immunization against enteric pathogens. Furthermore, no toxic effects of the administration of plant extracts containing SIP molecules from either N. clevelandii or cowpea were observed, suggesting that this approach is safe as well as efficacious. In addition, the correlation between in vitro neutralization and in vivo protection means that it is possible to assess the efficacy of any particular batch of plant extract prior to its administration to animals.
We chose to use a viral vector rather than a transgenic approach to express the ɛSIP molecules, as this can potentially give the high levels of expression necessary to achieve passive immunization. In addition, large amounts of material can be produced in a relatively short time (Porta and Lomonossoff, 2002). Prior to the work reported here, in terms of antibody production, virus‐based vectors have mainly been used to express scFvs in plants (McCormick et al., 1999; Ziegler et al., 2000), although there is a single report of the assembly of a full‐length antibody using this approach (Verch et al., 1998). The viral vectors used in the current study (PVX and CPMV) were capable of directing the expression of ɛSIP molecules which had the ability to bind to TGEV, although the expression levels achieved varied. In the case of expression from PVX, inclusion of the original leader peptide from the mouse immunoglobulin VH domain was found to be essential for the accumulation of detectable levels of ɛSIP. The presence of this leader peptide allows the expressed SIP molecules to be directed to the plant secretory pathway, where post‐translational modifications can take place, rather than accumulating in the cytoplasm.
To maximize ɛSIP accumulation using a CPMV vector, the ɛSIP coding sequence was flanked by a leader peptide at its N‐terminus and an ER retention signal (HDEL) at its C‐terminus. The ɛSIP sequence was expressed at the C‐terminus of the RNA‐2‐encoded polyprotein, and thus targeting to the secretory pathway should not affect any viral protein. The HDEL sequence was included because retention in the ER enhances the levels of accumulation of single‐chain antibodies in plants (Schouten et al., 1996). Furthermore, the presence of this sequence has been shown to be necessary to achieve high levels of expression of green fluorescent protein (GFP), which had been directed to the secretory pathway after expression from CPMV (L. Nicholson et al., unpublished data). The strategy was successful, as levels of ɛSIP sufficient to protect newborn pigs against TGEV accumulated in leaf tissue. The levels of antibody expression (approximately 2% total soluble protein) are similar to those previously reported for an ER‐retained version of an scFv expressed from a PVX vector (Ziegler et al., 2000), and considerably higher (about 15–20‐fold) than the levels obtained when ɛSIP was expressed from PVX without an ER retention signal. The fact that CPMV infects an edible plant, cowpea, is a further advantage of the system as it reduces potential concerns about administering plant tissue, such as that from Nicotiana species, which contains significant levels of alkaloids.
The demonstration that quantities of an antibody sufficient to afford protection to target animals through the oral supply of crude plant extracts can be produced using viral vectors, in particular CPMV, indicates that this approach could provide a general method for oral passive immunization. It will be very straightforward to express ɛSIP molecules with different specificities simply by substituting the scFv domain of the construct. In this regard, it is noteworthy that, although the plant tissue expressing the anti‐TGEV ɛSIP was supplied only orally, a significant decrease in virus titre was observed not only in enteric tissue, but also in the lungs. This could be a result of the prevention of virus shedding by the faecal–oral route or, alternatively, may reflect the fact that SIP molecules exhibit high tissue penetration and may therefore prevent the internal distribution of the virus to the respiratory organs. Whatever the reason for this observation, it suggests that the oral administration of SIP molecules may provide passive protection against respiratory as well as enteric pathogens, although further animal studies will be required to establish this unequivocally. Overall, the results presented here indicate that plantibodies are safe and efficacious molecules which can provide immediate protection against virus infections, as required in newborn animals or healthcare workers. Furthermore, their expression via viral vectors allows plant material expressing antibodies with different specificities to be rapidly produced, thus opening up the way to a new approach to control diseases such as severe acute respiratory syndrome coronavirus and other enteric and respiratory pathogens.
