Abstract
Porcine epidemic diarrhea virus (PEDV) causes an acute, highly contagious, and devastating viral enteric disease with a high mortality rate in suckling pigs. A large‐scale outbreak of PED occurred in China in 2010, with PEDV emerging in the United States in 2013 and spreading rapidly, posing significant economic and public health concerns. In this study, LC–MS/MS coupled to iTRAQ labeling was used to quantitatively identify differentially expressed cellular proteins in PEDV‐infected Vero cells. We identified 49 differentially expressed cellular proteins, of which 8 were upregulated and 41 downregulated. These differentially expressed proteins were involved in apoptosis, signal transduction, and stress responses. Based on these differentially expressed proteins, we propose that PEDV might utilize apoptosis and extracellular signal regulated kinases pathways for maximum viral replication. Our study is the first attempt to analyze the protein profile of PEDV‐infected cells by quantitative proteomics, and we believe our findings provide valuable information with respect to better understanding the host response to PEDV infection.
Keywords: Differentially expressed proteins, iTRAQ, Microbiology, Porcine epidemic diarrhea virus (PEDV), Quantitative proteomics
Abbreviations
- 14‐3‐3 gamma
tyrosine 3‐monooxy‐genase/tryptophan 5‐monooxygenase activation protein, gamma
- CPE
cytopathic effect
- ERK
extracellular signal regulated kinases
- HIV
human immunodeficiency virus
- hpi
h postinfection
- HSDL2
hydroxysteroid dehydrogenase like 2
- IFA
immunofluorescence assay
- IPA
Ingenuity Pathway Analysis
- PED
porcine epidemic diarrhea
- PEDV
porcine epidemic diarrhea virus
- SARS‐CoV
severe acute respiratory syndrome coronavirus
1. Introduction
Porcine epidemic diarrhea (PED) is an acute and highly contagious enteric disease of pigs. Typical clinical symptoms of PED include watery diarrhea, vomiting, dehydration, and high mortality in suckling pigs 1. This disease was first reported in feeder and fattening swine in the United Kingdom in 1971 2, after which the causative agent PED virus (PEDV) was identified as a coronavirus 3. Since then, outbreaks of PED have subsequently been documented in European and Asian countries, resulting in severe economic losses to the swine industry 1.
The prevalence of PED was relatively low until 2010, probably because a PED vaccine was widely used throughout China 4. However, since December 2010, large‐scale outbreaks of diarrhea with 80–100% morbidity and 50–90% mortality in suckling piglets have occurred in most Chinese provinces, affecting even vaccinated pigs 5, 6. Investigations indicated that these large‐scale outbreaks of diarrhea were caused by a variant of PEDV 7, 8. New PEDV outbreaks have also occurred in Vietnam, Thailand, Korea, and the United States, where PEDV was considered an exotic before 2013. The PEDV variant has emerged and spread rapidly in the United States since May 2013 9, 10. By the end of April 2014, PEDV outbreaks had been reported in 27 states of the United States, with 2692 confirmed cases (http://www.aasv.org/news/story.php?id = 6989), resulting in significant economic and public health concerns.
Despite years of intensive research, the mechanisms of PEDV pathogenesis and immunomodulation remain largely unknown 11. Emerging technologies, such as proteomic approaches, have become important tools for facilitating a comprehensive characterization of virus–host interactions involved in infection and pathogenesis 12, 13. Several quantitative proteomics approaches have been used to provide specific insights into the gene expression profiles of coronaviruses including severe acute respiratory syndrome coronavirus (SARS‐CoV) 14, infectious bronchitis coronavirus 15, and transmissible gastroenteritis coronavirus 16. Generally, in host cells coronavirus infection gives rise to alterations in transcriptional patterns, the cell cycle, the cytoskeleton, apoptosis pathways, the immune system, stress responses, and likely also causes inflammation 17. More data related to the host response to coronavirus infection should shed some light on potential targets for antiviral agents.
To date, proteomics has not yet been applied to analyze whole cell changes caused by PEDV infection. Since Hofmann and Wyler 18 reported for the first time that PEDV could be successfully propagated in Vero cell cultures, Vero cells have been widely used for PEDV isolation, propagation, vaccine production, and basic research. In this study, we used iTRAQ coupled with LC−MS/MS to quantitate and identify cellular proteins that were differentially expressed in Vero cells following infection with PEDV.
2. Materials and methods
2.1. Vero cell culture and PEDV preparation
African green monkey kidney (Vero) cells were purchased from the American Type Culture Collection (ATCC). Cells were cultured in DMEM (Invitrogen, Carlsbad, CA, USA) containing 10% FBS and 1% penicillin–streptomycin at 37°C/5% CO2. The Chinese PEDV strain AJ1102 (GenBank Accession No. JX188454) used in this study was isolated in 2011 from the intestine of a suckling piglet with acute diarrhea 5. Virus stock was harvested in Vero cells and virus titer was 106.4 PFU/mL determined by plaque assays.
2.2. Virus inoculation
Monolayers of Vero cells were grown to 80% confluence in 10 cm2 cell culture dishes. Culture medium was removed and monolayers gently washed with serum‐free DMEM prior to inoculation. Cells incubated in serum‐free DMEM containing 10 μg/mL trypsin (Promega) were inoculated with PEDV strain AJ1102 at a multiplicity of infection of 0.1. Uninfected cells served as the mock‐infected group. Infected and mock groups were collected as biological replicates from three independent experiments at 24 h postinfection (hpi). Viral propagation was confirmed by the observation of cytopathic effect (CPE) and indirect immunofluorescence assay (IFA) using a monoclonal antibody against PEDV nucleocapsid (N) protein. This monoclonal antibody was produced from hybridoma cells derived from Sp2/0 myeloma cells and spleen cells of BALB/c mice immunized with recombinant N protein of PEDV strain AJ1102.
