Abstract
Background
Most respiratory bacterial carriage studies in children are based on cross‐sectional samples or longitudinal studies with infrequent sampling points. The prospective Observational Research in Childhood Infectious Diseases birth cohort study intensively evaluated the community‐based epidemiology of respiratory viruses and bacteria during the first 2‐years of life. Here we report the bacteriologic findings.
Methods
Pregnant women in Brisbane, Australia were recruited between September 2010 and October 2012, and their healthy newborn children were followed for the first 2‐years of life. Parents kept a daily symptom diary for the study child, collected a weekly anterior nose swab and completed an illness burden diary when their child met pre‐defined illness criteria. Specimens were tested for respiratory bacteria by real‐time polymerase chain reaction (PCR) assays and those containing human genomic DNA, deemed as high‐quality, were analyzed.
Results
Altogether 8100 high‐quality nasal swab specimens from 158 enrolled children were analyzed. Streptococcus pneumoniae, Moraxella catarrhalis, and Haemophilus influenzae were detected in 42.4%, 38.9%, and 14.8% of these samples, respectively. Concomitant detection of bacteria was common. In contrast, Bordetella pertussis, B. parapertussis, Mycoplasma pneumoniae, Chlamydia pneumoniae, and Simkania negevensis were rarely identified. The prevalence of the three major bacteria was higher with increasing age and in the winter and spring months. Siblings and childcare attendance were the other risk factors identified.
Conclusions
We confirmed the feasibility of frequent nasal swabbing by parents for studying bacterial colonization. PCR detected the major respiratory tract bacteria with expected high frequencies, but atypical bacteria were found rarely in this cohort.
Keywords: bacteria, carriage, child, colonization, polymerase chain reaction
1. INTRODUCTION
Acute respiratory infections (ARI) are responsible for substantial global morbidity and are important causes of mortality in children from low‐income countries.1 Carriage of respiratory bacterial pathogens, such as Streptococcus pneumoniae, Moraxella catarrhalis, and Haemophilus influenzae, is a prerequisite for two important childhood ARIs; acute otitis media, which is the most frequently reported pediatric bacterial infection,2 and bacterial pneumonia, a leading cause of childhood deaths.1, 3 These bacteria are also carried in the upper airways of healthy young children who are considered the principal reservoirs for maintaining these organisms within populations.4 Similarly, so‐called atypical bacteria, like Mycoplasma pneumoniae, can also colonize the upper airways of healthy children and in some cases result in lower respiratory tract infections.5
Understanding the nature of upper airway bacterial colonization in young children is critical for determining the impact, including unintended consequences, of antibiotics and vaccines upon this ecological niche and for designing future therapeutic strategies to reduce the burden of ARIs in children. However, colonization is a highly dynamic process and most respiratory carriage studies in children are based upon cross‐sectional samples in various patient populations or longitudinal studies of healthy children with infrequent sampling points. We used a novel approach to describe the epidemiology, symptoms, and risk factors associated with carriage of S. pneumoniae, M. catarrhalis, and H. influenzae in a longitudinal cohort of young children with nasal samples collected weekly from birth until age 24‐months.
2. MATERIALS AND METHODS
2.1. Study population and setting
The Observational Research in Childhood Infectious Diseases (ORChID) study was conducted in the subtropical city of Brisbane, Queensland, Australia from September 2010 through October 2014 (http://clinicaltrials.gov: NCT01304914).6 In this prospective, community‐based birth cohort study, children were enrolled at their mothers’ antenatal visits and followed from birth until their second birthday. Recruitment was progressive over 2‐years at two metropolitan hospitals (one private and one government‐funded). To be eligible to participate, children needed to be healthy, without congenital abnormalities or underlying chronic disorders, and to be born at term (36‐42 weeks gestation). Participants were able to leave the study temporarily (such as during family holidays) and re‐join at a later date. The primary aim of the ORChID study was to describe the nature and timing of viral respiratory and gastrointestinal infections during the first 2‐years of life.6, 7, 8 Here, we undertook a secondary analysis of the ORChID database and sample bank to evaluate bacterial carriage. Parents provided written informed consent for their newborn infants at enrolment. The Children's Health Queensland, Royal Brisbane and Women's Hospital, and The University of Queensland Human Research Ethics Committees approved the study.
2.2. Data and symptom diary collection
After enrolment, data on demographic, social, and health history, pregnancy, and birth details were collected by parental interview. Subsequently, telephone interviews were conducted every 3‐months to update feeding, vaccination, and childcare attendance details. Childcare was categorized as formal (regulated care outside the child's home) or informal (non‐regulated care provided by family or friends). Parents received training to recognize respiratory symptoms. Parents used diary cards to record daily, pre‐defined respiratory symptoms, or diagnoses in a designed tick‐box format. Parents also recorded in a separate illness impact diary any health visits and antibiotic prescriptions for their child's ARI episodes.9 Both diaries were returned by mail each month to the research team.
