Abstract
| Summary | 16 | |
| I. | I. | 16 |
| II. | II. | 18 |
| III. | III. | 20 |
| IV. | IV. | 23 |
| Acknowledgements | 24 | |
| References | 24 |
Summary
This review discusses the varying roles that have been played by many plant‐viral regulatory sequences and proteins in the creation of plant‐based expression systems and virus particles for use in nanotechnology. Essentially, there are two ways of expressing an exogenous protein: the creation of transgenic plants possessing a stably integrated gene construction, or the transient expression of the desired gene following the infiltration of the gene construct. Both depend on disarmed strains of Agrobacterium tumefaciens to deliver the created gene construction into cell nuclei, usually through the deployment of virus‐derived components. The importance of efficient mRNA translation in the latter process is highlighted. Plant viruses replicate to sustain an infection to promote their survival. The major product of this, the virus particle, is finding increasing roles in the emerging field of bionanotechnology. One of the major products of plant‐viral expression is the virus‐like particle (VLP). These are increasingly playing a role in vaccine development. Similarly, many VLPs are suitable for the investigation of the many facets of the emerging field of synthetic biology, which encompasses the design and construction of new biological functions and systems not found in nature. Genetic and chemical modifications to plant‐generated VLPs serve as ideal starter templates for many downstream synthetic biology applications.
Keywords: loading, modification, protein expression, synthetic biology, transgenic, transient, virus particle
I. Transgenic gene expression
The production of numerous transgenic plants has relied heavily on plasmid vectors that have the ability to transfer the gene of interest into plant cell nuclei. Their propagation, not only in Agrobacterium (essential for plant infection), but in common laboratory hosts, such as Escherichia coli, for genetic manipulation has been essential for their development. These plasmids, referred to as binary vectors, were originally derived from the tumorigenic (Ti) strains of Agrobacterium, and are of minimal size before the addition of the gene of interest. They possess essential elements comprising defined left and right borders (LB and RB, Fig. 1a), sequences between which the gene of interest, with its promoter and terminator sequences, is located. The LB and RB sequences are vital for the transfer of the nucleic acid located between the border sequences into the host nucleus and, ultimately, its chromosome. Virulence factors encoded within the Ti plasmid, essential for the transfer of the genetic material into the nuclear genome, are located on other plasmids harboured by the Agrobacterium species used in the transformation experiment. Figure 1(a) highlights the many features of a transformation vector that are essential for the expression of a gene of interest in a plant. Many plant virus‐derived sequences, such as promoter sequences, translational enhancement sequences and RNA interference (RNAi) suppressor sequences (discussed in detail later), have been utilized in varying forms to enhance the expression of the transgenes. The gene or genes, and selectable markers, such as neomycin phosphotransferase II (NPTII), used for the selection of transformed cells, are, in most cases, driven by transcription from the Cauliflower mosaic virus (CaMV) 35S promoter sequence. This promoter sequence is constitutive and, in the CaMV replication cycle, is responsible for the generation of full‐length RNA copies of the DNA viral genome in CaMV‐infected cells. The 35S promoter was one of the first plant virus promoters to be characterized in detail and, to this day, is often the promoter of choice, unless tissue‐specific or inducible expression is desired. The natural host range for CaMV is that of the Brassica plant family, and thus it is not surprising that the activity of this promoter in transgenic tobacco (a member of the Solanaceae) is much reduced compared with its activity in CaMV‐infected plants. Thus, transcription rates from the 35S promoter are often improved through the use of tandem copies of the promoter.
Figure 1.
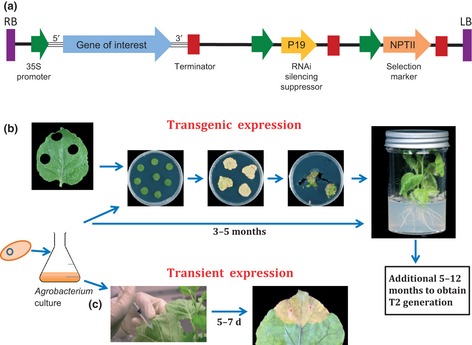
Relative location of plant virus‐derived components, between the left and right border sequences, of a binary expression vector, and an illustration of the timescale required to achieve protein expression by the use of transgenic or transient expression. (a) The gene of interest is cloned between the CaMV 35S promoter and the nopaline synthase (nos) transcription terminator and can incorporate 5′ and 3′ nonencoding plant viral sequences. Incorporation of the P19 or HC‐Pro (sometimes mutated versions) RNA interference (RNAi) suppression gene sequences will enhance gene expression. An antibiotic selection marker, such as the neomycin phosphotransferase II (NPTII) gene, must be included for the isolation of transgenic plants, as only the DNA sequence located between the left and right border elements is incorporated into the host genome. Once verified, the gene construct must be transformed into Agrobacterium for plant cell infection and expression. (b, c) Illustration of the experimental steps for successful gene expression in transgenic or transient expression after the initial infection of leaf material. Gene expression can be readily verified when using transient expression, typically within 2 wk, compared with well over 1 yr for stable transgenic expression.
The initial stage of infection with single‐stranded RNA plant viruses is the translation of the incoming RNA. Consequently, the genomic RNAs of such viruses are very efficient at recruiting host ribosomes and have also evolved highly efficient translational abilities. Thus, the nonencoding 5′ and 3′ ends of these viral RNAs are often employed to enhance the translation of genes in transgenic plants (Fig. 1a). In particular, the Tobacco etch virus (TEV) 5′ leader sequence has been shown to enhance indoleacetic acid‐lysine synthetase activity seven‐fold compared with transgenic plants without the TEV leader sequence (Savka et al., 2001). Transgenic plants expressing recombinant hepatitis B surface antigen virus‐like particles (VLPs) have also employed the TEV leader sequence to enhance expression (Mason et al., 1992). Indeed, VLPs of a number of viruses have been successfully expressed in transgenic plants with the use of both the 35S CaMV promoter and the TEV leader sequence (reviewed by Korban, 2002). The Tobacco mosaic virus (TMV) 5′ omega leader sequence enhances the expression of foreign gene transcripts in vitro and in vivo (Gallie et al., 1987b). Creager et al. (1999), celebrating the first century of TMV research at the Royal College of Physicians of Edinburgh, UK, in 1998, reported the commercial use of the TMV omega leader sequence for the transgenic expression of therapeutic human enzymes from field‐grown tobacco plants. The translational enhancement of both the TEV and TMV omega leader sequences for the expression of the human calcitonin gene in independent transgenic potato plants showed that the TMV omega element provided an increase of between two‐ and three‐fold of that which was achieved by the TEV leader sequence (Ofoghi et al., 2005). An expression system based on Cowpea mosaic virus (CPMV) utilizes mutated forms of the 5′ RNA‐2 leader (discussed below) to enhance expression in transgenic plants; the expression could be further enhanced by the simultaneous expression of a modified viral RNA silencing suppressor gene (Saxena et al., 2011).
The process by which stable transgenic plants are created is dependent on the successful transformation of an individual plant cell, which is then cultivated, selected and maintained on hormone‐containing growth medium (Fig. 1b). However, the selection of the desired transformed cell and its subsequent development to form callus tissue, differentiated structures, such as stem, roots and leaves, and, finally, a mature plant is often time‐consuming. Transgenic plants resulting from this procedure must be subjected to intensive characterization procedures, in particular for the selection of those lines with high expression levels. Lines of plants so identified must then undergo several growth cycles, including rounds of self‐fertilization, before sufficient quantities of true‐breeding seed are available to permit commercial protein production. The process may well take up to a year or more to achieve lines of plants with the desired expression characteristics. Facilities, such as glasshouses specifically designed for the containment of transgenic pollen, are employed for their growth to satisfy regulatory authorities. Often, it is not possible to achieve the successful transformation of a desired plant species ideally suited for the transgene. Consequently, Nicotiana species have become the plants of choice for most transgenes, because of their ease of transformation and regeneration. Paul & Ma (2011) have discussed, in depth, the range of promising product–platform combinations, and also emphasize synergies during production and in clinical trials of plant recombinant protein production. The potential uses of different plant species and/or expression in specific subcellular locations, such as chloroplasts, for transgenic recombinant protein production, in particular, for vaccine development, have been reviewed by Rybicki (2010).