Experimental procedures
Plasmids
Functional recombinant mAb 6A.C3 genes were originally described in Castilla et al. (1997). The source of the sequence of the anti‐TGEV ɛSIP for all the experiments was pcDNA‐6AC3‐huɛsip (Figure 1a; M. Bestagno et al., in preparation). The viral vectors used for the expression of the ɛSIP sequence were pGR106 (Jones et al., 1999) and pBinP‐NS‐1 (Liu et al., 2005) for the expression from PVX and CPMV RNA‐2, respectively. Full‐length CPMV RNA‐1 was supplied by plasmid pBinPS1NT (Liu and Lomonossoff, 2002).
Construction of viral vectors expressing an anti‐TGEV ɛ‐SIP
For cloning in pGR106, the ɛSIP gene in pcDNA‐hu6AC3‐ɛsip was amplified by PCR using CCATCGATCCATGGGCTGGAGC or CCATCGATCCATGGACATTGTG as the forward primer to produce ɛSIP constructs with and without the original murine leader peptide, respectively. In each case, a ClaI site (italic) was introduced upstream of the SIP‐specific sequence. In both cases, GCGTCGACCTAGCAGCCACC, containing a SalI site, was used as the reverse primer. The PCR products were digested with ClaI and SalI and ligated into ClaI/SalI‐digested pGR106 to give pGR106‐eSIP and pGR106‐eSIPnaked, respectively.
To create a CPMV‐based vector containing the sequence of anti‐TGEV ɛSIP, pcDNA‐6AC3‐huɛsip was modified by PCR‐based mutagenesis. To remove the intron in the leader sequence, to introduce a unique ApaI site at the 5′‐terminus of the coding sequence and to eliminate the original ATG start codon, the forward primer ACTCTAGCCAAGCTTGTCGGGGCCCGGCTGGAGCCTGATCCTCCTGTTCCTCGTCGCTGTGGCTACAGGTGTGCACTCGGACATTGTGATGACCC was used. The reverse primer GCTAACCGAGCTCGGTACCTAGAGTTCGTCGTGGCAGCCACCCCTCCTCG was used to fuse the sequence encoding an ER retention signal, HDEL, to the C‐terminus of the CH4 domain. In addition, an internal ApaI site in the CH4 domain was removed using the mutagenic primer GTCTCCTCCGGAGGCTCTGGCGGC to introduce a silent G to C change. The construct (pYP‐2) containing the modified version of ɛ‐SIP was digested with ApaI and EcoRV, and the 1.5‐kb fragment encoding the sequence of SIP was used to replace that of GFP in ApaI/StuI‐digested pBinP‐NS‐1 to give plasmid pBinP‐YP2.
The structure of the various ɛ‐SIP constructs was verified by sequence analysis, and appropriate Agrobacterium tumefaciens strains (GV3010 for pGR106‐eSIP and pGR106‐eSIPnaked, and LBA4404 for pBinP‐YP2) were transfected by electroporation.
Infection of plants
In all cases, plants were initially infected by agroinoculation. For pGR106‐eSIP and pGR106‐eSIPnaked, N. clevelandii was used as the host. Agrobacterium GV3101 cultures carrying pGR106 vectors were pelleted and resuspended to an optical density at 600 nm (OD600) of 0.5 in a solution containing 150 µm acetosyringone, 10 mm morpholinepropanesulphonic acid (MES) and 10 mm MgCl2, and incubated in this solution at room temperature for 2 h. Approximately 150 µL of the Agrobacterium suspension was applied with a syringe to the underside of three leaves of young N. clevelandii plants. Passaging was carried out by inoculating fresh N. clevelandii plants with sap from agroinoculated plants. In the case of pBinP‐YP2, cowpea (Vigna unguiculata) plants were co‐inoculated with bacteria containing pBinPS1NT to provide RNA‐1 for virus multiplication. For passaging, virus in the sap of agroinoculated leaves was first concentrated by precipitation with polyethylene glycol (Mechtcheriakova et al., 2006).