2.3. Nasal swab collection and laboratory methods
Research personnel collected anterior nasal swabs soon after birth and, following training and receiving written instructions, parents obtained swabs weekly thereafter. All swabs were collected using a plastic‐shaft, rayon‐budded swab in a transport tube with a foam pad reservoir soaked with viral transport medium (Virocult MW950, Medical Wire & Equipment, Wiltshire, England) as one of the primary aims of the ORChID study was to detect respiratory viruses. Swabs were sent by standard surface mail to the laboratory where they were stored at −80°C, at a median of 3 (interquartile range 2‐4) days after swabbing.
Since the swab's viral transport medium contained chloramphenicol and amphotericin B, polymerase chain reaction (PCR) assays rather than culture were used to detect the bacterial pathogens of interest. For nucleic acid extraction, each swab was resuspended in 2 mL of phosphate buffered saline from which 200 μL was used for extraction. Samples were extracted on the QIAxtractor automated high‐throughput extraction platform using DX reagents following the manufacturer's instructions (Qiagen, Australia). Total DNA and RNA were eluted into 150 μL of elution buffer. Each sample was spiked with whole equine herpesvirus (EHV) to assess nucleic acid extraction quality and presence of potential PCR inhibitors. Any extract having a >3 cycle threshold (Ct) difference to that of the expected value by EHV real‐time PCR assay was considered to have failed quality control and the sample was re‐extracted. Nasal swab specimen quality was determined by screening for human DNA using an endogenous retrovirus‐3 (ERV3) assay as described previously.10 Swabs with ERV3 Ct values >38 were deemed lower‐quality and they, and associated person‐time, were excluded from analyses.
Swabs were batch‐tested for five bacterial (S. pneumoniae, M. catarrhalis, H. influenzae, Bordetella pertussis, B. parapertussis) and three atypical bacterial (M. pneumoniae, Chlamydophila pneumoniae, Simkania negevensis) pathogens using validated real‐time PCR assays (Supplemental Digital Content 1, Table). Batch testing methods and results for 17 respiratory viruses from the ORChID cohort were available and have been described previously.6, 7 Ct values <40 for individual bacteria were considered positive.11
2.4. Definitions
A new bacterial detection episode (BDE) for each bacterial species occurred when either: (i) a new bacterium was first detected; (ii) the same bacterium was detected after two negative intervening high‐quality swabs; or (iii) the same bacterium was detected at least 30‐days after the last positive swab. ARIs were categorized hierarchically as either: Lower respiratory tract infection (LRTI) or Upper respiratory tract infection (URTI). An infection was an LRTI when parents reported rattly breathing, moist cough, shortness of breath, wheeze, or doctor‐diagnosed pneumonia. An infection was a URTI when parents reported nasal discharge/congestion, dry cough, or a doctor‐diagnosed otitis media.7, 9
2.5. Analysis
Acquisition rates for bacterial detections were investigated for only high‐quality child swabs. Acquisition rate per child‐year was estimated based on the definition of BDE using Poisson regression with the logarithm of individual study follow‐up time as an offset. The time periods of consecutive positive swabs where the participant was not at risk of a new BDE were removed from the denominator when calculating acquisition rates. The univariable association between potential risk factors and bacterial detection was examined using mixed‐effects logistic regression, taking into account the repeated measurements of the subjects through the use of an autoregressive model with lag one, that is, the previous sample result was included as a covariable. Potential risk factors investigated were both fixed (sex, type of delivery, gestational age at birth, family history of atopy [asthma or eczema in parents or siblings], tobacco smoke exposure, presence of siblings in their usual residence, maternal education status) and time‐varying (age, breastfeeding, season of swab sampling, type of childcare attendance). A final multivariable model was constructed with age (in 4 categories), sex and the variables with consistent differences in univariable analysis (smoking, siblings, maternal education, childcare, season) included as covariables. The association between potential risk factors and bacterial carriage was estimated rather than acquisition, because no bacterial subtype data were available (serotypes or genotypes). All analyses are exploratory and were not pre‐specified in the protocol. Data were analyzed using Stata v12.1 (StataCorp, TX).
3. RESULTS
The characteristics of the 158 enrolled cohort children, of whom 154 provided daily symptom data, are shown in Table 1. Overall, 11 192 nasal swab samples were submitted, of which 3025 lower‐quality swabs were removed, as were 67 swabs submitted after the child's second birthday, leaving 8100 swabs from 157 children (151 with accompanying symptom data) for analysis (Figure 1). The number of high‐quality swabs per child ranged from 1 to 102 (median 57, interquartile range 32‐74) during a total follow‐up time of 245 person‐years (median 717‐days per subject, interquartile range 449‐726). The median interval between successive swabs was 7‐days (interquartile range 7‐12).
Table 1.
Characteristics of the 158 ORChID study participants
| Characteristics (n = 158 unless stated otherwise) | Frequency (%) or mean (±SD) |
|---|---|
| Sex (male) | 75 (47.5) |
| Gestational age (weeks) | 39.8 (±1.3) |
| Vaginal delivery | 107 (67.7) |
| First born | 106 (67.1) |
| First degree relative with asthma or eczema a | 83 (52.5) |
| Maternal education (n = 157) | |
| Primary/high school | 20 (12.8) |
| Diploma/certificate | 38 (24.2) |
| University degree | 99 (63.1) |
| Tobacco smoke exposure at birth | |
| Mother (n = 156) | 5 (3.2) |
| Other household member (n = 155) | 17 (11.0) |
| Child vaccination status to age 18‐months b | |
| Completed primary PCV series | 152 (96.2) |
| Exclusive breast feeding | |
| At birth (n = 149) | 142 (95.3) |
| At 3‐months (n = 142) | 97 (68.3) |
| At 6‐months (n = 133) | 5 (3.8) |
| Childcare | |
| At 6‐months (n = 133) | 33 (24.8) |
| At 9‐months (n = 123) | 48 (39.0) |
| At 12‐months (n = 115) | 72 (62.6) |
| At 15‐months (n = 110) | 87 (79.1) |
Abbreviation: PCV, pneumococcal conjugate vaccine; SD, standard deviation.