The yield of the desired protein of interest in transgenic plants is, in many cases, rather low. A major reason for this is that a transgenic plant is often a battleground between the gene construct, in its quest for a high yield of the transgene, and the plant defence systems, which attempt to eliminate or reduce this unwanted passenger that is alien to its metabolism. Plants have developed a sophisticated mechanism, referred to as RNA silencing, by which unwanted host mRNA species are reduced or eliminated by a specific degradative pathway leading to the production of small interfering RNAs (siRNAs). By this means, a level of gene expression and regulation is achieved by the plant post‐transcription. Similarly, this process also offers some protection to the plant in the event of a virus infection. Thus, in order to be successful, plant viruses have the ability to disrupt RNA silencing often by the expression of specific viral products, known as silencing suppressors, which interfere at various stages of the silencing pathway. These include the HC‐Pro gene found in potyviruses and the P19 gene of Tomato bushy stunt virus (TBSV). When incorporated into gene constructs (Fig. 1a), the yield of a transgenic product can be improved as the mRNA encoding it is stabilized. However, developmental problems often arise when wild‐type suppressors are deployed, often leading to a reduction in the number of transformed plants generated from transformation experiments. Thus, mutant forms of P19 have now superseded the use of wild‐type P19 for transgenic plant production (Saxena et al., 2011; Garabagi et al., 2012).
II. Transient gene expression
Gene expression can be achieved by the inoculation or, more accurately, the infiltration of an Agrobacterium culture containing a transformation plasmid into plant leaves (Fig. 1c). As for a transgenic plant study, the fast‐growing and highly susceptible common laboratory host Nicotiana benthamiana is frequently used, although other plant species, such as lettuce, have been employed. Potentially, the requirement of having to select and use a particular plant species, just because it is amenable to transformation, but is unfavourable for the expression of the exogenous gene under investigation, is no longer a constraint. Similar to the creation of a transgenic plant, transient expression mediated by Agrobacterium plasmid vectors is dependent on the transfer of genetic material into the nucleus of the cell. However, for transient expression to succeed, it is not necessary for the DNA between the LB and RB to be incorporated into the host chromosome, but only for its transcription to occur at some stage after the transfer into the nucleus. Thus, transient expression is dependent on the same regulatory signals that are necessary for successful expression in a transgenic plant. Again, use is often made of the CaMV 35S promoter, and the incorporation of RNA silencing suppressors, such as HC‐Pro or P19, into gene constructs will enhance expression. Targeting signals, although not of plant virus origin, for directing gene expression to selected tissues, such as the endoplasmic reticulum or chloroplasts etc., may also be included. Acetosyringone, a natural plant hormone induced during a wounding response, is often included with Agrobacterium in the infiltration solution (Sheikholeslam & Weeks, 1987). Its use increases the efficiency of T‐DNA transfer into the nucleus, thereby allowing gene expression in almost the entire leaf tissue. Transient gene expression by syringe infiltration offers a rapid and reliable procedure to test several gene constructs over a relatively short space of time, usually 5–7 d. The technique depends on the wounding of the leaf surface, usually with a syringe needle, followed by the injection of the Agrobacterium solution via a blunt‐ended syringe into the wounded surface. Once a particular culture has been characterized for its expression, the process, if desired, can be scaled up by performing vacuum infiltrations. Here, whole plants are immersed in the Agrobacterium solution and subjected to negative pressure, whereupon the air in the intercellular space is replaced with the bacterial solution. Ultimately, this process can be automated for commercial applications for the production of plant‐derived pharmaceuticals.
An example of a highly effective transient expression system making use of viral components is the CPMV‐HT system. This makes use of the ability of a modified 5′ untranslated region (UTR) from CPMV RNA‐2 to enhance translation when placed in front of a gene construct (Sainsbury & Lomonossoff, 2008). Expression is further enhanced by placing the 3′ UTR from CPMV RNA‐2 after the coding sequence and co‐expressing the P19 silencing suppressor from TBSV to stabilize the transcribed RNA. These features have been incorporated into the pEAQ‐HT series of transient expression vectors (Sainsbury et al., 2009). Here, the gene of interest is placed in a ‘CPMV‐HT cassette’, which consists of the CaMV 35S promoter and nopaline synthase (nos) terminator surrounding the modified 5′ UTR (HT) and 3′ UTR of CPMV RNA‐2, between which are located multiple restriction endonuclease cloning sites. A series of Gateway‐compatible versions of this plasmid are also available and offer the choice of either amino or carboxyl histidine (His)‐tagged protein expression. A description of the principles of Gateway cloning is given in Esposito et al. (2009). To demonstrate the potential of pEAQ‐HT as a vector for the production of plant‐derived pharmaceuticals, the anti‐human immunodeficiency virus (anti‐HIV) monoclonal antibody, 2G12, was expressed in plants and shown to have similar anti‐HIV activity to Chinese hamster ovary cell‐produced 2G12 (Sainsbury et al., 2010a).
Many VLPs have been successfully expressed in plant leaves utilizing pEAQ‐HT‐derived binary vectors. VLPs are authentic copies of virions or particles, but are unable to replicate and generate viral symptoms of infection when inoculated into the host cell. However, they may still be able to encapsidate nucleic acid, not necessary of viral origin, as a consequence of the formation of the VLP. Hence, their potential use as safe vaccines merits them worthy of investigation (Kushnir et al., 2012). For some viruses, such as CPMV, it is possible to produce empty VLPs (eVLPs) that are entirely devoid of nucleic acid. As a consequence of CPMV infection, empty but naturally occurring top component particles are readily synthesized alongside the RNA‐1 and RNA‐2 independently encapsidated bottom and middle components. The presence of the top component at c. 10% of the viral yield is considered to be a by‐product, the result of failure to encapsidate viral nucleic acid by the virus replicating machinery. Large amounts of CPMV eVLPs are readily formed in infiltrated leaves by the transient expression of the virus capsid precursor protein, VP60, together with the CPMV 24K proteinase (Saunders et al., 2009). Here, protein expression of both is driven solely by HT expression. An elevated level of eVLP formation, if desired, can be achieved by the co‐expression of both proteins when cloned onto one single binary vector. Thus, CPMV eVLP expression is a consequence of the interaction between a protein substrate, VP60, and its viral‐encoded 24K proteinase, whereupon 60 copies of VP60 are cleaved to form the large (L) and small (S) coat proteins found in a virus particle. Transient and independent expression of the mature L and S coat proteins fails to yield mature eVLPs. Similarly, plants created for the transgenic simultaneous expression of both L and S capsid proteins fail to form particles (P. Saxena & G. P. Lomonossoff, unpublished). This strongly suggests that the enzymatic processing of the capsid precursor protein, or several closely associated capsid precursor proteins, is vital for particle formation and is not dependent on the simple coming together of the preformed L and S capsid components. The in planta expression, via the pEAQ‐HT vector, of the single coat protein component of the animal virus hepatitis core protein resulted in VLP formation. Here, these VLPs are formed by the interaction of at least 120 or 180 copies of a single protein. It has also proved possible to synthesize the complex Bluetongue VLPs (BTV VLPs) in plants by the transient expression of the four capsid proteins (Thuenemann et al., 2013). Presumably, particle formation is achieved by the self‐interaction of the relevant capsid components at the correct molar ratio. There are two distinct protein layers in a BTV particle. VP3 and VP7 form the inner layer or core, whereas VP2 and VP5 are assembled on the core layer to form mature VLPs. Formation of just the core layer is possible with the expression of just VP3 and VP7.
As an alternative to the development of Agrobacterium‐mediated expression vectors, transient expression has also been achieved through the development of replicating plant virus expression vectors. These vectors are usually initially introduced into the plants by agroinfiltration, but achieve their high expression levels through their ability to replicate and spread within the plant. Initially, many virus‐based vectors were based on wild‐type viruses, with the gene to be expressed being added as an addition to the complement of viral genes. However, such vectors suffer from problems of biocontainment; thus, more recently, plant viruses have been genetically modified to create a deconstructed virus vector, in which viral replication ability is preserved, allowing for the expression of an exogenous gene, but the ability of the vector to spread in the environment is curtailed. An example of such a system is where the virus genes required for virus replication, assembly and movement are supplied in trans with the gene of interest under investigation being supplied on a separate plasmid vector (for a review of deconstructed viral systems, see Hefferon, 2012). Expression of the gene in question is dependent on an active, but usually restricted, virus infection. Thus, a deconstructed vector utilizing the Rep gene of the geminivirus Bean yellow dwarf virus (Huang et al., 2009) has been developed, and the expression of norovirus‐like particles has been achieved in plants when this coat protein is cloned between the duplicated copies of the geminiviral large intergenic region (LIR) on a complementary plasmid. Here, interaction between the Rep protein and the LIR sequences occurs in the cell nucleus in order to generate mRNA of the gene of interest necessary for cytoplasmic expression.