2.3. Protein extraction, digestion, and labeling with iTRAQ reagents
Cultures of PEDV‐infected and control cells, including three independent biological replicates, were collected using a cell scraper, centrifuged (300 × g, 10 min), and washed twice with ice‐cold PBS containing 1 mM pervanadate and 1 mM sodium fluoride. Cell pellets were resuspended in 500 μL of lysis buffer (7 M urea, 2 M thiourea, 4% CHAPS, 40 mM Tris‐HCl, pH 8.5) containing 1 mM PMSF, 2 mM EDTA, and 10 mM DTT. Further protein solubilization was achieved by sonicating, with cellular debris removed by centrifugation (25 000 × g, 20 min, 4°C). The protein concentration in supernatants was quantified using the Bradford protein assay. We digested 100 μg of each sample at 37°C for 12 h with sequencing grade modified trypsin (Promega, Madison, WI, USA). Mock‐infected samples were labeled with iTRAQ 113 (IT113), iTRAQ 116 (IT116), or iTRAQ 119 (IT119), and PEDV‐infected samples were labeled with iTRAQ 114 (IT114), iTRAQ 118 (IT118), or iTRAQ 121 (IT121). The labeled digests were then pooled and vacuum‐dried.
2.4. SCX chromatography
Peptide mixtures were loaded onto an UltremexSCX column (4.6 × 250 mm) and separated by SCX chromatography using a 20AB HPLC system (Shimadzu, Kyoto, Japan). The labeled peptides were eluted with 4 mL of buffer A (25 mM NaH2PO4 in 25% (ACN), pH 2.7) for 10 min, followed by a linear gradient of 5–35% and 35–80% buffer B (25 mM NaH2PO4, 1 M KCl in 25% ACN, pH 2.7) for 11 and 1 min at 1 mL/min, respectively. The process of elution was monitored by the absorbance at 214 nm. A total of 20 fractions that were desalted with a StrataX desalting column were collected and lyophilized in a vacuum concentrator.
2.5. MS/MS analysis
The equal amounts of digested proteins were dissolved in buffer A (2% ACN, 0.1% formic acid) at the final concentration of 0.25 μg/μL and analyzed in an LC‐MS/MS system. After that, 9 μL supernatant was loaded on a Symmetry C18 trapping column (5 μm particles, 180 μm inner diameter × 20 mm; Waters) with the nanoACQUITY Sample Manager (Waters) and the peptides were eluted from the trapping column over a BEH 130 C18 column (1.7 μm particles, 100 μm inner diameter × 100 mm). The samples were loaded at 2 μL/min for 15 min, and then gradient run at 300 nL/min were from 5 to 35% buffer B (98% ACN, 0.1% formic acid) in 40 min followed by a wash for 5 min at 80% B and a 2‐min equilibration step. MS/MS was performed with a Triple TOF 5600 mass spectrometer (AB Sciex, Concord, ON). The ion source was a NanoSpray III source (AB Sciex), and the radiator was a needle made of quartz (New Objectives, Woburn, MA, USA). During data acquisition, the spray voltage was set to 2.5 kV, the temperature of the interface heater was 150°C, with the curtain gas 30 pounds per square inch. Survey scans were acquired in 250 ms and 30 product ion scans were collected under the condition of exceeding the threshold of 120 counts per second (counts/s). Total cycle time was fixed to 3.3 s. Dynamic exclusion was set for half of peak width (18 s), and the fragmentation energy was set to 35 ± 5 eV.
2.6. Data analysis
The acquired MS raw data files were converted into MGF files by using 5600 ms converter and the MGF files were further searched. MS/MS data were searched against the monkey sequence database from the NCBInr database (release April 2013, containing 29 736 sequences) and the proteins were identified using MASCOT search engine (Matrix Science, London, UK; version 2.3.02). The search was conducted using trypsin as a specific enzyme. A maximum of one missed cleavage was accepted and Gln→pyro‐Glu (N‐term Q), Oxidation (M), iTRAQ8plex (Y) were chosen as variable modifications. The data acquired from the Triple TOF 5600 mass spectrometer were searched with a fragment mass tolerance of 0.1 Da and a peptide mass tolerance of 0.05 Da. The charge states of peptides were set to +2 and +3. An automatic decoy database search was carried out in MASCOT through choosing the decoy checkbox in which a random sequence of database was generated and tested for raw spectra and the real database. Peptide spectra matches were confirmed through percolator based on q‐values at a false discovery rate of 0.6% 19. Peptide identifications were grouped into proteins according to the law of parsimony and filtered to 0.6% false discovery rate 20. Each of the confident protein identification involves at least two peptides and only unique peptides were used to quantify proteins. The quantitative protein ratios were weighted and normalized by the median ratio in MASCOT. For accurate comparison between samples, fold change > 1.5 or <0.667 and a p‐value < 0.05 were considered as significantly different expressions according to the t‐test. Bioinformatics analysis was conducted using Ingenuity Pathway Analysis software (IPA, http://www.ingenuity.com), GO, and UniProt.
2.7. IFA
Vero cells were seeded on glass coverslips (NEST Biotechnology) that had been placed in 24‐well plates until cultures were 70–80% confluent. Cells were infected with PEDV (multiplicity of infection = 0.1) and fixed with cold 4% paraformaldehyde at 12, 18, 24, 30, and 36 hpi, respectively. The PEDV‐infected cells were detected using a mouse monoclonal antibody directed against the PEDV N protein and FITC‐conjugated goat anti‐mouse IgG antibody (Sigma). Cell nuclei were counterstained with 0.01% DAPI (Invitrogen). The fluorescent images were examined by using a confocal laser scanning microscope (LSM 510 Meta, Carl Zeiss).