Parent or sibling.
Vaccination status for all cohort subjects at 18‐months of age was confirmed by cross checking with the Australian Immunisation Register. The PCV program began in Queensland in 2005 with the 7‐valent PCV using 3 primary doses without a booster (3 + 0 schedule, vaccination at ages 2, 4 and 6‐months). In mid‐2011, the 13‐valent PCV replaced the 7‐valent PCV. In contrast, influenza vaccines are not part of the standard Australian vaccine schedule and only 7 children received this vaccine during their first 2‐years of life.
Figure 1.
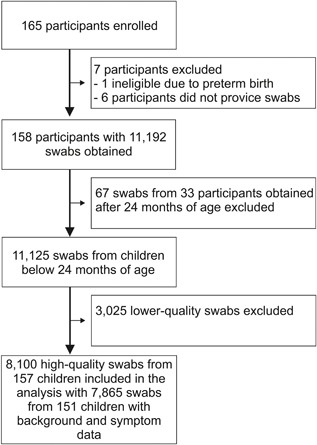
Flow chart of the ORChID study
Overall, the prevalence of the major respiratory bacterial pathogens in the 8100 high‐quality nasal swabs were: S. pneumoniae (42.4%), M. catarrhalis (38.9%), and H. influenzae (14.8%). Altogether, 58.4% of nasal swabs were positive for at least one of these pathogens. Co‐detection of bacterial pathogens was common, occurring in 29.7% of swabs (Table 2 and Figure 2). The prevalence of the major respiratory bacterial pathogens increased with age and during the winter and spring months of June through November (Supplemental Digital Content 2, Figure). The estimated acquisition rates of all new BDEs per 100 child‐years were for S. pneumoniae 8.9 (95% confidence interval (CI) 8.3‐9.6), M. catarrhalis 5.9 (5.5‐6.5), and H. influenzae 2.8 (2.6‐3.2), respectively.
Table 2.
Prevalence of major respiratory bacterial pathogens by clinical status in weekly‐collected nasal swabs from 151 children in the ORChID cohort
| All nasal swabs % | Swab positive for any respiratory virus a % | During LRTI b % | During URTI b % | During acute otitis media % | |
|---|---|---|---|---|---|
| Pathogen | N = 8100 | N = 2550 | N = 591 | N = 1379 | N = 60 |
| Streptococcus pneumoniae | 42.4 | 55.7 | 57.2 | 56.2 | 65.0 |
| Moraxella catarrhalis | 38.9 | 55.2 | 68.7 | 59.1 | 60.0 |
| Haemophilus influenzae | 14.8 | 24.7 | 27.9 | 27.5 | 35.0 |
| Positive for any of the 3 bacterial pathogens | 58.4 | 75.4 | 84.4 | 77.7 | 91.7 |
| Multiple pathogens | 29.7 | 46.2 | 54.7 | 47.9 | 50.0 |
Abbreviations: LRTI, lower respiratory tract infection; URTI, upper respiratory tract infection.
17 respiratory viruses tested by polymerase chain reaction assays (human rhinovirus, respiratory syncytial virus A and B, influenza A and B, parainfluenza 1‐3, human metapneumovirus, human coronaviruses (OC43, NL63, 229E, HKU1), adenovirus, human polyomaviruses (KI, WU) and human bocavirus‐1).7
data based on daily symptom diary cards available for time periods of 7865/8100 high‐quality swabs taken.
Figure 2.
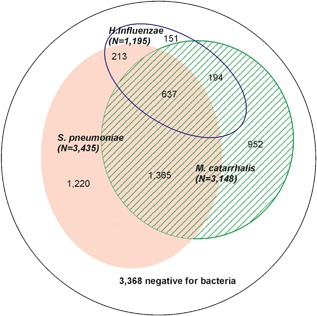
Venn diagram on swabs positive for at least one bacterium (N = 4732) in the high‐quality swabs (N = 8100)
In contrast, B. pertussis (n = 1), B. parapertussis (n = 4), and the atypical bacteria C. pneumoniae (n = 2), M. pneumoniae (n = 0), and S. negevensis (n = 0), were detected rarely or not at all. A separate analysis confirmed lower‐quality swabs provided significantly lower detections for the three major bacterial pathogens (Supplemental Digital Content 3, Table).