Single‐stranded RNA plant viruses have also been developed as deconstructed virus vectors. A TMV‐based vector in which the genomic RNA is split into 5′ and 3′ modules has been used extensively (MagniCON system) (Marillonnet et al., 2004; Gleba et al., 2005). Recently, for the production of a functional immunoglobulin G (IgG) (Giritch et al., 2006), vectors derived from both TMV and Potato virus X (PVX) were developed. Gateway‐compatible TMV vectors, the TMV‐Gate vectors (Kagale et al., 2012), have recently been developed and offer, in addition to a high level of the expressed protein, the ability to express proteins with either N‐ or C‐terminal fusions to a broad series of epitope tags and fluorescent proteins.
III. Exploitation of viral shells as templates for synthetic biological applications
The advent of techniques for the manipulation of the genomes of RNA viruses, coupled with structural studies of their capsids to high resolution and the recent use of heterologous protein expression systems, has allowed plant virus particles to be at the forefront of the exploitation for applications in both bio‐ and nanotechnology. Plant virus capsids are particularly suited to this application in view of their stability to both temperature and the use of organic solvents. Furthermore, for many plant viruses, efficient in vitro assembly systems have been established. Particles can be produced either by infection of appropriate plants or by expression of the coat protein in either heterologous systems, such as E. coli, yeast or insect cells, or plants using transient expression approaches such as the pEAQ‐HT vector system described above. The major difference between the approaches is that the use of infection results in the production of particles containing the viral genome; such particles are, themselves, infectious. The use of heterologous systems results in the production of particles which either encapsidate host RNA molecules (e.g. Cowpea chlorotic mottle virus (CCMV) (Zhao et al., 1995), Turnip crinkle virus (TCV) (K. Saunders & P. G. Lomonossoff, unpublished)) or are empty (CPMV; Saunders et al., 2009); such particles are not infectious and are termed ‘virus‐like particles’ (VLPs). Plant virus particles have been exploited in three different ways: modification of the outer capsid surface (genetically, chemically or a combination of the two); exploitation of the inner cavity; and the incorporation of particles into supramolecular structures.
1. Modifications to the outer surface
The first examples of the modification of the outer surface of virus particles involved the genetic modification of the coat protein (Lomonossoff & Johnson, 1996). The original motivation for this work was to modify particles to express antigenic peptides; such modified particles (chimaeras) could potentially serve as novel subunit vaccines (Montague et al., 2011). Subsequently, the alternative approach of chemically modifying particles was explored. Several of the amino acids within viral coat protein subunits have side chains which are suitable for chemical modification. These include the carboxyl groups of aspartic and glutamic acid, the ε‐amino group of lysine, the thiol group of cysteine and the hydroxyl group of tyrosine. When such side chains are exposed on the outer surface of the virus particle, they are addressable by a number of chemical reactants, allowing the virus particles to be modified in vitro. This allows the introduction of a greater range of moieties than is possible to introduce genetically. It is also possible to combine genetic and chemical modification of the virus surface by the introduction or elimination of defined amino acids, thereby modulating the reactivity of the virus particles. It is also possible to genetically insert peptides, which catalyse certain reactions, such as the specific deposition of minerals. The capsids of a number of plant viruses with varying morphologies have been modified, both genetically and chemically, on their exterior surfaces, thus allowing for their display. The main prerequisite for genetic modification is that the presence of the foreign sequence does not interfere with the ability of the modified coat protein to assemble into virions or VLPs. For chemical modification, it is important that the reaction conditions are not so harsh that they disrupt the virus structure and that some information is available about the numbers and types of addressable groups. For these reasons, attention, to date, has focused on those viruses which are known to be robust and for which there is at least some information available about the topology of the coat protein in the assembled virions.
CPMV was the first plant virus to be developed as a system for the display of foreign peptides (Usha et al., 1993; Porta et al., 1994, 1996) and has subsequently been used extensively for chemical modification (Chatterji et al., 2002; Steinmetz et al., 2009a). All the initial work was carried out on particles produced via the infection approach and involved the insertion of sequences into the coat protein region of an infectious cDNA clone of the virus. A number of sites on the coat proteins were identified as suitable for the insertion of foreign peptides. Work on the production of chimaeras for vaccine purposes culminated in the demonstration of protective immunity in target animals (Dalsgaard et al., 1997; Langeveld et al., 2001) and the ability to correlate the structure adopted by a peptide with its immunological properties (Lin et al., 1996; Taylor et al., 2000). For further information, the reader is referred to specialist reviews on the subject (Lomonossoff & Hamilton, 1999; Cañizares et al., 2005; Lomonossoff, 2005; Sainsbury et al., 2010b). More recently, the ability to express peptides on the CPMV surface has been exploited to introduce peptides which are capable of, or promote, subsequent chemical modification (Shah et al., 2009; Steinmetz et al., 2009b).
As an alternative to genetic modification, chemical modification of CPMV capsids has been investigated extensively. Wild‐type CPMV particles have five exposed lysines and eight or nine exposed carboxylates (from aspartic and glutamic acid residues) per asymmetric unit (the asymmetric unit consists of one copy each of the L and S protein). Thus, each particle should have 300 addressable amine and 480–540 addressable carboxyl groups per virus particle. There are also exposed tyrosines, but no cysteines. All the naturally occurring addressable groups have been exploited to produce chemically modified particles. Probably the most frequently utilized group has been the ε‐amino group of surface‐exposed lysines. Initial studies revealed that c. 240 dye molecules could be attached per particle under forcing conditions (Wang et al., 2002a,b), suggesting that all exposed lysines can be modified. The exposed lysines have also been used extensively to couple biotin to the virus surface to enable the particles to bind to avidin (Medintz et al., 2005; Steinmetz et al., 2006a), and this introduced binding ability has been exploited for the creation of supramolecular structures (see part 3 ‘Creation of supramolecular structures’). Lysines have also been modified with ferrocenecarboxylate to produce redox‐active nanoparticles bearing c. 240 ferrocene moieties per particle (Steinmetz et al., 2006a). The redox‐active particles resulting from these studies may lead to the development of electron‐transfer mediators in redox catalysis, amperometric biosensors and, eventually, nanoelectronic devices, such as molecular batteries. As an alternative to addressing lysines, Steinmetz et al. (2006b) demonstrated that it is possible to couple the redox‐active compound viologen via the surface‐exposed carboxyl groups of aspartic and glutamic acids. Although examination of the virus surface indicated that each particle should have 480–540 addressable carboxyl groups, only 180 viologen moieties per particle were added. Carboxylates have also been used to introduce ferrocenes (Aljabali et al., 2010a). The aromatic side chain of surface‐exposed tyrosine residues has also been investigated as a site for modification (Meunier et al., 2004).
In addition to utilizing the side chains of naturally occurring amino acids on the CPMV surface, genetic modification can be employed to either remove or add reactive sites. For example, Chatterji et al. (2004) created a series of mutants in which the exposed lysines were sequentially substituted with arginines. The results showed that all the lysine residues identified as being exposed are, indeed, addressable, and contribute to the overall reactivity of the virus particles. Thiol‐addressable CPMV mutants with cysteine residues on the exterior surface were generated by the insertion of cysteine residues at specific points on the virus surface (Wang et al., 2002c). Gold nanoparticles attached to the surface of cysteine‐substituted CPMV particles have been interconnected using molecular wires to create a three‐dimensional conducting network (Blum et al., 2005). The cysteine‐substituted mutants have also proven to be useful for the conjugation of a number of other moieties (Strable & Finn, 2009). A more complete description of the modifications, which have been chemically introduced on CPMV particles, can be found in Steinmetz & Evans (2007) and Strable & Finn (2009).
All the studies already described have been conducted using particles, either wild‐type or genetically modified, produced by the infection of plants. The recent observation, reviewed above, that the co‐inoculation of plants with the VP60 precursor to the L and S coat protein and the 24K viral proteinase results in the production of empty (RNA‐free) CPMV capsids (Saunders et al., 2009) provides a means to generate large quantities of CPMV eVLPs. Thus, many future studies involving the modification of the outer surface may well be conducted using particles produced in this manner, rather than by infection, as the particles produced in this manner are noninfectious and present no biohazard. Furthermore, the empty particles produced in this manner could potentially be loaded with foreign ‘cargo’ (see below).