2.8. Western blot analysis
We chose to analyze hydroxysteroid dehydrogenase like 2 (HSDL2), tyrosine 3‐monooxygenase/tryptophan 5‐monooxygenase activation protein (14‐3‐3) gamma, and HSPB1 (HSP27) by Western blotting as significant changes in their protein expression levels were observed. Cell lysates for PEDV‐ and mock‐infected cultures were harvested at 24 hpi and protein concentrations determined. Equivalent quantities of cell lysates were from the three independent biological replicates denatured in 5× sample loading buffer by heating at 100°C for 10 min and separated by 12% SDS‐PAGE. Separated proteins were electroblotted onto 0.45 μm PVDF membranes (Millipore, Billerica, MA, USA) and blocked with 10% w/v skim milk in Tris‐buffered saline containing Tween 20 for 2 h at room temperature. Membranes were incubated with a rabbit polyclonal antibody against HSDL2, 14‐3‐3 gamma, and HSP27 (Proteintech, China) at 4°C overnight. After washing three times with Tris‐buffered saline containing Tween 20, membranes were incubated with the appropriate HRP‐conjugated secondary antibodies (Beyotime, China) at 37°C for 1 h. Detection was performed using Clarity ECL reagents (Bio‐Rad, Hercules, CA, USA).
3. Results
3.1. PEDV propagation in Vero cells
The kinetics of PEDV propagation in Vero cells was determined by monitoring CPE and viral protein expression, as well as viral titers at 12, 18, 24, 30, and 36 hpi. As shown in Fig. 1A, minimal CPE was visible at 18 hpi and obvious CPE could be observed at 30 hpi; at 36 hpi, almost all cells fell off. The results of IFA for detecting the expression of PEDV N protein showed that the majority of cells were infected at 24 hpi (Fig. 1B). The one‐step growth curve showed that viral titer reached to 6.1 log10 at 24 hpi, peaked at 6.8 log10 at 30 hpi, and then gradually declined (Fig. 1C). To make sure higher proportion of the infected cells and to avoid excessive CPE, we thus chose 24 hpi as the time point under our infection conditions for further proteomic analysis.
Figure 1.
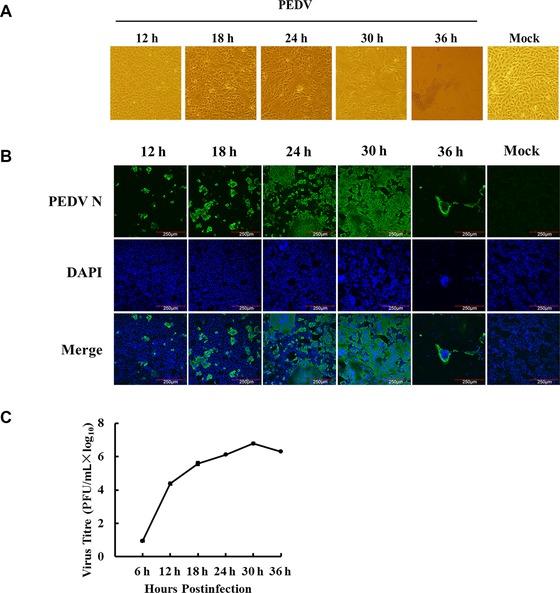
PEDV infection in Vero cells. (A) The cytopathic effects (CPE) of Vero cells at 12, 18, 24, 30, and 36 h after PEDV infection, and mock‐infected cells at 36 h as a control. (B) Dynamics of PEDV proliferation by immunofluorescence staining in the infected Vero cells at 12, 18, 24, 30, and 36 h postinfection, respectively, and mock‐infected cells at 36 h as a control. (C) One‐step growth curve of PEDV strain AJ1102 in Vero cells.
3.2. Analysis of the differentially regulated proteins
Protein extracts were prepared from PEDV‐ and mock‐infected Vero cells, and then treated as described in Section 2. We detected and quantified 3418 proteins, of which 8 were significantly upregulated and 41 downregulated based on our criteria for the identification of differentially expressed proteins. Among these 49 proteins, 45 proteins have at least two unique peptides, while 4 proteins (mitogen‐activated protein kinase 3‐like (MAPK3), adenylyl cyclase associated protein 1 isoform 2, alpha‐enolase isoform 3, transportin‐2‐like) have only one unique peptide and one nonunique peptide. The annotated tandem mass spectra and the detailed information regarding protein assignments based on single‐peptide assignments of the four proteins are provided as Supporting Information Data 1.
The molecular functional classes and subcellular locations of the 49 differentially expressed proteins were analyzed using UniProt and the GO database (Supporting Information Table 1). The eight upregulated proteins in infected cells were localized to the membrane (one molecule), mitochondria (three molecules), nucleus (two molecules), membrane and mitochondria (one molecule), and cytoplasm and nucleus (one molecule). The 41 downregulated proteins were located in the cytoplasm and nucleus (12 molecules), cytoplasm (6 molecules), ribosome (5 molecules), unknown cellular location (4 molecules), cytoskeleton (4 molecules), nucleus (3 molecules), membrane (2 molecules), cytoplasm and membrane (2 molecules), cytoplasm, extracellular and nucleus (2 molecules), and mitochondrion and nucleus (1 molecule).
Based on gene identifications of the proteins in Table 1, protein GI numbers and ratios (infection/control) were imported into the IPA tool. Their functional classification and interacting pathways were analyzed and reconstructed based on the underlying biological evidence from the literature. The analysis of function classification by IPA showed that the 8 significantly upregulated and 41 downregulated proteins could be divided among four distinct functional sets: diseases and disorders; molecular and cellular functions; physiological system development; and functions including toxicity functions (Fig. 2 and Supporting Information Table 2). These differentially regulated proteins are involved in biological processes relevant to gene expression, cell cycle, cell signaling, cell death and survival, cellular function, and maintenance.