3.1. Association with clinical indicators
Bacterial prevalence was higher in symptomatic children (Table 2). Higher prevalence of bacteria was detected in swabs taken during respiratory symptoms, especially when acute otitis media was diagnosed. This was observed also in swabs negative for any virus (Supplemental Digital Content 4, Table). Conversely, the prevalence of respiratory symptoms was more common in bacteria‐positive (34%) compared to bacteria‐negative swabs (12%); this difference was observed both in virus‐positive (53% symptomatic in bacteria‐positives vs 35% in bacteria‐negatives) and virus‐negative (21% symptomatic in bacteria‐positives vs 7% in bacteria‐negatives) swabs. Of the virus‐negative, bacteria‐positive group, 14.9%, 6.2%, and 0.8% had symptoms of an URTI, LRTI, and acute otitis media, respectively.
A total of 269 antibiotic courses were recorded in 220 illness impact diary cards from 90 children. Exposure to antibiotics did not reduce the high bacterial detection by PCR during the illness episodes.
3.2. Risk factors for carriage
Table 3 shows the risk factors evaluated, the distributions of risk factors, and the prevalence of the bacterial findings in each stratum. Increasing age, winter and spring seasons, siblings, and attendance in formal childcare were independently associated with each of the bacterial detections. Lower maternal education was also associated with most bacterial detections. Presence of siblings seemed to modify the effect of formal childcare on bacterial detection. Children with siblings experienced high carriage prevalence earlier and the entry into formal childcare had a minor additive risk on bacterial detection (adjusted odds ratio (aOR) 2.0, 95%CI 1.4‐2.8 for S. pneumoniae carriage). Alternatively, children without siblings had very high carriage proportions associated with formal childcare (aOR 3.8, 95%CI 2.8‐5.0 for S. pneumoniae carriage) compared to children not entering formal childcare (Supplemental Digital Content 5, Figure). A similar pattern with even higher differences between the aORs was seen for the other bacterial species (data not shown).
Table 3.
Prevalence of bacterial detection in 157 children who returned high‐quality swabs, and association with participant characteristics
| Factor | Children in the stratum, N (N total = 157) | Number of swabs (N = 8100) | S. pneumoniae (42.4%) | Fully Adjusted b | M. catarrhalis (38.9%) | Fully adjusted | H. influenzae (14.8%) | Fully adjusted |
|---|---|---|---|---|---|---|---|---|
| Age (months) | ||||||||
| 0‐<3 | 157 | 1176 | 229 (19.5) | Ref | 179 (15.2) | Ref | 39 (3.3) | Ref |
| 3‐<6 | 144 | 1134 | 317 (27.9) | 1.4 (1.1, 1.7) | 282 (24.9) | 1.6 (1.2, 2.2) | 89 (7.8) | 2.0 (1.3, 3.2) |
| 6‐<12 | 136 | 2149 | 1020 (47.5) | 2.3 (1.7, 3.0) | 919 (42.8) | 3.0 (2.2, 4.0) | 271 (12.6) | 2.0 (1.3, 3.2) |
| 12‐<24 | 120 | 3641 | 1869 (51.3) | 2.0 (1.3, 3.0) | 1768 (48.6) | 3.6 (2.1, 5.3) | 796 (21.9) | 2.4 (1.3, 4.4) |
| Sex | ||||||||
| Male | 76 | 3612 | 1504 (41.6) | Ref | 1382 (38.3) | Ref | 489 (13.5) | Ref |
| Female | 82 | 4488 | 1931 (43.0) | 1.0 (0.8, 1.3) | 1766 (39.3) | 1.1 (0.8, 1.4) | 706 (15.7) | 1.1 (0.8, 1.6) |
| Siblings | ||||||||
| None | 102 | 5285 | 1989 (37.6) | Ref | 1811 (34.3) | Ref | 660 (12.5) | Ref |
| At least one | 55 | 2815 | 1446 (51.4) | 1.9 (1.5, 2.4) | 1337 (47.5) | 2.0 (1.5, 2.7) | 535 (19.0) | 2.4 (1.6, 3.4) |
| Maternal education status | ||||||||
| University degree | 99 | 5358 | 2153 (40.2) | Ref | 2039 (38.1) | Ref | 715 (13.3) | Ref |
| Diploma/Certificate | 37 | 1820 | 778 (42.8) | 1.2 (0.9, 1.6) | 664 (36.5) | 0.9 (0.7, 1.3) | 275 (15.1) | 1.3 (0.8, 1.9) |
| Primary/high school | 20 | 914 | 503 (55.0) | 1.6 (1.1, 2.3) | 444 (48.6) | 1.4 (0.9, 2.1) | 205 (22.4) | 1.6 (1.0, 2.8) |
| Tobacco smoke exposure | ||||||||
| No exposure | 136 | 7183 | 2943 (41.0) | Ref | 2717 (37.8) | Ref | 1001 (13.9) | Ref |
| Either parent smokes | 19 | 850 | 445 (52.3) | 1.2 (0.8, 1.7) | 393 (46.2) | 1.0 (0.7, 1.6) | 182 (21.4) | 1.2 (0.7, 2.0) |
| Childcare attendance a | ||||||||
| No childcare | 156 | 4107 | 1211 (29.5) | Ref | 1058 (25.8) | Ref | 272 (6.6) | Ref |
| Informal childcare | 42 | 998 | 376 (37.7) | 1.2 (0.9, 1.5) | 288 (28.9) | 0.9 (0.7, 1.2) | 84 (8.4) | 0.8 (0.5, 1.2) |
| Formal childcare | 89 | 2995 | 1848 (61.7) | 2.4 (2.0, 3.0) | 1802 (60.2) | 3.0 (2.4, 3.7) | 839 (28.0) | 2.8 (2.1, 3.8) |
| Season | ||||||||
| Summer | 145 | 1811 | 672 (37.1) | Ref | 442 (24.4) | Ref | 156 (8.6) | Ref |
| Autumn | 146 | 1989 | 785 (39.5) | 1.0 (0.9, 1.2) | 659 (33.1) | 1.6 (1.3, 1.9) | 231 (11.6) | 1.3 (1.0, 1.7) |
| Winter | 141 | 2248 | 1044 (46.4) | 1.3 (1.1, 1.5) | 1167 (51.9) | 3.3 (2.7, 4.0) | 435 (19.3) | 2.3 (1.8, 2.9) |
| Spring | 141 | 2052 | 934 (45.5) | 1.2 (1.0, 1.4) | 880 (42.9) | 2.1 (1.7, 2.5) | 373 (18.2) | 2.1 (1.6, 2.7) |
Childcare was categorized as formal (regulated care outside the child's home) or informal (non‐regulated care provided by family or friends).