The virus CCMV is a tripartite virus which has a capsid consisting of 180 identical coat protein subunits which form a spherical particle of 28 nm in diameter. Like CPMV, the CCMV capsid displays addressable lysines and carboxylates derived from aspartic and glutamic acid. Amine‐ and carboxy‐selective chemistry has been used to selectively attach fluorescent dyes to the virus surface, with c. 540 lysine residues and 560 carboxylates being addressable (Gillitzer et al., 2002). It also proved possible to genetically introduce two solvent‐exposed cysteines per coat protein into the virus capsid. Probing of the resultant particles with thiol‐selective dyes showed that approximately one‐third of the introduced thiols could be addressed. Subsequently, a large diversity of ligands, including intact IgG antibodies, were chemically linked to the exterior surface of CCMV, clearly illustrating that chemical modification is a generic approach to surface modification of the virus capsid (Gillitzer et al., 2002; Suci et al., 2007a,b). Further details of the chemical attachment of molecules to the surface of CCMV can be found in Steinmetz & Evans (2007) and Young et al. (2008).
TMV particles consist of a single molecule of genomic RNA encapsidated by 2130 copies of the 17.5‐kDa coat protein arranged with helical symmetry to form rigid particles of 300 nm in length. The subunits are largely α‐helical, with the N‐ and C‐termini being exposed on the outer virus surface. Most attempts to express foreign peptides via genetic fusion have focused on the C‐terminus of the coat protein in view of its exposed location. TMV‐based peptide presentation systems have been developed, in which the coat protein subunits are modified to express foreign peptides (Fitchen et al., 1995; Turpen et al., 1995). Koo et al. (1999) showed that mice immunized with a TMV chimaera expressing a peptide from the spike protein of the coronavirus, Murine hepatitis virus (MHV), at the C‐terminus of the coat protein, were protected from subsequent challenge with the virus. However, a problem with the use of direct fusions of peptides at or near the C‐terminus of the TMV coat protein is that the size of the inserts which can be tolerated seems to be quite small, the largest reported insert at this site being 23 amino acids (Bendahmane et al., 1999). However, modifying the C‐terminus of the coat protein can alleviate this problem. Werner et al. (2006) found that a functional fragment of protein A, of 133 amino acids in length, could be displayed on the surface of a close relative of TMV, the tobamovirus Turnip vein clearing virus (TVCV), if the sequence was fused to the C‐terminus of the coat protein via a 15‐amino‐acid linker. Given the success of TMV‐based systems for peptide presentation, there has been considerable interest in the commercial development of the technology. For a description of such developments, the reader is referred to recent reviews by McCormick & Palmer (2008) and Smith et al. (2009).
The outer surface of wild‐type TMV particles is somewhat devoid of chemically reactive amino acids, such as cysteine and lysine. To overcome the lack of reactive amino acid chains on the virus surface, several mutants displaying reactive cysteine or lysine residues on the solvent‐exposed exterior of the virus have been made, allowing decoration via thiol‐ or amine‐selective chemistry (Demir & Stowell, 2002; Yi et al., 2005, 2007). However, in many cases, the presence of these added residues adversely affects the virus yield. To counteract this, Smith et al. (2006) screened a random collection of TMV mutants which had an additional four amino acids, including a single lysine, inserted near the N‐terminus of the coat protein. By selecting those mutants which grew well, the authors were able to identify a particular mutant which could be used for the chemical coupling of a variety of epitopes (McCormick & Palmer, 2008).
PVX has filamentous particles consisting of c. 1260 coat protein subunits encapsidating a single RNA molecule. Although an atomic resolution structure of the coat protein subunits is not available, the overall architecture of the viral particles is known (Kendall et al., 2008). It has proven possible to genetically fuse peptides to the surface‐ exposed N‐terminus of either a proportion or all of the subunits. To achieve partial modification, the sequence of the foot and mouth disease virus (FMDV) 2A catalytic peptide was inserted between the peptide and the N‐terminus of the coat protein (Santa Cruz et al., 1996), such that both wild‐type and N‐terminally modified subunits can be produced from the same construct. This approach has the advantage that it potentially permits the expression of longer peptides, including whole proteins, than would be the case if all the subunits were modified. Using the ability of the 2A cleavage strategy to permit the fusion of whole proteins, Smolenska et al. (1998) expressed a single‐chain antibody on the particle surface and showed that it retained its binding specificity. Carette et al. (2007) subsequently expressed the enzyme lipase B from Candida antarctica on the surface of the virus. These authors showed that the virus‐anchored lipase molecules were catalytically active, and suggested that it could act as an anchored biocatalyst. Using an alternative approach of modifying all the subunits, Marusic et al. (2001) expressed a highly conserved hexapeptide epitope from gp41 of HIV‐1 on PVX particles. Mice immunized with the chimaeric particles produced high levels of HIV‐1‐specific IgG and IgA. To examine the possibility of chemically modifying PVX, Steinmetz et al. (2010) conducted a detailed study of the reactivity of functional groups present on the surface of the particle. Each of the 1260 PVX coat protein subunits contains 11 lysine residues, 10 aspartic acid residues and 10 glutamic acid residues, all of which could potentially be modified if they were surface exposed. Preliminary data indicated that none of the carboxylates were addressable under these conditions; by contrast, lysine residues could be modified with an average of just over one lysine per subunit being modified.
2. Utilizing the interior of plant virus particles
The interiors of plant virus capsids potentially provide a nanosized environment for the packaging of foreign materials. There are essentially two approaches that can be taken to encapsulate foreign molecules within the capsid. In the first approach, the foreign molecules are incorporated into the particles during the capsid assembly process. In the second, the foreign molecules are introduced into preassembled particles. Most research has concentrated on the use of the enclosed space of a variety of icosahedral viruses; however, the internal channel of TMV, which is open at both ends, has also been used for some specific purposes. In yet another type of application, the ability of the virus coat protein to package specific RNA molecules has been exploited.
The plant virus that has been used most extensively for interior modification is CCMV. Empty (RNA‐free) particles of CCMV can be produced in vitro through the assembly of coat protein subunits. CCMV particles have a particular advantage for the encapsulation of foreign molecules as they undergo a pH‐ and cation‐dependent structural transition that can be used to control the loading and release of such material. At pH values above 6.5 and in the absence of divalent cations, the CCMV capsid undergoes a reversible swelling which increases the diameter of the particles by c. 10% and leads to the formation of 60 pores (Speir et al., 1995). The interior surface of wild‐type particles carries a high positive charge density because of the presence of nine basic residues (arginine and lysine) in the amino‐terminal region of each subunit. These positively charged residues normally interact with the negatively charged viral RNA. The positively charged interior surface and the availability of pores have been used to promote mineralization within the preformed capsid to produce defined inorganic nanoparticles of anionic polyoxometallate salts (Douglas & Young, 1998). The resulting nanoparticles were constrained in both size and shape by the interior dimensions of the CCMV virion. Using heterologous expression, it is possible to produce CCMV particles with an altered interior charge. When the interior was made acidic, it proved possible to catalyse the formation of cationic transition metal oxides inside the particles when they were incubated with the appropriate cations (Douglas et al., 2002). Another potentially useful feature of CCMV is the ability of its coat protein to assemble into structures distinct from the normal virion. For example, it is possible to produce particles containing 60 or 120 subunits, as opposed to 180, by making deletions in the N‐terminus of the coat protein (Tang et al., 2006). Brome mosaic virus (BMV), in the same genus as CCMV, has also been investigated for its ability to encapsulate foreign materials. For example, the BMV coat protein has been shown to be able to assemble around preformed gold nanoparticles provided that there is a citrate layer between the gold and the protein surface (Dragnea et al., 2003). Using a similar approach, Huang et al. (2007) assembled BMV capsids around iron oxide nanotemplates. When the iron oxide core was larger than the inner cavity of native BMV, capsids larger than native BMV particles were obtained. The particles containing the iron oxide were superparamagnetic, suggesting that they could have applications in magnetic imaging and biosensing.
The coat protein subunits of two other self‐assembling icosahedral plant viruses, Red clover necrotic mosaic virus (RCNMV) and Hibiscus chlorotic ringspot virus (HCRSV), have been investigated for their ability to encapsulate foreign material. Both have particles consisting of 180 identical subunits arranged with icosahedral symmetry. Virus particles of RCNMV are stabilized by an internal protein–RNA cage and their assembly is initiated with the recognition of an origin of assembly site on the viral RNA by the coat protein (Sit et al., 1998). By attaching an artificial origin of assembly sequence to a gold nanoparticle, it proved possible to achieve the in vitro encapsidation of the gold particle by the viral coat protein (Loo et al., 2006, 2007). Using this approach, it proved possible to encapsidate a range of gold core sizes. As suggested for BMV, the resultant material could be used for biosensing purposes, and RCNMV has been proposed as a targeted particle for cancer treatment (Franzen & Lommel, 2009). In the case of HCRSV, empty particles can be produced by the disassembly/reassembly of virions produced by infection. Anionic polymers, such as polystyrenesulfonic acid and polyacrylic acid, but not neutrally charged dextran molecules, could be successfully loaded into these empty particles (Ren et al., 2006). Ren et al. (2007) made use of this phenomenon to co‐encapsidate the anti‐cancer drug doxorubicin with polystyrenesulfonic acid into HCRSV particles. To target the particles to cancerous cells, folic acid was conjugated to lysine residues on the outer virus surface. The resultant particles improved the uptake and cytotoxicity of doxorubicin to ovarian cancer cells, suggesting that modified plant virus capsids may provide the basis for targeted drug delivery in cancer chemotherapy.