Table 1.
Statistically significant differentially expressed proteins identified by iTRAQ analysis of Vero cells infected with PEDV
| Protein name | Accession no. | Ratios (infection/control) | Peptides | Sequence coverage (%) | Functions |
|---|---|---|---|---|---|
| Proteins present in increased abundance in PEDV‐infected cells | |||||
| Glucosidexylosyltransferase 1 isoform 1 | gi|109096177 | 2.042 | 2 | 4.5 | UDP‐xylosyltransferase activity |
| Hydroxysteroid dehydrogenase like 2 isoform 4 | gi|109110554 | 1.979 | 4 | 17.7 | Oxidoreductase activity |
| Metaxin‐1 like | gi|297280200 | 1.654 | 2 | 5.1 | Protein transport |
| Single‐stranded DNA‐binding protein, mitochondrial isoform 1 | gi|109068491 | 1.568 | 6 | 44.6 | Single‐stranded DNA binding |
| Transportin‐2 like | gi|297276226 | 1.557 | 2 | 1.7 | Protein transport |
| Phosphate carrier protein, mitochondrial isoform 1 | gi|297263324 | 1.547 | 2 | 5.5 | Phosphate ion carrier activity |
| Pentatricopeptide repeat containing protein 1 like | gi|297287945 | 1.539 | 2 | 3.1 | tRNA processing |
| Small nuclear ribonucleoprotein E like isoform 2 | gi|109018742 | 1.507 | 2 | 25 | Histone 3′‐end processing |
| Proteins present in decreased abundance in PEDV‐infected cells | |||||
| Acetyl‐CoA acetyltransferase, cytosolic like | gi|297292010 | 0.666 | 2 | 10.9 | Acetyl‐CoAC‐acetyltransferase activity |
| NSFL1 cofactor p47 like isoform 7 | gi|109092580 | 0.664 | 6 | 23.1 | Lipid binding |
| 39S ribosomal protein L24, mitochondrial like isoform 1 | gi|109017398 | 0.656 | 3 | 17.1 | Structural constituent of ribosome |
| Ran‐specific GTPase‐activating protein like | gi|297260581 | 0.654 | 3 | 22.5 | GDP‐dissociation inhibitor activity |
| 14‐3‐3 Protein eta isoform 2 | gi|109093926 | 0.651 | 6 | 30.5 | Adapter protein implicated in the regulation of signaling pathways |
| Protein‐l‐isoaspartate (d‐aspartate) O‐methyltransferase like | gi|297284352 | 0.651 | 3 | 21.1 | Protein‐l‐isoaspartate (d‐aspartate) O‐methyltransferase activity |
| Dipeptidyl peptidase 3 like, partial | gi|109109554 | 0.649 | 2 | 36.9 | Dipeptidyl‐peptidase activity |
| Small acidic protein like | gi|109107167 | 0.648 | 3 | 21.9 | Unknown |
| Gamma interferon inducible protein 16 like isoform 2 | gi|109017483 | 0.648 | 3 | 6.3 | Double‐stranded DNA binding |
| Caspase‐8 | gi|297264681 | 0.643 | 2 | 10 | Induced cell death |
| High‐mobility group protein B1 like | gi|109098276 | 0.642 | 3 | 17.7 | DNA binding, bending |
| 14‐3‐3 Protein theta isoform 1 | gi|109075480 | 0.639 | 7 | 37.1 | Adapter protein implicated in the regulation of signaling pathways |
| HSP beta‐1 | gi|109066218 | 0.637 | 4 | 31.2 | Response to stress |
| Gamma‐enolase isoform 1 | gi|109095369 | 0.634 | 7 | 23.7 | Magnesium ion binding |
| UV excision repair protein RAD23 homolog B isoform 5 | gi|109110657 | 0.632 | 5 | 21.8 | Nucleotide‐excision repair |
| SH3 domain binding glutamic acid rich like protein like | gi|297304256 | 0.632 | 2 | 19.3 | Protein disulfide oxidoreductase activity |
| Gelsolin isoform 13 | gi|109110383 | 0.631 | 8 | 15.9 | Calcium ion binding |
| 60S ribosomal protein L24 isoform 2 | gi|109032850 | 0.631 | 3 | 19.1 | Structural constituent of ribosome |
| Multiple coagulation factor deficiency protein 2 like isoform 1 | gi|297265949 | 0.63 | 2 | 28.1 | Vesicle‐mediated transport |