Immunological detection of ɛSIP expression in leaf tissue
Total protein was extracted from frozen powdered leaves using a buffer containing 50 mm Tris‐acetate pH 8.0, 10 mm potassium acetate and 1 mm ethylenediaminetetraacetic acid (EDTA). The extracts were filtered through Miracloth and centrifuged to remove further debris. Samples were denatured by heating in sodium dodecylsulphate (SDS) sample buffer (30 mm Tris‐HCl pH 6.8, 1.5% SDS, 10% (v/v) glycerol, 0.1 mg/mL bromophenol blue) in the presence or absence of 2.5% (v/v) β‐mercaptoethanol to achieve reducing or non‐reducing conditions. Approximately 25 µg of crude protein (as estimated by the Bradford assay) was separated on a 10% sodium dodecylsulphate‐polyacrylamide gel electrophoresis (SDS‐PAGE) gel and electroblotted on to polyvinylidene difluoride (PVDF) membrane (Millipore Immobilon P, Bedford, MA) overnight. The membrane was probed with anti‐human ɛ‐chain goat antibodies conjugated with peroxidase (KPL), diluted 1 : 5000 in phosphate‐buffered saline (PBS) containing 0.1% Tween‐20 (PBS/T). Bound antibodies were detected by enhanced chemiluminescence (ECL) using reagents supplied by Amersham Biosciences (Buckinghamshire, UK).
In vitro analysis of SIP activity
Infected plant tissue was collected at several times post‐inoculation and macerated with 2 mL/g ice‐cold 0.05 m sodium phosphate buffer (pH 7.2) to break open the cells. Cell debris was removed by centrifugation at high speed for 20 min at 4 °C, and the supernatant containing total soluble plant proteins was collected. The ability of SIP molecules in the plant extracts to bind to TGEV was determined by ELISA and virus neutralization assays, following previously reported procedures (Correa et al., 1988). Purified TGEV virions were adsorbed to ELISA plates before the extracts (50 µL) were added to the wells. TGEV‐bound SIPs were detected with a goat anti‐human IgE antibody (Nordic, Tilburg, the Netherlands), diluted 1 : 500 in PBS/T containing 0.3% bovine serum albumin, with horseradish peroxidase‐conjugated rabbit anti‐goat antibody (Sigma, St Louis, MO), diluted 1 : 1000 in PBS, as second antibody. Neutralization assays were performed by combining 50 µL from a TGEV (PUR46‐MAD) preparation of known titre with an equal volume of plant extract. At least three serial dilutions of the extract were analysed in each assay. After incubation for 30 min at 37 °C, the mixture was added to confluent monolayers of swine testis (ST) cells grown on 24‐well tissue culture dishes. After the adsorption of TGEV to the cells for 45 min at 37 °C, the medium was removed and replaced by overlay medium, and the cells were incubated for a further 48 h at 37 °C. The monolayer was then fixed and stained with crystal violet to visualize TGEV plaques. The neutralization index was calculated as the logarithm of the ratio of virus plaques in the absence of antibody to virus plaques after incubation with the antibody.
In vivo analysis of SIP activity
Between 70 and 90 g of infected tissue was snap‐frozen in liquid nitrogen, ground to fine powder and extracted in 2 mL/g ice‐cold 0.05 m sodium phosphate buffer (pH 7.2). After removal of cell debris, the cleared extract was frozen at −80 °C and lyophilized. About 2.6 g of lyophilized material was obtained from each sample; 0.03 g of each sample was reconstituted with 1 mL of phosphate buffer and the in vitro SIP activity was assayed. To assess the ability of the plant‐expressed SIP to confer protection, 0.3 g of lyophilized plant tissue dissolved in 4 mL of water and TGEV PUR46 C11 [107 plaque‐forming units (pfu) per animal] were added to milk, and the mixture was supplied to 2‐day‐old piglets via an oral cannula. The piglets were subsequently fed, via baby bottles, with milk mixed with 0.3 g of lyophilized plant tissue dissolved in 4 mL of water. The antibody‐containing mixture was administered twice more on the day of challenge and three times on the following 2 days post‐challenge. Piglets were killed 2 and 3 days post‐challenge, a time at which the virus levels reach a peak in TGEV‐infected piglets (Sánchez et al., 1999), and virus titres in gut and lung tissue were determined as described by Jiménez et al. (1986). Protection against TGEV by plant extracts was expressed as the ratio of pfu obtained after the administration of extracts from plants infected with the wild‐type vectors to that obtained when the corresponding SIP‐expressing extracts were supplied. As positive controls, the protection afforded by the administration of 4 mL of a 1 : 10 dilution of ascitic fluid containing mAb 6A.C3 compared with ascitic fluid not containing the antibody, and that afforded by 4 mL of supernatants from Sp2/0 cells expressing anti‐TGEV ɛSIP compared with supernatants expressing a SIP of irrelevant specificity, were assessed.