Until recently, the interior cavity of CPMV particles has not been amenable to loading with foreign molecules as no in vitro assembly system has been available. The production of virions via the infection of plants results in the majority of particles containing the viral RNA. An initial attempt to produce RNA‐free, loadable particles involved the treatment of wild‐type CPMV at high pH to eliminate the encapsidated virion RNAs (Ochoa et al., 2006). The potential utility of the resultant RNA‐free particles was demonstrated by showing that cysteine residues on the inner capsid surface, which are normally occluded by the viral RNA, could be labelled with a reporter dye (Wen et al., 2012). An alternative approach is to produce empty particles in plants by the co‐expression of VP60 and the 24K proteinase discussed previously (Saunders et al., 2009), and to test whether such particles can be loaded with foreign materials. It has recently been demonstrated that, when CPMV eVLPs are produced by this means, they are capable of being loaded with cobalt or iron oxide (Aljabali et al., 2010b). The presence of the metal within the particles allows them to be visualized by electron microscopy in the absence of negative stain.
TMV particles are hollow cylinders with an internal diameter of 4 nm. The interior channel is lined with aspartic and glutamic acid residues and these have been labelled with a variety of small molecules, such as biotin (Schlick et al., 2005). Nanowires with lengths up to 100 nm and diameters of 4 nm have been synthesized within the TMV capsid channel (Tsukamoto et al., 2007), and the formation of small isolated nanoparticles of silver and nickel within the channel has also been reported (Dujardin et al., 2003). The encapsidation of TMV RNA by its coat protein is known to proceed from a defined sequence on the viral RNA, the origin of assembly. It has been known for some time that the attachment of this sequence to a heterologous RNA will promote encapsidation of the foreign RNA by the coat protein (Gallie et al., 1987a). Smith et al. (2007) exploited this phenomenon to deliver RNA encoding the nonstructural proteins from Semliki forest virus into mammalian cells. They showed that the encapsidated RNA was uncoated, translated within the cells and stimulated an immune response in mice.
3. Creation of supramolecular structures
A major aim of synthetic biology is to incorporate biologically derived components into small‐scale devices. In the case of virus‐based bionanotechnology, this involves the incorporation of modified viruses or VLPs into supramolecular structures, often by binding the particle to surfaces. An early example of the incorporation of a plant virus into a supramolecular structure was reported by Lvov et al. (1994), who incorporated the icosahedral particles of Carnation mottle virus (CarMV) into an alternating multilayered thin film. With the advent of methods for the genetic and chemical modification of particles, the range of structures that can be built up has steadily become more sophisticated.
CCMV particles have been immobilized on to surfaces with a view to constructing arrays. Immobilization has been achieved either by the absorption of cysteine‐containing CCMV particles on to a gold surface (Klem et al., 2003) or via electrostatic interactions of the negatively charged capsids on to positively charged surfaces (Suci et al., 2005, 2006). Furthermore, multilayers consisting of CCMV particles immobilized on a solid support can be constructed using electrostatic interactions or the biotin–streptavidin interaction. The ability to construct thin films of CCMV could be coupled with the multivalent display of various molecules on the capsid surface and with the ability of the virus to encapsidate and release materials from the capsids in a pH‐dependent manner. This could potentially lead to the development of semi‐permeable functionalized membranes or controlled release coatings.
The first approach to the incorporation of modified CPMV particles into a larger structure involved the immobilization of particles expressing His residues (Medintz et al., 2005). In addition to producing continuous layers, single virus particle arrays have also been constructed (Cheung et al., 2003, 2006, 2010; Smith et al., 2003). One method of creating three‐dimensional structures is to build up successive two‐dimensional structures using a layer‐by‐layer (LbL) approach. To test the feasibility of this with CPMV, particles were covalently labelled with two different ligands: biotin to allow self‐assembly via interaction with streptavidin, and fluorescent labels to enable the particles to be imaged (Steinmetz et al., 2006a). Attachment of the different functionalities was achieved via the modification of lysine side chains. The immobilization of the CPMV particles on a solid support was achieved using either direct binding of cysteine‐added mutants to a gold surface or, indirectly, by binding biotinylated particles mediated via streptavidin.
The adsorption properties of TMV on various surfaces, such as gold, mica, glass and silicon wafers, has been investigated (Knez et al., 2004), and a technique for rapid and large‐scale assembly of thin film coatings and ordered fibres consisting of aligned TMV particles has also been reported (Kuncicky et al., 2006). Yi et al. (2005, 2007) partially disassembled the coat protein from TMV particles to expose the RNA at the 5′ end of the rods. Oriented assembly of TMV on solid supports was then achieved in a controlled manner via nucleic acid hybridization using complementary oligonucleotides, and the immobilization of fluorescently labelled TMV onto electrodes was also demonstrated.
IV. Conclusion and future prospects
Successful plant‐based gene expression was achieved initially by the creation of transgenic plants, driven primarily by the development of Agrobacterium binary plasmids and by the characterization of the CaMV 35S promoter. Several additional RNA plant‐viral regulatory sequences, such as translational enhancer sequences and genes encoding suppressors of silencing, were subsequently characterized and used to create highly efficient expression systems. Concurrently, the past two decades have seen tremendous advances in the manipulation of plant virus particles, both genetically and chemically, and investigations into the potential uses of such modified particles. These two aspects of the use of plant virus‐derived components have recently come together with the use of a plant transient expression system (pEAQ‐HT) to create empty particles of CPMV. However, at present, all studies have been conducted at an academic level. Thus, a major challenge in the future will be the deployment of the technical advances in both biotechnology and nanotechnology into the arena of synthetic biology. For example, although it has been demonstrated that chimaeric plant virus particles can stimulate protective immunity in experimental animals, this technology has not been approved for use outside the laboratory. The same is true for the potential imaging agents based on the incorporation of foreign materials within particles. In a similar way, a major step in the adoption of plant virus particles in nanotechnology or synthetic biological applications will require the demonstration that some type of device with unusual or highly desirable properties can be produced cost‐effectively.
Acknowledgements
This work was supported by the UK Biotechnology and Biological Sciences Research Council (BBSRC) Institute Strategic Programme Grant ‘Understanding and Exploiting Plant and Microbial Secondary Metabolism’ (BB/J004596/1) and the John Innes Foundation.