Adjusted for age, sex, tobacco smoke exposure, older siblings, maternal education, childcare, and season.
4. DISCUSSION
In this community‐based birth cohort we observed the expected prevalence for upper airway carriage by the most common respiratory bacterial pathogens, S. pneumoniae, M. catarrhalis, and H. influenzae in children from birth until 24‐months of age using weekly nasal swabs collected by parents and sent by mail to the laboratory. Age, having siblings, childcare outside the home and season were the most important risk factors for bacterial carriage. In contrast, B. pertussis, B. parapertussis, and atypical bacterial pathogens were detected rarely.
Our collection methods vary from the World Health Organization standardized method for evaluating pneumococcal carriage,12 which recommend nasopharyngeal swabs (NPS), storing samples in tubes containing skim‐milk tryptone glucose glycerol transport media and detecting by culture. However, nasal swabs are also reported to be comparable to NPS for detecting S. pneumoniae,13, 14 M. catarrhalis,14 and H. influenzae 13, 14 in children with ARI symptoms. Moreover, parent‐collected nasal swabs and healthcare worker‐collected NPS had similar detection rates for S. pneumoniae in healthy children,15 although yields for M. catarrhalis and H. influenzae in parent‐collected nasal swabs were slightly lower. Furthermore, compared to healthcare worker‐collected NPS from children during ARI episodes, we have observed similar yields by PCR of H. influenzae, and higher yields of S. pneumoniae and M. catarrhalis, from parent‐collected anterior nasal swabs mailed into the laboratory (Lambert, unpublished RESPECT data; http://clinicaltrials.gov: NCT00966069). In addition, a recent 16S rDNA sequencing study in hospitalized infants with bronchiolitis found good within‐individual correlations between Haemophilus and Moraxella in nasopharyngeal aspirate and mailed nasal swab specimens.16 Finally, while the oropharynx rather than nasopharynx has been suggested as the optimal sampling site for H. influenzae, this observation may be biased by inadequate differentiation between H. influenzae and H. haemolyticus, the latter being common in the oropharynx.12, 17
Another difference with the WHO method was storing nasal swabs in antibiotic‐containing viral transport medium and identifying bacteria by real‐time PCR instead of culture. However, this method has been used before with high yields of bacterial detection consistent with other studies in similar populations.11 Indeed, PCR may offer advantages of increased detection sensitivity when target bacterial loads are low or dominated by other bacteria,18, 19 or after antimicrobial exposure as observed in our data.
Despite differences in methods our results agree with earlier prospective studies. Prevalence by age was slightly higher than observed during the 1990s in Finland,20 but similar to recent studies from the United States,21 and lower than reported from Peru18 and Thailand.22 Although, our study was conducted during an established infant pneumococcal conjugate vaccine (PCV) vaccination program, PCVs affect the vaccine‐type carriage, but not the overall carriage of all serotypes, and therefore, should not impact the current findings despite serotype data being unavailable. Additionally, epidemiologic patterns of seasonality, age‐dependent prevalence, and risk factors were consistent with previous studies.20, 22, 23, 24
In concordance with earlier studies, we also observed the association of bacterial detection with respiratory symptoms.25 Furthermore, although symptoms were more prevalent in virus‐positives, the association of bacterial detection with symptoms was also seen in virus‐negatives. The longitudinal design of the ORChID study with frequent sampling offers possibilities for detailed analyses on temporal and causal relationships of viruses and bacteria, and their interactions, with symptoms.