Acknowledgements
W.M. and A.M.J. contributed equally to this work and should be considered joint first authors. This work was funded under the EC Framework 5 Quality of Life Programme (Contract Nos QLK2‐CT‐2000‐00739 and QLK2‐CT‐2002‐01050). The research was also supported by grant BIO2004‐02687 from the Spanish MEC and by grants from the Comisión Interministerial de Ciencia y Tecnología (CICYT) of Spain. CPMV was propagated under DEFRA licence No. 185B/4836(6/2004).
References
- Batista, F.D. , Efremov, D.G. and Burrone, O.R. (1996) Characterization of a second secreted IgE isoform and identification of an asymmetric pathway of IgE assembly. Proc. Natl. Acad. Sci. USA, 93, 3399–3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsi, L. , Balza, E. , Bestagno, M. , Castellani, P. , Carnemolla, B. , Biro, A. , Leprini, A. , Sepúlveda, J. , Burrone, O. , Neri, D. and Zardi, L. (2002) Selective targeting of tumoral vasculature: comparison of different formats of an antibody (L19) to the ED‐B domain of fibronectin. Int. J. Cancer, 102, 75–85. [DOI] [PubMed] [Google Scholar]
- Castilla, J. , Sola, I. and Enjuanes, L. (1997) Interference of coronavirus infection by expression of immunoglobulin G (IgG) or IgA virus‐neutralizing antibodies. J. Virol. 71, 5251–5258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castilla, J. , Pintado, B. , Sola, I. , Sánchez‐Morgado, J.M. and Enjuanes, L. (1998) Engineering passive immunity in transgenic mice secreting virus‐neutralizing antibodies in milk. Nat. Biotechnol. 16, 349–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa, I. , Jiménez, G. , Suñé, C. , Bullido, M.J. and Enjuanes, L. (1988) Antigenic structure of the E2 glycoprotein from transmissible gastroenteritis coronavirus. Virus Res. 10, 77–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enjuanes, L. and Van Der Zeijst, B.A.M. (1995) Molecular basis of TGE coronavirus epidemiology In: The Coronaviridae (Siddell S.G., ed.), pp. 337–376. New York: Plenum Press. [Google Scholar]
- Gebauer, F. , Posthumus, W.A.P. , Correa, I. , Suñé, C. , Sanchez, C.M. , Smerdou, C. , Lenstra, J.A. , Meloen, R. and Enjuanes, L. (1991) Residues involved in the formation of the antigenic sites of the S protein of transmissible gastroenteritis coronavirus. Virology, 183, 225–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez, G. , Correa, I. , Melgosa, M.P. , Bullido, M.J. and Enjuanes, L. (1986) Critical epitopes in transmissible gastroenteritis virus neutralization. J. Virol. 60, 131–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, L. , Hamilton, A.J. , Voinnet, O. , Thomas, C.L. , Maule, A.J. and Baulcombe, D.C. (1999) RNA–DNA interactions and DNA methylation in post‐transcriptional gene silencing. Plant Cell, 11, 2291–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, E. , Pedraza, A. , Bestagno, M. , Mancardi, S. , Sanchez, R. and Burrone, O.R. (1997) Mammalian cell expression of dimeric small immune proteins (SIP). Protein Eng. 10, 731–736. [DOI] [PubMed] [Google Scholar]
- Liu, L. and Lomonossoff, G.P. (2002) Agroinfection as a rapid method for propagating cowpea mosaic virus‐based constructs. J. Virol. Methods, 105, 343–348. [DOI] [PubMed] [Google Scholar]
- Liu, L. , Cañizares, M.C. , Monger, W. , Perrin, Y. , Tsakiris, E. , Porta, C. , Shariat, N. , Nicholson, L. and Lomonossoff, G.P. (2005) Cowpea mosaic virus‐based systems for the production of antigens and antibodies in plants. Vaccine, 23, 1788–1792. [DOI] [PubMed] [Google Scholar]