| Protein TFG isoform 2 | gi|109032777 | 0.628 | 4 | 19.8 | Signal transducer activity |
| Phosphoglyceratemutase 1 isoform 2 | gi|109100081 | 0.624 | 6 | 37 | Phosphoglyceratemutase activity |
| Myotrophin isoform 3 | gi|109068363 | 0.623 | 2 | 20.3 | Structural constituent of cytoskeleton |
| Annexin A3 isoform 3 | gi|109074379 | 0.622 | 5 | 19.5 | Phospholipase A2 inhibitor activity |
| Protein phosphatase 1 regulatory subunit 7 like isoform 1 | gi|109101715 | 0.62 | 4 | 14.1 | Protein phosphatase type 1 regulator activity |
| Hepatoma‐derived growth factor like nucleotide binding | gi|297281928 | 0.609 | 2 | 28.4 | Transcription regulation |
| Far upstream element binding protein 2 | gi|297275911 | 0.597 | 9 | 16.6 | mRNA trafficking |
| 40S ribosomal protein S24 like isoform 1 | gi|109037800 | 0.59 | 2 | 17.7 | Structural constituent of ribosome |
| NudC domain containing protein 3 | gi|109066663 | 0.586 | 3 | 12.9 | Unknown |
| Gephyrin isoform 2 | gi|109083983 | 0.577 | 3 | 7.2 | Structural constituent of cytoskeleton |
| 60S ribosomal protein L27a like | gi|109065734 | 0.572 | 3 | 20.9 | Structural constituent of ribosome |
| 14‐3‐3 Protein beta/alpha | gi|297259739 | 0.551 | 5 | 23.6 | Adapter protein implicated in the regulation of signaling pathways |
| 60S ribosomal protein L36 like | gi|109081357 | 0.549 | 2 | 20 | Structural constituent of ribosome |
| Synapse‐associated protein 1 like | gi|297305163 | 0.538 | 2 | 14.5 | Unknown |
| 14‐3‐3 Protein gamma like isoform 1 | gi|109066214 | 0.537 | 5 | 24.3 | Protein kinase C inhibitor activity |
| Tubulin gamma‐1 chain | gi|109115505 | 0.537 | 3 | 10.4 | Structural constituent of cytoskeleton |
| Destrin isoform 1 | gi|109092999 | 0.533 | 4 | 33.8 | Structural constituent of cytoskeleton |
| Prostaglandin E synthase 3 like isoform 4 | gi|109097318 | 0.492 | 4 | 35.6 | Chaperone cofactor dependent protein refolding |
| Adenylyl cyclase associated protein 1 isoform 2 | gi|297278362 | 0.479 | 7 | 26.8 | Establishment or maintenance of cell polarity |
| Coiled‐coil domain containing protein 124 like | gi|109123948 | 0.449 | 2 | 14.4 | Unknown |
| Mitogen‐activated protein kinase 3 like (MAPK3) | gi|297283792 | 0.366 | 2 | 5.6 | Signal transduction |
| Alpha‐enolase isoform 3 | gi|109066574 | 0.277 | 13 | 30.6 | Transcription corepressor activity |
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
Figure 2.
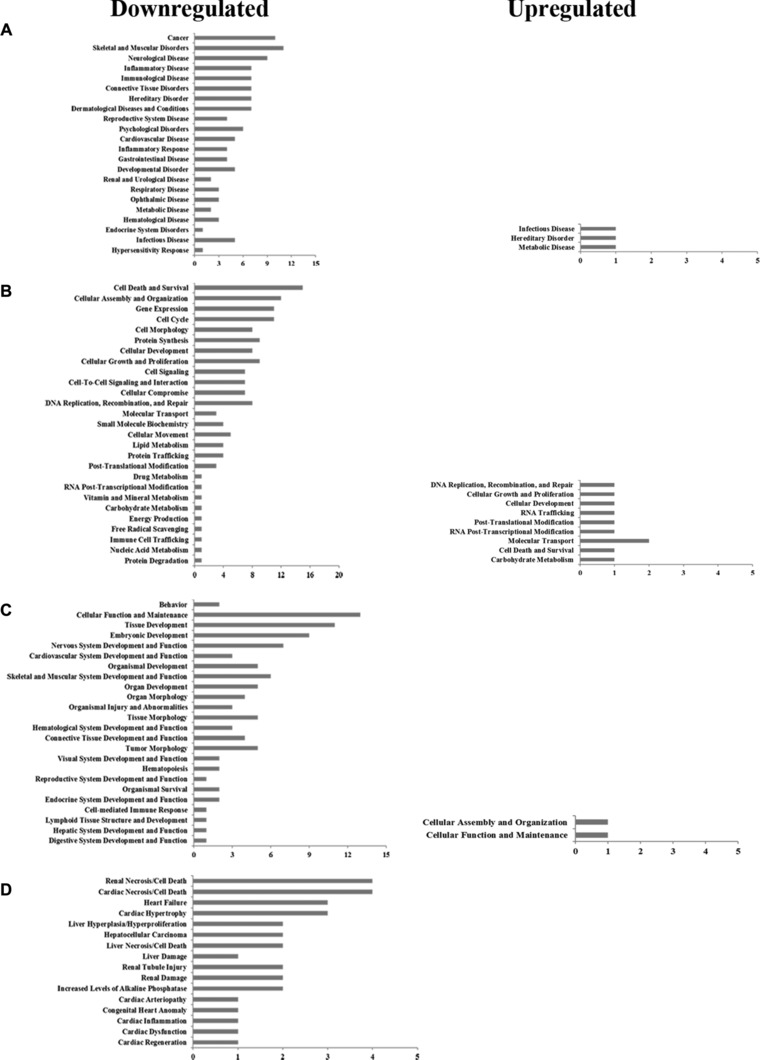
Functional characterization of up‐ and downregulated proteins. (A) Diseases and disorders. (B) Molecular and cellular functions. (C) Physiological system development and functions. (D) Toxicity functions. More information is available in Supporting Information Table 2.
With goal of exploring the potential protein network connections for the differentially regulated proteins in detail, IPA tool was used. The differentially regulated proteins were mapped to three specific functional networks. Each network had at least five “focus” proteins (significantly up‐ or downregulated proteins). The three networks of interest correspond to cancer, cell death and survival, cell‐to‐cell signaling, and interaction (Fig. 3A); cancer, cardiovascular disease, and hereditary disorder (Fig. 3B); RNA post‐transcriptional modification, cellular developmental, cellular growth, and proliferation (Fig. 3C).
Figure 3.