An interesting finding was the high proportion of bacterial co‐detections, compared to those based on marginal prevalences, in children sharing the same risk factors.26, 27 Bacteria may act synergistically in the nasopharynx, although competition can also occur.28 PCR detection is probably much more efficient at detecting multiple bacteria than conventional cultures. Nevertheless, the concomitant carriage was especially striking for H. influenzae, which was detected alone in only a very small proportion of samples. Meanwhile, B. pertussis and B. parapertussis are reported uncommonly in infants with ARI symptoms.29 The incidence of B. pertussis and B. parapertussis infections in children aged <3‐years in a large pertussis vaccine trial with active follow‐up was 29 and 2.1 per 1000 person‐years, respectively.30, 31 This was higher than the estimated national Australian incidence of 4 per 1000 person‐years identified through passive surveillance, despite considerably increased reported cases since 2010 and adopting PCR for diagnosis.32 Thus, having only 1‐2 children with positive B. pertussis and B. parapertussis swabs, respectively, was not unexpected. Additionally, M. pneumoniae, C. pneumoniae, and S. negevensis occur more commonly in older children,33, 34 and large numbers of positive children in this age group were not anticipated. Due to the low numbers of positives, the suitability of our detection methodology for atypical pathogens remains uncertain.
This study confirms previous findings that the main risk factors for carrying respiratory bacterial pathogens are age, having siblings, formal childcare attendance, and winter‐spring season with frequent viral infections.20, 22, 23, 24, 25, 34, 35 Of these, formal childcare attendance is likely the main modifiable risk factor for reducing the burden of respiratory tract carriage and infections. The interaction of childcare attendance with having siblings suggests that children without siblings might be protected if other options for formal childcare attendance were available during the most vulnerable age periods.
The strengths of the ORChID study include its prospective design, the comprehensive surveillance provided by daily symptom diaries, and weekly sample collections from birth until age 24‐months. Enrolment over two calendar years and follow‐up of 4‐years allowed for different seasonal and year‐to‐year epidemiologic patterns. Considering its intensive nature, cohort retention was good with >70% followed for at least 18‐months, and 68% of the maximum expected swabs were collected with a median interval of 1‐week as stated in the protocol.6 Routine laboratory quality assurance included assaying samples for the human DNA marker, ERV3, to ensure human cells were present. While important for viral detection,10 it seems also relevant for bacteria as we observed considerably higher bacterial detection rates in samples containing ERV‐3. Consequently, only high‐quality nasal swabs were used to avoid compromising sensitivity in the analysis.
There are, however, important limitations to consider. We did not validate upper airway bacterial detection by nasal swab and PCR against the gold standard of NPS culture. However, our current analyses using previously validated PCR targets resulted in expected bacterial prevalence rates. The specificity of the PCR assays are high, yet the ply‐PCR may cross‐react with oral streptococci.36 We therefore re‐analyzed 34 ply‐PCR positives with lytA PCR, of which 31 (91%) remained positive (data not shown). Differentiating between H. influenzae and the closely related respiratory tract commensal, H. haemolyticus can also be difficult, although the latter is uncommonly detected in the nasal cavity and the hpd#3 target gene in our PCR assay is both sensitive and specific for differentiating between the two species.17, 37 Storing nasal swabs in the refrigerator and sending by mail before freezing 4 to 11‐days post‐sampling results in a 10‐13% decrease in sensitivity for detecting bacteria by PCR, with some variation according to the bacterial species.11 However, in our study transport times were much shorter (median 3‐days, interquartile range, 2‐4). Another limitation is that the assays lacked serotype/genotype determination for the bacterial detections. Evaluating bacterial species only, underestimates acquisition rates by different bacterial subtypes. Therefore, in the current analysis we focused upon the prevalence when evaluating the overall burden of bacterial carriage in young children. Finally, the ORChID study was conducted in one major subtropical city in Australia and as often occurs with studies of this nature involved smaller, socially‐advantaged families. Nonetheless, the findings are valid and prevalence of bacterial detection agrees with cohort studies from other settings18, 20, 21, 22, 23, 24 as do the rates of ARI and virus detection.7, 9
In conclusion, we have used a novel method to evaluate bacterial carriage in children using nasal swabs optimized initially for viral detection. This method, however, is suitable also for bacterial detection and is especially feasible when frequent sampling is required. Future analyses will explore interactions between nasal viruses, bacteria, and ARI symptoms, while subtyping will allow carriage dynamics to be explored in greater detail.
CONFLICTS OF INTEREST
Arto A. Palmu has received research funding from GlaxoSmithKline Vaccines for a nationwide effectiveness trial of 10‐valent pneumococcal conjugate vaccine and from Wyeth‐Lederle Vaccines and Pediatrics (currently Pfizer Inc.) for the FinOM trial. Michael D. Nissen is currently a full‐time employee of GlaxoSmithKline, Vaccines, 23 Rochester Park, Singapore 139234, Singapore. Other authors have no conflicts of interest to declare.
Supporting information
Additional supporting information may be found online in the Supporting Information section at the end of the article.
Supporting Figure S1.
Supporting Figure S2.
Supporting Table S1.
Supporting Table S2.
Supporting Table S3.
ACKNOWLEDGMENTS
This study was supported by an Australian National Health and Medical Research Council (NHMRC) project grant (GNT: 615700) and a Children's Hospital Foundation Queensland (CHFQ) program grant (CHFQ: 5006). The sabbatical leave at Griffith University for AAP was supported by the National Institute for Health and Welfare, Finland. SBL received an NHMRC Early Career Fellowship and a CHFQ Mid‐career Fellowship. MS is the recipient of a NHMRC Dora Lush Clinical Scholarship and a CHFQ PhD Clinical Scholarship. KLS received a NHMRC Career Development Fellowship.