- Ma, J.K. , Hikmat, B.Y. , Wycoff, K. , Vine, N.D. , Chargelegue, D. , Yu, L. , Hein, M.B. and Lehner, T. (1998) Characterisation of a recombinant plant monoclonal secretory antibody and preventative immunotherapy in humans. Nat. Med. 4, 601–604. [DOI] [PubMed] [Google Scholar]
- McCormick, A.A. , Kumagai, M.H. , Hanley, K. , Turpen, T.H. , Hakim, I. , Grill, L.K. , Tusé, D. , Levy, S. and Levy, R. (1999) Rapid production of specific vaccines for lymphoma by expression of the tumor‐derived single‐chain Fv epitopes in tobacco plants. Proc. Natl. Acad. Sci. USA, 96, 703–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechtcheriakova, I.A. , Eldarov, M.A. , Nicholson, L. , Shanks, M. , Skryabin, K.G. and Lomonossoff, G.P. (2006) The use of viral vectors to produce hepatitis B virus core particles in plants. J. Virol. Methods, 131, 10–15. [DOI] [PubMed] [Google Scholar]
- Porta, C. and Lomonossoff, G.P. (2002) Viruses as vectors for the expression of foreign sequences in plants. Biotechnol. Genet. Eng. Rev. 19, 245–291. [DOI] [PubMed] [Google Scholar]
- Sánchez, C.M. , Izeta, A. , Sánchez‐Morgado, J.M. , Alonso, S. , Sola, I. , Balasch, M. , Plana‐Durán, J. and Enjuanes, L. (1999) Targeted recombination demonstrates that the spike gene of transmissible gastroenteritis coronavirus is a determinant of its enteric tropism and virulence. J. Virol. 73, 7607–7618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schouten, A. , Roosien, J. , Van Engelen, F.A. , De Jong, G.A.M. , Borst‐Vrenssen, A.W.M. , Zilverentant, J.F. , Bosch, D. , Stiekema, W.J. , Gommers, F.J. , Schots, A. and Bakker, J. (1996) The C‐terminal KDEL sequence increases the expression level of a single‐chain antibody designed to be targeted to both the cytosol and the secretory pathway in transgenic tobacco. Plant Mol. Biol. 30, 781–793. [DOI] [PubMed] [Google Scholar]
- Sola, I. , Castilla, J. , Pintado, J. , Sánchez‐Morgado, J.M. , Whitelaw, C.B.A. and Enjuanes, L. (1998) Transgenic mice secreting coronavirus neutralizing antibodies in milk. J. Virol. 72, 3762–3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoger, E. , Sack, M. , Fischer, R. and Christou, P. (2002) Plantibodies: application, advantages and bottlenecks. Curr. Opin. Biotechnol. 13, 161–166. [DOI] [PubMed] [Google Scholar]
- Stoger, E. , Ma, J.K. , Fischer, R. and Christou, P. (2005) Sowing the seeds of success: pharmaceutical proteins from plants. Curr. Opin. Biotechnol. 16, 167–173. [DOI] [PubMed] [Google Scholar]
- Suñe, C. , Jiménez, G. , Correa, I. , Bullido, M.J. , Gebauer, F. , Smerdou, C. and Enjuanes, L. (1990) Mechanisms of transmissible gastro‐enteritis coronavirus neutralisation. Virology, 177, 559–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verch, T. , Yusibov, V. and Koprowski, H. (1998) Expression and assembly of a full‐length monoclonal antibody in plants using a plant virus vector. J. Immunol. Methods, 230, 69–75. [DOI] [PubMed] [Google Scholar]
- Zeitlin, L. , Olmsted, S.S. , Moench, T.R. , Co, M.S. , Martinell, B.J. , Paradkhar, V.M. , Russell, D.R. , Queen, C. , Cone, R.A. and Whaley, K.J. (1998) A humanized monoclonal antibody produced in transgenic plants for immunoprotection of the vagina against genital herpes. Nat. Biotechnol. 16, 1361–1364. [DOI] [PubMed] [Google Scholar]
- Ziegler, A. , Cowan, G.H. , Torrance, L. , Ross, H.A. and Davies, H.V. (2000) Facile assessment of cDNA constructs for the expression of functional antibodies in plants using the potato virus X vector. Mol. Breed. 6, 327–335. [Google Scholar]