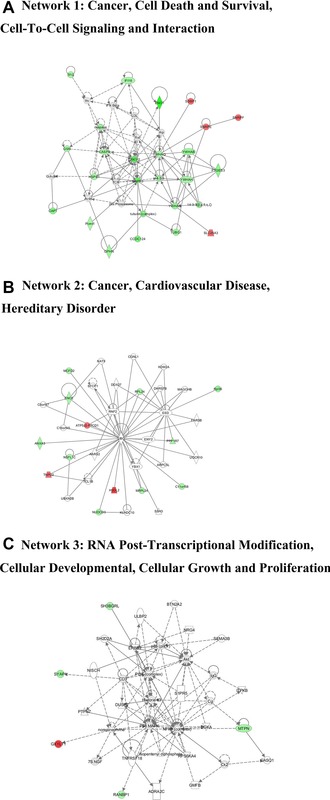
Specific network analysis of proteins significantly altered in PEDV‐infected cells. Significantly upregulated proteins are shown in red, significantly downregulated proteins in green, and proteins that were not identified in this study but are involved in the networks are shown in white. The color shade indicates the magnitude of the change in protein expression. Shapes are indicative of the molecular class, and lines with arrows indicate molecular relationships (Supporting Information Fig. 1).
3.3. Validation of protein identification and quantitation
We analyzed expression levels of HSDL2 (upregulated), 14‐3‐3 gamma (downregulated), and HSP27 (downregulated) in PEDV‐infected Vero cells (Fig. 4) by Western blotting. The proteins were selected on the basis of interest and on their different ratios. The ratios of these three proteins between infected and uninfected cells were consistent with those determined using the iTRAQ labeled LC‐MS/MS system.
Figure 4.
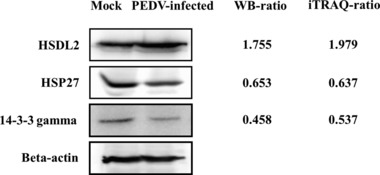
Confirmation of differentially expressed proteins by Western blotting. Analysis of HSDL2, 14‐3‐3 gamma, and HSP27 expression levels in PEDV‐infected and control cells by Western blotting. The WB‐ratio (Western blotting ratio, PEDV‐infected/mock) was calculated based on the relative intensity of each band in PEDV‐infected cells to the corresponding band in mock‐infected cells. Beta‐actin was used as a loading control. The iTRAQ‐ratio (infection/control) obtained by MS analysis are shown on the right.
4. Discussion
Proteomics has become an important methodology for determining cellular protein interactions and host cellular pathophysiological processes following virus infection 12, 21. To date, no analysis has been reported for the differential proteomes of host cells infected with PEDV. Based on our findings, the expressions of 49 different proteins were significantly altered in PEDV‐infected cells. These proteins were involved in a variety of processes including apoptosis, signal transduction, and stress responses of host cells. Our data are overview of the altered proteins during a host response to PEDV infection and should provide clues regarding PEDV pathogenesis.
The apoptosis of host cells plays a major role in regulating the pathogenesis of many infectious diseases 22. Apoptosis triggered by virus infection directly leads to viral pathogenesis; however, blocking apoptosis can avoid premature death of the infected cells, permitting a high titer of virus replication or a persistent infection 17. In our analysis, two apoptosis‐related genes, (14‐3‐3 and caspase‐8) were identified following PEDV infection. The 14‐3‐3 proteins constitute a family of conserved adaptor and scaffold proteins present in all eukaryotic organisms. Previous reports showed that 14‐3‐3 proteins modulate the cell cycle and apoptosis at multiple steps, leading to an inhibition of the onset of apoptosis 23, 24. Our results demonstrate that decreased expression of four 14‐3‐3 proteins (eta, theta, beta/alpha, gamma) was concomitant with PEDV infection, and thus predicted to promote apoptosis of infected cells. Caspase‐8 is the initiator caspase that is activated by the correlation between external apoptotic elements and cell surface molecules, playing fundamental roles in initiating the apoptotic cascade response 25. The downregulation of caspase‐8 expression in PEDV‐infected cells suggests a mechanism that attenuates virus‐induced apoptosis at the early infection phase, thus favoring a virus to replicate to a higher titer. In the case of SARS‐CoV, stimulation of both apoptotic and antiapoptotic elements has been seen, allowing for rapid multiplication of progeny virus before cell lysis 26. The interplay between apoptosis‐related genes in PEDV infection might contribute to it achieving maximum viral replication before cell death.
Viruses are known to develop the ability to manipulate a variety of host cellular signal transduction pathways to facilitate virus survival. Among these pathways is the extracellular signal regulated kinases (ERKs) pathway, comprising Raf, MEK1/2, and ERK1/2. These play central roles in the modulation of cell survival, differentiation, and proliferation 27. Many viruses activate the ERK pathway for higher levels of cellular and viral gene production. By activating ERK1/2, enterovirus 71 prevents activation of the proapoptotic molecules caspase‐9 and Bcl‐2 associated death promoter 28. Porcine reproductive and respiratory syndrome virus infection activates ERK1/2 at the early infection phase in PAM cells, with ERK activation essential for porcine reproductive and respiratory syndrome virus replication in vitro 29. Some studies have shown a negative correlation between ERK activity and viral replication. The Ebola virus glycoprotein GP specifically inhibits ERK1/2 to cause increased cell toxicity and viral titers 30. Yoshizuka et al. reported that overexpression of the human immunodeficiency virus (HIV) type 1 (HIV‐1) Vpr protein downregulated gene expression and phosphorylation of ERK1/2 in the ERK/MAPK pathway. It was speculated that the decrease in ERK1/2 activation is important for HIV replication and pathogenesis 31. SARS‐CoV N induces apoptosis of COS‐1 monkey kidney cells in the absence of growth factors through downregulation of the ERK pathway 32. ERK1/2 regulation of viral replication functions occurs at specific points during the replication cycle 33. The manipulation of the ERK pathway is a ubiquitous characteristic during viral infection and is not limited to specific viruses or viral events 34. In our study, ERK1 was significantly downregulated during PEDV infection. The specific mechanism by which PEDV manipulates the ERK pathway for maximal viral replication is currently under investigation in our laboratory.