References
- Aljabali AAA, Barclay JE, Butt JN, Lomonossoff GP, Evans DJ. 2010a. Redox‐active ferrocene‐modified Cowpea mosaic virus nanoparticles. Dalton Transactions 39: 7569–7574. [DOI] [PubMed] [Google Scholar]
- Aljabali AAA, Sainsbury F, Lomonossoff GP, Evans DJ. 2010b. Cowpea mosaic virus unmodified virus‐like particles can be loaded with metal and metal oxide. Small (Weinheim an der Bergstrasse, Germany) 6: 818–821. [DOI] [PubMed] [Google Scholar]
- Bendahmane M, Koo M, Karrer E, Beachy RN. 1999. Display of epitopes on the surface of Tobacco mosaic virus: impact of charge and isoelectric point of the epitope on virus–host interactions. Journal of Molecular Biology 290: 9–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum AS, Soto CM, Wilson CD, Brower TL, Pollack SK, Schull TL, Chatterji A, Lin T, Johnson JE, Amsinck C et al 2005. An engineered virus as a scaffold for three‐dimensional self‐assembly on the nanoscale. Small (Weinheim an der Bergstrasse, Germany) 1: 702–706. [DOI] [PubMed] [Google Scholar]
- Cañizares MC, Lomonossoff GP, Nicholson L. 2005. Development of Cowpea mosaic virus‐based vectors for the production of vaccines in plants. Expert Review Vaccines 4: 687–697. [DOI] [PubMed] [Google Scholar]
- Carette N, Engelkamp H, Akpa E, Pierre SJ, Cameron NR, Christianen PCM, Maan JC, Thies JC, Weberskirch R, Rowan AE et al 2007. A virus‐based catalyst. Nature Nanoletters 2: 226–229. [DOI] [PubMed] [Google Scholar]
- Chatterji A, Burns LL, Taylor SS, Lomonossoff GP, Johnson JE, Lin T, Porta C. 2002. Cowpea mosaic virus: from the presentation of antigenic peptides to the display of active biomaterials. Intervirology 45: 362–370. [DOI] [PubMed] [Google Scholar]
- Chatterji A, Ochoa W, Paine M, Ratna BR, Johnson JE, Lin T. 2004. New addresses on an addressable virus nanoblock: uniquely reactive Lys residues on Cowpea mosaic virus. Chemistry and Biology 11: 855–863. [DOI] [PubMed] [Google Scholar]
- Cheung CL, Chung SW, Chatterji A, Lin T, Johnson JE, Hok S, Perkins J, De Yoreo JJ. 2006. Physical controls on directed virus assembly at nanoscale chemical templates. Journal of the American Chemical Society 128: 10801–10807. [DOI] [PubMed] [Google Scholar]
- Cheung CL, Rubinstein AI, Peterson EJ, Chatterji A, Sabirianov RF, Mei WN, Lin T, Johnson JE, De Yoreo JJ. 2010. Steric and electrostatic complementarity in the assembly of two‐dimensional virus arrays. Langmuir 26: 3498–3505. [DOI] [PubMed] [Google Scholar]
- Cheung L, Camarero JA, Woods BW, Lin T, Johnson JE, De Yoreo JJ. 2003. Fabrication of assembled virus nanostructures on templates of chemoselective linkers formed by scanning probe nanolithography. Journal of the American Chemical Society 125: 6848–6849. [DOI] [PubMed] [Google Scholar]
- Creager ANH, Scholthof K‐BG, Citovsky V, Scholthof H. 1999. Tobacco mosaic virus: pioneering research for a century. The Plant Cell 11: 301–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalsgaard K, Uttenthal Å, Jones TD, Xu F, Merryweather A, Hamilton WDO, Langeveld JPM, Boshuizen RS, Kamstrup S, Lomonossof GP et al 1997. Plant‐derived vaccine protects target animals against a virus disease. Nature Biotechnology 15: 248–252. [DOI] [PubMed] [Google Scholar]
- Demir M, Stowell MHB. 2002. A chemoselective biomolecular template for assembling diverse nanotubular materials. Nanotechnology 13: 541–544. [Google Scholar]
- Douglas T, Strable E, Willits D, Aitouchen A, Libera M, Young M. 2002. Protein engineering of a viral cage for constrained nanomaterials synthesis. Advance Materials 14: 415–418. [Google Scholar]
- Douglas T, Young M. 1998. Host‐guest encapsulation of materials by assembled virus protein cages. Nature 393: 152–155. [Google Scholar]
- Dragnea B, Chen C, Kwak ES, Stein B, Kao C. 2003. Gold nanoparticles as spectroscopic enhancers for in vitro studies on single viruses. Journal of the American Chemical Society 125: 6374–6375. [DOI] [PubMed] [Google Scholar]
- Dujardin E, Peet C, Stubbs G, Culver JN, Mann S. 2003. Organization of metallic nanoparticles using Tobacco mosaic virus templates. Nano Letters 3: 413–417. [Google Scholar]
- Esposito D, Garvey LA, Chakiath CS. 2009. Gateway cloning for protein expression. Methods in Molecular Biology 498: 31–54. [DOI] [PubMed] [Google Scholar]
- Fitchen J, Beachy RN, Hein MB. 1995. Plant virus expressing hybrid coat protein with added murine epitope elicits autoantibody response. Vaccine 13: 1051–1057. [DOI] [PubMed] [Google Scholar]
- Franzen S, Lommel SA. 2009. Targeting cancer with ‘smart bombs’: equipping plant virus nanoparticles for a ‘seek and destroy’ mission. Nanomedicine (London) 4: 575–588. [DOI] [PubMed] [Google Scholar]
- Gallie DR, Sleat DE, Watts JW, Turner PC, Wilson TM. 1987a. In vivo uncoating and efficient expression of foreign mRNAs packaged in TMV‐like particles. Science 236: 1122–1124. [DOI] [PubMed] [Google Scholar]
- Gallie DR, Sleat DE, Watts JW, Turner PC, Wilson TMA. 1987b. The 5′‐leader sequence of tobacco mosaic virus RNA enhances the expression of foreign gene transcripts in vitro and in vivo . Nucleic Acids Research 15: 3257–3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garabagi F, Gilbert E, Loos A, McLean MD, Hall JC. 2012. Utility of the P19 suppressor of gene‐silencing protein for production of therapeutic antibodies in Nicotiana expression hosts. Plant Biotechnology Journal 10: 1118–1128. [DOI] [PubMed] [Google Scholar]
- Gillitzer E, Willits D, Young M, Douglas T. 2002. Chemical modification of a viral cage for multivalent presentation. Chemical Communications 2002: 2390–2391. [DOI] [PubMed] [Google Scholar]
- Giritch A, Marillonnent S, Engler C, van Eldik G, Botterman J, Klimyuk V, Gleba Y. 2006. Rapid high‐yield expression of full‐size IgG antibodies in plants coinfected with noncompeting viral vectors. Proceedings of the National Academy of Sciences, USA 103: 14701–14706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleba Y, Klimyuk V, Marillonnet S. 2005. Magnifection – a new platform for expressing recombinant vaccines in plants. Vaccine 23: 2042–2048. [DOI] [PubMed] [Google Scholar]
- Hefferon KL. 2012. Plant virus expression vectors set the stage as production platforms for biopharmaceutical proteins. Virology 433: 1–6. [DOI] [PubMed] [Google Scholar]
- Huang X, Bronstein LM, Retrum J, Dufort C, Tsvetkova I, Aniagyei S, Stein B, Stucky G, McKenna B, Remmes N et al 2007. Self‐assembled virus‐like particles with magnetic cores. Nano Letters 7: 2407–2416. [DOI] [PubMed] [Google Scholar]
- Huang Z, Chen Q, Hjelm B, Arntzen CJ, Mason H. 2009. A DNA replicon system for rapid high‐level production of virus‐like particles in plants. Biotechnology and Bioengineering 103: 706–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagale S, Uzuhashi S, Wigness M, Bender T, Yang W, Borhan MH, Rozwadowski K. 2012. TMV‐Gateway vectors: gateway compatible tobacco mosaic virus based expression vectors for functional analysis of proteins. Scientific Reports 2: 874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendall A, McDonald M, Bian W, Bowles T, Baumgarten SC, Shi J, Stewart PL, Bullitt E, Gore D, Irving TC et al 2008. Structure of flexible filamentous plant viruses. Journal of Virology 82: 9546–9554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klem MT, Willits D, Young M, Douglas T. 2003. 2‐D array formation of genetically engineered viral cages on Au surfaces and imaging by atomic force microscopy. Journal of the American Chemical Society 125: 10806–10807. [DOI] [PubMed] [Google Scholar]
- Knez M, Sumser MP, Bittner AM, Wege C, Jeske H, Hoffmann DM, Kuhnke K, Kern K. 2004. Binding the Tobacco mosaic virus to inorganic surfaces. Langmuir 20: 441–447. [DOI] [PubMed] [Google Scholar]