Palmu AA, Ware RS, Lambert SB, et al. Nasal swab bacteriology by PCR during the first 24‐months of life: A prospective birth cohort study. Pediatric Pulmonology. 2019;54:289–296. 10.1002/ppul.24231
For cooperative studies, the institution where research was primarily done should be indicated: Menzies Health Institute Queensland, Griffith University, Gold Coast, Queensland.
Identify meetings, if any, at which the paper was presented: The results have been partly presented at the 10th Congress of the World Society for Pediatric Infectious Diseases (WSPID) in Shenzhen, China, December 2‐5, 2017 and in 11th International Symposium on Pneumococci and Pneumococcal Diseases (ISPPD) in Melbourne, Australia, 15‐19th April 2018.
REFERENCES
- 1.GBD 2015 LRI Collaborators. Estimates of the global, regional, and national morbidity, mortality, and aetiologies of lower respiratory tract infections in 195 countries: a systematic analysis for the Global Burden of Disease Study 2015. Lancet Infect Dis. 2017; 17:1133–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Schilder AG, Chonmaitree T, Cripps AW, et al. Otitis media. Nat Rev Dis Primers. 2016; 2:16063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. DeAntonio R, Yarzabal JP, Cruz JP, Schmidt JE, Kleijnen J. Epidemiology of community‐acquired pneumonia and implications for vaccination of children living in developing and newly industrialized countries: a systematic literature review. Hum Vaccin Immunother. 2016; 12:2422–2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Simell B, Auranen K, Käyhty H, et al. The fundamental link between pneumococcal carriage and disease. Expert Rev Vaccines. 2012; 11:841–855. [DOI] [PubMed] [Google Scholar]
- 5. Spuesens EB, Fraaij PL, Visser EG, et al. Carriage of Mycoplasma pneumoniae in the upper respiratory tract of symptomatic and asymptomatic children: an observational study. PLoS Med. 2013; 10:e1001444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lambert SB, Ware RS, Cook AL, et al. Observational research in childhood infectious diseases (ORChID): a dynamic birth cohort study. BMJ Open. 2012; 2:e002134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sarna M, Lambert SB, Sloots TP, et al. Viruses causing lower respiratory symptoms in young children: findings from the ORChID birth cohort. Thorax. 2018; 73:969–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ye S, Whiley DM, Ware RS, et al. Detection of viruses in weekly stool specimens collected during the first 2 years of life: a pilot study of five healthy Australian infants in the rotavirus vaccine era. J Med Virol. 2017; 89:917–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sarna M, Ware RS, Sloots TP, Nissen MD, Grimwood K, Lambert SB. The burden of community‐managed acute respiratory infections in the first 2‐years of life. Pediatr Pulmonol. 2016; 51:1336–1346. [DOI] [PubMed] [Google Scholar]
- 10. Alsaleh AN, Whiley DM, Bialasiewicz S, et al. Nasal swab samples and real‐time polymerase chain reaction assays in community‐based, longitudinal studies of respiratory viruses: the importance of sample integrity and quality control. BMC Infect Dis. 2014; 14:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. O'Grady KA, Whiley DM, Torzillo PJ, Sloots TP, Lambert SB. Mailed versus frozen transport of nasal swabs for surveillance of respiratory bacteria in remote Indigenous communities in Australia. BMC Infect Dis. 2013; 13:543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Satzke C, Turner P, Virolainen‐Julkunen A, et al. Standard method for detecting upper respiratory carriage of Streptococcus pneumoniae: updated recommendations from the World Health Organization Pneumococcal Carriage Working Group. Vaccine. 2013; 32:165–179. [DOI] [PubMed] [Google Scholar]
- 13. Rapola S, Salo E, Kiiski P, Leinonen M, Takala AK. Comparison of four different sampling methods for detecting pharyngeal carriage of Streptococcus pneumoniae and Haemophilus influenzae in children. J Clin Microbiol. 1997; 35:1077–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. van den Bergh MR, Bogaert D, Dun L, et al. Alternative sampling methods for detecting bacterial pathogens in children with upper respiratory tract infections. J Clin Microbiol. 2012; 50:4134–4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Coughtrie AL, Whittaker RN, Begum N, et al. Evaluation of swabbing methods for estimating the prevalence of bacterial carriage in the upper respiratory tract: a cross sectional study. BMJ Open. 2014; 4:e005341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Luna PN, Hasegawa K, Ajami NJ, et al. The association between anterior nares and nasopharyngeal microbiota in infants hospitalized for bronchiolitis. Microbiome. 2018; 6:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hare KM, Binks MJ, Grimwood K, et al. Culture and PCR detection of Haemophilus influenzae and Haemophilus haemolyticus in Australian Indigenous children with bronchiectasis. J Clin Microbiol. 2012; 50:2444–2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chien YW, Vidal JE, Grijalva CG, et al. Density interactions among Streptococcus pneumoniae, Haemophilus influenzae and Staphylococcus aureus in the nasopharynx of young Peruvian children. Pediatr Infect Dis J. 2013; 32:72–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wyllie AL, Wijmenga‐Monsuur AJ, van Houten MA, et al. Molecular surveillance of nasopharyngeal carriage of Streptococcus pneumoniae in children vaccinated with conjugated polysaccharide pneumococcal vaccines. Sci Rep. 2016; 6:23809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Syrjänen RK, Kilpi TM, Kaijalainen TH, Herva EE, Takala AK. Nasopharyngeal carriage of Streptococcus pneumoniae in Finnish children younger than 2 years old. J Infect Dis. 2001; 184:451–459. [DOI] [PubMed] [Google Scholar]