Activation of the heat‐shock response might be a specific function that a virus exerts to guarantee appropriate synthesis of viral proteins 35. In our study, HSP27 was significantly downregulated in Vero cells following PEDV challenge. HSP27, a ubiquitous stress‐inducible cellular protein, is a multifunctional protein that plays a role in the inhibition of caspase‐independent apoptosis, actin cytoskeletal remodeling, and might also participate in protein degradation 36, 37. It has been implicated that HSP27 is upregulated during enterovirus 71, feline herpesvirus, and infectious bursal disease virus infections 38, 39, 40. Based on our results, HSP27 was downregulated during PEDV infection, similar to previous reports regarding adenoviruses 41, classical swine fever virus 42, and porcine circovirus type 2 21. HSP27 is an intracellular antiviral factor against hepatitis B virus through the induction of type I interferon and downstream antiviral effectors 43 and inhibits caspase‐independent apoptosis. We suspected that decreased expression of HSP27 might have a specific role in host cells during viral infection, however that specific mechanism requires further investigation.
In summary, the proteomic changes in PEDV‐infected Vero cells were characterized using iTRAQ combined with LC‐MS/MS. To the best of our knowledge, this is the first time proteomics has been used to explore the pathogen–host protein interaction network in PEDV‐infected Vero cells. Since the PEDV variant emerged in China and the United States, PEDV has received increasing attention, and more researchers have undertaken the study of PEDV. Our analyses of the proteins discussed above were descriptive, and further functional investigations are required to elucidate the pathogenic mechanisms and cellular responses to PEDV infection. Such molecular insights into differentially expressed cellular proteins would be helpful in studying the infectivity and pathogenesis of PEDV.
The authors have declared no conflict of interest.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary Data 1
Figure s1
Table 1. Information for each significantly altered protein (P < 0.05) identified in PEDV‐infected cells
Table 2. Functional characterization of up‐ and down‐regulated proteins in PEDV‐infected cells
Acknowledgement
This work was supported by the Key Technology R&D Programme of China (2015BAD12B02), the Natural Science Foundation of Hubei Province (2014CFA009), the National Natural Sciences Foundation of China (31402239), and the Fundamental Research Funds for the Central Universities (2013PY043).
Colour Online: See the article online to view Figs. 1 and 3 in colour.
5 References
- 1. Junwei, G. , Baoxian, L. , Lijie, T. , Yijing, L. , Cloning and sequence analysis of the N gene of porcine epidemic diarrhea virus LJB/03. Virus Genes 2006, 33, 215–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wood, E. N. , An apparently new syndrome of porcine epidemic diarrhoea. Vet. Rec. 1977, 100, 243–244. [DOI] [PubMed] [Google Scholar]
- 3. Chasey, D. , Cartwright, S. F. , Virus‐like particles associated with porcine epidemic diarrhoea. Res. Vet. Sci. 1978, 25, 255–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Li, W. , Li, H. , Liu, Y. , Pan, Y. et al., New variants of porcine epidemic diarrhea virus, China, 2011. Emerg. Infect. Dis. 2012, 18, 1350–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bi, J. , Zeng, S. , Xiao, S. , Chen, H. , Fang, L. , Complete genome sequence of porcine epidemic diarrhea virus strain AJ1102 isolated from a suckling piglet with acute diarrhea in China. J. Virol. 2012, 86, 10910–10911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang, X. M. , Niu, B. B. , Yan, H. , Gao, D. S. et al., Genetic properties of endemic Chinese porcine epidemic diarrhea virus strains isolated since 2010. Arch. Virol. 2013, 158, 2487–2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen, J. , Liu, X. , Shi, D. , Shi, H. et al., Complete genome sequence of a porcine epidemic diarrhea virus variant. J. Virol. 2012, 86, 3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sun, R. Q. , Cai, R. J. , Chen, Y. Q. , Liang, P. S. et al., Outbreak of porcine epidemic diarrhea in suckling piglets, China. Emerg. Infect. Dis. 2012, 18, 161–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen, Q. , Li, G. , Stasko, J. , Thomas, J. T. et al., Isolation and characterization of porcine epidemic diarrhea viruses associated with the 2013 disease outbreak among swine in the United States. J. Clin. Microbiol. 2014, 52, 234–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Stevenson, G. W. , Hoang, H. , Schwartz, K. J. , Burrough, E. R. et al., Emergence of Porcine epidemic diarrhea virus in the United States: clinical signs, lesions, and viral genomic sequences. J. Vet. Diagn. Invest. 2013, 25, 649–654. [DOI] [PubMed] [Google Scholar]
- 11. Cho, W. K. , Kim, H. , Choi, Y. J. , Yim, N. H. et al., Epimedium koreanum Nakai water extract exhibits antiviral activity against porcine epidermic diarrhea virus in vitro and in vivo. Evid. Based Complement. Alternat. Med. 2012, 2012, 985151. doi: 10.1155/2012/985151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Maxwell, K. L. , Frappier, L. , Viral proteomics. Microbiol Mol Biol Rev. 2007, 71, 398–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Munday, D. C. , Surtees, R. , Emmott, E. , Dove, B. K. et al., Using SILAC and quantitative proteomics to investigate the interactions between viral and host proteomes. Proteomics 2012, 12, 666–672. [DOI] [PubMed] [Google Scholar]