- Koo M, Bendahmane M, Lettieri GA, Paoletti AD, Lane TE, Fitchen JH, Buchmeier MJ, Beachy RN. 1999. Protective immunity against murine hepatitis virus (MHV) induced by intranasal or subcutaneous administration of hybrids of tobacco mosaic virus that carries an MHV epitope. Proceedings of the National Academy of Sciences, USA 96: 7774–7779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korban SS. 2002. Targeting and expression of antigenic proteins in transgenic plants for production of edible oral vaccines. In vitro Cellular and Developmental Biology. Plant 38: 231–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuncicky DM, Naik RR, Velev OD. 2006. Rapid deposition and long‐range alignment of nanocoatings and arrays of electrically conductive wires from tobacco mosaic virus. Small (Weinheim an der Bergstrasse, Germany) 2: 1462–1466. [DOI] [PubMed] [Google Scholar]
- Kushnir N, Streatfield S, Yusibov V. 2012. Virus‐like particles as a highly efficient vaccine platform: diversity of targets and production systems and advances in clinical development. Vaccine 31: 58–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langeveld JP, Brennan FR, Martinez‐Torrecuadrada JL, Jones TD, Boshuizen RS, Vala C, Casal JI, Kamstrup S, Dalsgaard K, Meloen RH et al 2001. Inactivated recombinant plant virus protects dogs from a lethal challenge with canine parvovirus. Vaccine 19: 3661–3670. [DOI] [PubMed] [Google Scholar]
- Lin T, Porta C, Lomonossoff G, Johnson JE. 1996. Structure‐based design of peptide presentation on a viral surface: the crystal structure of a plant/animal virus chimaera at 2.8Å resolution. Folding and Design 1: 179–187. [DOI] [PubMed] [Google Scholar]
- Lomonossoff GP. 2005. Antigen delivery systems: use of recombinant plant viruses In: Mestecky J, Bienenstock J, Lamm ME. et al, eds. Mucosal immunology, 3rd edn Waltham, MA: Elsevier, 1061–1072. [Google Scholar]
- Lomonossoff GP, Hamilton WDO. 1999. Cowpea mosaic virus‐based vaccines. Current Topics in Microbiology and Immunology 240: 177–189. [DOI] [PubMed] [Google Scholar]
- Lomonossoff GP, Johnson JE. 1996. Use of macromolecular assemblies as expression systems for peptides and synthetic vaccines. Current Opinion in Structural Biology 6: 176–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo L, Guenther RH, Basnayake VR, Lommel SA, Franzen S. 2006. Controlled encapsidation of gold nanoparticles by a viral protein shell. Journal of the American Chemical Society 128: 4502–4503. [DOI] [PubMed] [Google Scholar]
- Loo L, Guenther RH, Lommel SA, Franzen S. 2007. Encapsidation of nanoparticles by red clover necrotic mosaic virus. Journal of the American Chemical Society 129: 11111–11117. [DOI] [PubMed] [Google Scholar]
- Lvov Y, Haas H, Decher G, Moehwald H, Mikhailov A, Mtchedlishvily B, Morgunova E, Vainshtein B. 1994. Successive deposition of alternate layers of polyelectrolytes and a charged virus. Langmuir 10: 4232–4236. [Google Scholar]
- Marillonnet S, Giritch A, Gils M, Kandzia R, Klimyuk V, Gleba Y. 2004. In planta engineering of viral RNA replicons: efficient assembly by recombination of DNA modules delivered by Agrobacterium . Proceedings of the National Academy of Sciences, USA 101: 6852–6857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marusic C, Rizza P, Lattanzi L, Mancini C, Spada M, Balardelli F, Benvenuot E, Capone I. 2001. Chimeric plant virus particles as immunogens for inducing murine and human immune responses against Human immunodeficiency virus type 1. Journal of Virology 75: 8434–8439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason HS, Lam DM‐K, Arntzen CJ. 1992. Expression of hepatitis B surface antigen in transgenic plants. Proceedings of the National Academy of Sciences, USA 89: 11745–11749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick A, Palmer KE. 2008. Genetically engineered Tobacco mosaic virus as nanoparticle vaccines. Expert Review of Vaccines 7: 33–41. [DOI] [PubMed] [Google Scholar]
- Medintz IL, Sapsford KE, Konnert JH, Chatterji A, Lin T, Johnson JE, Mattoussi H. 2005. Decoration of discretely immobilized cowpea mosaic virus with luminescent quantum dots. Langmuir 21: 5501–5510. [DOI] [PubMed] [Google Scholar]
- Meunier S, Strable E, Finn MG. 2004. Crosslinking of and coupling to viral capsid proteins by tyrosine oxidation. Chemistry and Biology 11: 319–326. [DOI] [PubMed] [Google Scholar]
- Montague NP, Thuenemann EC, Saxena P, Saunders K, Lenzi P, Lomonossoff GP. 2011. Recent advances of cowpea mosaic virus‐based particle technology. Human Vaccines 7: 383–390. [DOI] [PubMed] [Google Scholar]
- Ochoa W, Chatterji A, Lin T, Johnson JE. 2006. Generation and structural analysis of reactive empty particles derived from an icosahedral virus. Chemistry and Biology 13: 771–778. [DOI] [PubMed] [Google Scholar]
- Ofoghi H, Moazami N, Ivanov I. 2005. Comparison of tobacco etch virus and tobacco mosaic virus enhances for expression of human calcitonin gene in transgenic potato plant. Key Engineering Materials 277–279: 7–11. [Google Scholar]
- Paul M, Ma JK‐C. 2011. Plant‐made pharmaceuticals: leading products and production platforms. Biotechnology and Applied Biochemistry 58: 58–67. [DOI] [PubMed] [Google Scholar]
- Porta C, Spall VE, Lin T, Johnson JE, Lomonossoff GP. 1996. The development of cowpea mosaic virus as a potential source of novel vaccines. Intervirology 39: 79–84. [DOI] [PubMed] [Google Scholar]
- Porta C, Spall VE, Loveland J, Johnson JE, Barker PJ, Lomonossoff GP. 1994. Development of Cowpea mosaic virus as a high‐yielding system for the presentation of foreign peptides. Virology 202: 949–955. [DOI] [PubMed] [Google Scholar]
- Ren Y, Wong S‐M, Lim L‐Y. 2006. In vitro‐reassembled plant virus‐like particles for loading of polyacids. Journal of General Virology 87: 2749–2754. [DOI] [PubMed] [Google Scholar]
- Ren Y, Wong S‐M, Lim L‐Y. 2007. Folic acid‐conjugated protein cages of a plant virus: a novel delivery platform for doxorubicin. Bioconjugate Chemistry 18: 836–843. [DOI] [PubMed] [Google Scholar]
- Rybicki EP. 2010. Plant‐made vaccines for human and animals. Plant Biotechnology Journal 8: 620–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainsbury F, Cañizares MC, Lomonossoff GP. 2010a. Cowpea mosaic virus: the plant virus‐based biotechnology workhorse. Annual Review of Phytopathology 48: 437–455. [DOI] [PubMed] [Google Scholar]
- Sainsbury F, Lomonossoff GP. 2008. Extremely high‐level and rapid transient protein production in plants without the use of viral replication. Plant Physiology 148: 1212–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainsbury F, Sack M, Stadlmann J, Quendler H, Fischer R, Lomonossoff GP. 2010b. Rapid transient production in plants by replicating and non‐replicating vectors yields high quality functional anti‐HIV antibody. PLoS ONE 5: e13976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainsbury F, Thuenemann EC, Lomonossoff GP. 2009. pEAQ: versatile expression vectors for easy and quick transient expression of heterologous proteins in plants. Plant Biotechnology Journal 7: 682–693. [DOI] [PubMed] [Google Scholar]
- Santa Cruz S, Chapman S, Roberts AG, Roberts IM, Prior DA, Oparka KJ. 1996. Assembly and movement of a plant virus carrying a green fluorescent protein overcoat. Proceedings of the National Academy of Sciences, USA 93: 6286–6290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders K, Sainsbury F, Lomonossoff GP. 2009. Efficient generation of cowpea mosaic virus empty virus‐like particles by the proteolytic processing of precursors in insect cells and plants. Virology 393: 329–337. [DOI] [PubMed] [Google Scholar]
- Savka MA, Black RC, Binns AN. 2001. Tobacco etch virus leader sequence enhances inducible indoleacetic acid‐lysine synthetase activity in transgenic plants. Plant Physiology and Biochemistry 39: 631–641. [Google Scholar]
- Saxena P, Hsieh Y‐C, Alvarado VY, Sainsbury F, Saunders K, Lomonossoff GP, Scholthof HB. 2011. Improved foreign gene expression in plants using a virus‐encoded suppressor of RNA silencing modified to be developmentally harmless. Plant Biotechnology Journal 9: 703–712. [DOI] [PubMed] [Google Scholar]
- Schlick TL, Ding ZB, Kovacs EW, Francis MB. 2005. Dual‐surface modification of the Tobacco mosaic virus. Journal of the American Chemical Society 127: 3718–3723. [DOI] [PubMed] [Google Scholar]