- 21. Xu Q, Casey JR, Newman E, Pichichero ME. Otitis‐prone children have immunologic deficiencies in naturally acquired nasopharyngeal mucosal antibody response after Streptococcus pneumoniae colonization. Pediatr Infect Dis J. 2016; 35:54–60. [DOI] [PubMed] [Google Scholar]
- 22. Turner P, Turner C, Jankhot A, et al. A longitudinal study of Streptococcus pneumoniae carriage in a cohort of infants and their mothers on the Thailand‐Myanmar border. PLoS ONE. 2012; 7:e38271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gray BM, Converse GM, Dillon HC, Jr . Epidemiologic studies of Streptococcus pneumoniae in infants: acquisition, carriage, and infection during the first 24 months of life. J Infect Dis. 1980; 142:923–933. [DOI] [PubMed] [Google Scholar]
- 24. Gray BM, Turner ME, Dillon HC, Jr . Epidemiologic studies of Streptococcus pneumoniae in infants. The effects of season and age on pneumococcal acquisition and carriage in the first 24 months of life. Am J Epidemiol. 1982; 116:692–703. [DOI] [PubMed] [Google Scholar]
- 25. Rodrigues F, Foster D, Nicoli E, et al. Relationship between rhinitis symptoms, respiratory viral infections and nasopharyngeal colonization with Streptococcus pneumoniae, Haemophilus influenzae and Staphylococcus aureus in children attending daycare. Pediatr Infect Dis J. 2013; 32:227–232. [DOI] [PubMed] [Google Scholar]
- 26. Jacoby P, Watson K, Bowman J, et al. Modelling the co‐occurrence of Streptococcus pneumoniae with other bacterial and viral pathogens in the upper respiratory tract. Vaccine. 2007; 25:2458–2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lewnard JA, Givon‐Lavi N, Huppert A, et al. Epidemiological markers for interactions among Streptococcus pneumoniae, Haemophilus influenzae, and Staphylococcus aureus in upper respiratory tract carriage. J Infect Dis. 2016; 213:1596–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lewnard JA, Huppert A, Givon‐Lavi N, et al. Density, serotype diversity, and fitness of Streptococcus pneumoniae in upper respiratory tract cocolonization with nontypeable Haemophilus influenzae. J Infect Dis. 2016; 214:1411–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Walsh PF, Kimmel L, Feola M, et al. Prevalence of Bordetella pertussis and Bordetella parapertussis in infants presenting to the emergency department with bronchiolitis. J Emerg Med. 2011; 40:256–261. [DOI] [PubMed] [Google Scholar]
- 30. Greco D, Salmaso S, Mastrantonio P, et al. A controlled trial of two acellular vaccines and one whole‐cell vaccine against pertussis. Progetto Pertosse Working Group. N Engl J Med. 1996; 334:341–348. [DOI] [PubMed] [Google Scholar]
- 31. Mastrantonio P, Stefanelli P, Giuliano M, et al. Bordetella parapertussis infection in children: epidemiology, clinical symptoms, and molecular characteristics of isolates. J Clin Microbiol. 1998; 36:999–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.WHO SAGE Pertussis Working Group. WHO SAGE Pertussis Working Group. Background Paper. SAGE April 2014. 2014. Available at: http://www.who.int/immunization/sage/meetings/2014/april/1_Pertussis_background_FINAL4_web.pdf?ua=. Accessed July 6th, 2018.
- 33. Weigl JA, Puppe W, Meyer CU, et al. Ten years' experience with year‐round active surveillance of up to 19 respiratory pathogens in children. Eur J Pediatr. 2007; 166:957–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Heiskanen‐Kosma T, Paldanius M, Korppi M. Simkania negevensis may be a true cause of community acquired pneumonia in children. Scand J Infect Dis. 2008; 40:127–130. [DOI] [PubMed] [Google Scholar]
- 35. Jacoby P, Carville KS, Hall G, et al. Crowding and other strong predictors of upper respiratory tract carriage of otitis media‐related bacteria in Australian Aboriginal and non‐Aboriginal children. Pediatr Infect Dis J. 2011; 30:480–485. [DOI] [PubMed] [Google Scholar]
- 36. Carvalho Mda G, Tondella ML, McCaustland K, et al. Evaluation and improvement of real‐time PCR assays targeting lytA, ply, and psaA genes for detection of pneumococcal DNA. J Clin Microbiol. 2007; 45:2460–2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Binks MJ, Temple B, Kirkham LA, et al. Molecular surveillance of true nontypeable Haemophilus influenzae: an evaluation of PCR screening assays. PLoS ONE. 2012; 7:e34083. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional supporting information may be found online in the Supporting Information section at the end of the article.
Supporting Figure S1.
Supporting Figure S2.
Supporting Table S1.
Supporting Table S2.
Supporting Table S3.