- Shah SN, Steinmetz NF, Aljabali AA, Lomonossoff GP, Evans DJ. 2009. Environmentally benign synthesis of virus‐templated, monodisperse, iron–platinum nanoparticles. Dalton Transactions 28: 8479–8480. [DOI] [PubMed] [Google Scholar]
- Sheikholeslam SN, Weeks DP. 1987. Acetosyringone promotes high efficiency transformation of Arabidopsis thaliana explants by Agrobacterium tumefaciens . Plant Molecular Biology 8: 291–298. [DOI] [PubMed] [Google Scholar]
- Sit TL, Vaewhongs AA, Lommel SA. 1998. RNA‐mediated trans‐activation of transcription from a viral RNA. Science 281: 829–832. [DOI] [PubMed] [Google Scholar]
- Smith JC, Lee KB, Wang Q, Finn MG, John JE, Mrksich M, Mirkin CA. 2003. Nanopatterning the chemospecific immobilization of Cowpea mosaic virus capsid. Nano Letters 3: 883–886. [Google Scholar]
- Smith ML, Corbo T, Bernales J, Lindbo JA, Pogue GP, Palmer KE, McCormick AA. 2007. Assembly of trans‐encapsidated recombinant viral vectors engineered from tobacco mosaic virus and Semliki Forest virus and their evaluation as immunogens. Virology 358: 321–333. [DOI] [PubMed] [Google Scholar]
- Smith ML, Fitzmaurice WP, Turpen TH, Palmer KE. 2009. Display of peptides on the surface of Tobacco mosaic virus particles. Current Topics in Microbiology and Immunology 332: 13–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ML, Lindbo JA, Dillard‐Telm S, Brosio PM, Lasnik AB, McCormick AA, Nguyen LV, Palmer KE. 2006. Modified tobacco mosaic virus particles as scaffolds for display of protein antigens for vaccine applications. Virology 348: 475–488. [DOI] [PubMed] [Google Scholar]
- Smolenska L, Roberts IM, Learmonth D, Porter AJ, Harris WJ, Wilson TMA, Santa‐Cruz S. 1998. Production of a functional single chain antibody attached to the surface of a plant virus. FEBS Letters 441: 379–382. [DOI] [PubMed] [Google Scholar]
- Speir JA, Munshi S, Wang G, Baker TS, Johnson JE. 1995. Structures of the native and swollen forms of Cowpea chlorotic mottle virus determined by X‐ray crystallography and cryo‐electron microscopy. Structure 3: 63–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz NF, Calder G, Lomonossoff GP, Evans DJ. 2006a. Plant viral capsids as nanobuilding blocks: construction of arrays on solid supports. Langmuir 22: 10032–10037. [DOI] [PubMed] [Google Scholar]
- Steinmetz NF, Evans D. 2007. Utilisation of plant viruses in bionanotechnology. Organic and Biomolecular Chemistry 5: 2891–2902. [DOI] [PubMed] [Google Scholar]
- Steinmetz NF, Lin T, Lomonossoff GP, Johnson JE. 2009a. Structure‐based engineering of an icosahedral virus for nanomedicine and nanotechnology. Current Topics in Microbiology and Immunology 327: 23–58. [DOI] [PubMed] [Google Scholar]
- Steinmetz NF, Lomonossoff GP, Evans DJ. 2006a. Decoration of Cowpea mosaic virus with multiple, redox‐active, organometallic complexes. Small (Weinheim an der Bergstrasse, Germany) 2: 530–533. [DOI] [PubMed] [Google Scholar]
- Steinmetz NF, Lomonossoff GP, Evans DJ. 2006b. Cowpea mosaic virus for material fabrication: addressable carboxylate groups on a programmable nanoscaffold. Langmuir 22: 3488–3490. [DOI] [PubMed] [Google Scholar]
- Steinmetz NF, Mertens ME, Taurog RE, Johnson JE, Commaneur U, Fischer T, Manchester M. 2010. Potato virus X as a novel platform for potential biomedical applications. Nano Letters 10: 305–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz NF, Shah NS, Barclay JE, Rallapalli G, Lomonossoff GP, Evans DJ. 2009b. Virus‐templated silica nanoparticles. Small (Weinheim an der Bergstrasse, Germany) 5: 813–816. [DOI] [PubMed] [Google Scholar]
- Strable E, Finn MG. 2009. Chemical modification of viruses and virus‐like particles. Current Topics in Microbiology and Immunology 332: 1–21. [DOI] [PubMed] [Google Scholar]
- Suci PA, Berglund DL, Liepold L, Brumfield S, Pitts B, Davison W, Oltrogge L, Hoyt KO, Codd S, Stewart PS et al 2007a. High‐density targeting of a viral multifunctional nanoplatform to a pathogenic, biofilm‐forming bacterium. Chemistry and Biology 14: 387–398. [DOI] [PubMed] [Google Scholar]
- Suci PA, Klem MT, Arce FT, Douglas T, Young M. 2006. Assembly of multilayer films incorporating a viral protein cage architecture. Langmuir 22: 8891–8896. [DOI] [PubMed] [Google Scholar]
- Suci PA, Klem MT, Douglas T, Young M. 2005. Influence of electrostatic interactions on the surface adsorption of a viral protein cage. Langmuir 21: 8686–8893. [DOI] [PubMed] [Google Scholar]
- Suci PA, Varpness Z, Gillitzer E, Douglas T, Young M. 2007b. Targeting and photodynamic killing of a microbial pathogen using protein cage architectures functionalized with a photosensitizer. Langmuir 23: 12280–12286. [DOI] [PubMed] [Google Scholar]
- Tang J, Johnson JM, Dryden KA, Young MJ, Zlotnick A, Johnson JE. 2006. The role of subunit hinges and molecular “switches” in the control of viral capsid polymorphism. Journal of Structural Biology 154: 59–67. [DOI] [PubMed] [Google Scholar]
- Taylor KM, Lin T, Porta C, Mosser AG, Giesing HA, Lomonossoff GP, Johnson JE. 2000. Influence of three‐dimensional structure on the immunogenicity of a peptide expressed on the surface of a plant virus. Journal of Molecular Recognition 13: 71–82. [DOI] [PubMed] [Google Scholar]
- Thuenemann EC, Meyers A, Verwey J, Rybicki E, Lomonossoff GP. 2013. A method for the rapid production of heteromultimeric protein complexes in plants: assembly of protective bluetongue virus‐like particles. Plant Biotechnology Journal, in press. [DOI] [PubMed] [Google Scholar]
- Tsukamoto R, Muraoka M, Seki M, Tabata H, Yamashita I. 2007. Synthesis of CoPt and FePt3 nanowires using the central channel of Tobacco mosaic virus as a biotemplate. Chemistry of Materials 19: 2389–2391. [Google Scholar]
- Turpen TH, Reinl SJ, Charoenvit Y, Hoffman SL, Fallarme V, Grill LK. 1995. Malarial epitopes expressed on the surface of recombinant Tobacco mosaic virus. Biotechnology 13: 53–57. [DOI] [PubMed] [Google Scholar]
- Usha R, Rohll JB, Spall VE, Shanks M, Maule AJ, Johnson JE, Lomonossoff GP. 1993. Expression of an animal virus antigenic site on the surface of a plant virus particle. Virology 197: 366–374. [DOI] [PubMed] [Google Scholar]
- Wang Q, Kaltgrad E, Lin T, Johnson JE, Finn MG. 2002a. Natural supramolecular building blocks: wild‐type Cowpea mosaic virus. Chemistry and Biology 9: 805–811. [DOI] [PubMed] [Google Scholar]
- Wang Q, Lin T, Johnson JE, Finn MG. 2002c. Natural supramolecular building blocks: cysteine‐added mutants of cowpea mosaic virus. Chemistry and Biology 9: 813–819. [DOI] [PubMed] [Google Scholar]
- Wang Q, Lin T, Tang L, Johnson JE, Finn MG. 2002b. Icosahedral virus particles as addressable nanoscale building blocks. Angewandte Chemie (International. Ed in English) 41: 459–462. [DOI] [PubMed] [Google Scholar]
- Wen AM, Shukla S, Saxena P, Aljabali AAA, Yildiz I, Dey S, Mealy JE, Yang AC, Evans DJ, Lomonossoff GP et al 2012. Interior engineering of a viral nanoparticle and its tumor homing properties. Biomacromolecules 13: 3990–4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner S, Marillonnet S, Hause G, Klimyuk V, Gleba Y. 2006. Immunoabsorbent nanoparticles based on a tobamovirus displaying protein A. Proceedings of the National Academy of Sciences, USA 103: 17678–17683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi HM, Nisar S, Lee SY, Powers MA, Bentley WE, Payne GF, Ghodssi R, Rubloff GW, Harris MT, Culver JN. 2005. Patterned assembly of genetically modified viral nanotemplates via nucleic acid hybridization. Nano Letters 5: 1931–1936. [DOI] [PubMed] [Google Scholar]
- Yi HM, Rubloff GW, Culver JN. 2007. TMV microarrays: hybridization‐based assembly of DNA‐programmed viral nanotemplates. Langmuir 23: 2663–2667. [DOI] [PubMed] [Google Scholar]
- Young M, Willits D, Uchida M, Douglas T. 2008. Plant viruses as biotemplates for materials and their use in nanotechnology. Annual Review of Phytopathology 46: 361–384. [DOI] [PubMed] [Google Scholar]
- Zhao X, Fox JM, Olson NH, Baker TS, Young MJ. 1995. In vitro assembly of cowpea chlorotic mottle virus from coat protein expressed in Escherichia coli and in vitro‐transcribed viral cDNA. Virology 207: 486–494. [DOI] [PubMed] [Google Scholar]