- 14. Zeng, R. , Ruan, H. Q. , Jiang, X. S. , Zhou, H. et al., Proteomic analysis of SARS associated coronavirus using two‐dimensional liquid chromatography mass spectrometry and one‐dimensional sodium dodecyl sulfate‐polyacrylamide gel electrophoresis followed by mass spectroemtric analysis. J. Proteome Res. 2004, 3, 549–555. [DOI] [PubMed] [Google Scholar]
- 15. Sun, J. , Han, Z. , Shao, Y. , Cao, Z. et al., Comparative proteome analysis of tracheal tissues in response to infectious bronchitis coronavirus, Newcastle disease virus, and avian influenza virus H9 subtype virus infection. Proteomics 2014, 14, 1403–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang, X. , Shi, H. Y. , Chen, J. F. , Shi, D. et al., Identification of cellular proteome using two‐dimensional difference gel electrophoresis in ST cells infected with transmissible gastroenteritis coronavirus. Proteome Sci. 2013, 11, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Enjuanes, L. , Almazan, F. , Sola, I. , Zuniga, S. , Biochemical aspects of coronavirus replication and virus‐host interaction. Annu. Rev. Microbiol. 2006, 60, 211–230. [DOI] [PubMed] [Google Scholar]
- 18. Hofmann, M. , Wyler, R. , Propagation of the virus of porcine epidemic diarrhea in cell culture. J. Clin. Microbiol. 1988, 26, 2235–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wright, J. C. , Collins, M. O. , Yu, L. , Kall, L. et al., Enhanced peptide identification by electron transfer dissociation using an improved Mascot Percolator. Mol. Cell. Proteomics 2012, 11, 478–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nesvizhskii, A. I. , Aebersold, R. , Interpretation of shotgun proteomic data: the protein inference problem. Mol. Cell. Proteomics 2005, 4, 1419–1440. [DOI] [PubMed] [Google Scholar]
- 21. Zhang, X. , Zhou, J. , Wu, Y. , Zheng, X. et al., Differential proteome analysis of host cells infected with porcine circovirus type 2. J. Proteome Res. 2009, 8, 5111–5119. [DOI] [PubMed] [Google Scholar]
- 22. Johns, H. L. , Bensaude, E. , La Rocca, S. A. , Seago, J. et al., Classical swine fever virus infection protects aortic endothelial cells from pIpC‐mediated apoptosis. J. Gen. Virol. 2010, 91, 1038–1046. [DOI] [PubMed] [Google Scholar]
- 23. van Hemert, M. J. , Steensma, H. Y. , van Heusden, G. P. , 14‐3‐3 proteins: key regulators of cell division, signalling and apoptosis. Bioessays 2001, 23, 936–946. [DOI] [PubMed] [Google Scholar]
- 24. Xing, H. , Zhang, S. , Weinheimer, C. , Kovacs, A. , Muslin, A. J. , 14‐3‐3 proteins block apoptosis and differentially regulate MAPK cascades. EMBO J. 2000, 19, 349–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cohen, G. M. , Caspases: the executioners of apoptosis. Biochem. J. 1997, 326(Pt 1), 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tang, B. S. , Chan, K. H. , Cheng, V. C. , Yuen, K. Y. , Comparative host gene transcription by microarray analysis early after infection of the Huh7 cell line by SARS coronavirus and human coronavirus 229E. Hong Kong Med. J. 2009, 15(Suppl 9), 23–26. [PubMed] [Google Scholar]
- 27. Fang, J. Y. , Richardson, B. C. , The MAPK signalling pathways and colorectal cancer. Lancet Oncol. 2005, 6, 322–327. [DOI] [PubMed] [Google Scholar]
- 28. Wong, W. R. , Chen, Y. Y. , Yang, S. M. , Chen, Y. L. , Horng, J. T. , Phosphorylation of PI3K/Akt and MAPK/ERK in an early entry step of enterovirus 71. Life Sci. 2005, 78, 82–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lee, Y. J. , Lee, C. , Porcine reproductive and respiratory syndrome virus replication is suppressed by inhibition of the extracellular signal‐regulated kinase (ERK) signaling pathway. Virus Res. 2010, 152, 50–58. [DOI] [PubMed] [Google Scholar]
- 30. Zampieri, C. A. , Fortin, J. F. , Nolan, G. P. , Nabel, G. J. , The ERK mitogen‐activated protein kinase pathway contributes to Ebola virus glycoprotein‐induced cytotoxicity. J. Virol. 2007, 81, 1230–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yoshizuka, N. , Yoshizuka‐Chadani, Y. , Krishnan, V. , Zeichner, S. L. , Human immunodeficiency virus type 1 Vpr‐dependent cell cycle arrest through a mitogen‐activated protein kinase signal transduction pathway. J. Virol. 2005, 79, 11366–11381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Surjit, M. , Liu, B. , Jameel, S. , Chow, V. T. , Lal, S. K. , The SARS coronavirus nucleocapsid protein induces actin reorganization and apoptosis in COS‐1 cells in the absence of growth factors. Biochem. J. 2004, 383, 13–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cai, Y. , Liu, Y. , Zhang, X. , Suppression of coronavirus replication by inhibition of the MEK signaling pathway. J. Virol. 2007, 81, 446–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Moser, L. A. , Schultz‐Cherry, S. , Suppression of astrovirus replication by an ERK1/2 inhibitor. J. Virol. 2008, 82, 7475–7482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Glotzer, J. B. , Saltik, M. , Chiocca, S. , Michou, A. I. et al., Activation of heat‐shock response by an adenovirus is essential for virus replication. Nature 2000, 407, 207–211. [DOI] [PubMed] [Google Scholar]
- 36. Vidyasagar, A. , Wilson, N. A. , Djamali, A. , Heat shock protein 27 (HSP27): biomarker of disease and therapeutic target. Fibrogenesis Tissue Repair 2012, 5, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ciocca, D. R. , Oesterreich, S. , Chamness, G. C. , McGuire, W. L. , Fuqua, S. A. , Biological and clinical implications of heat shock protein 27000 (Hsp27): a review. J. Natl. Cancer Inst. 1993, 85, 1558–1570. [DOI] [PubMed] [Google Scholar]
- 38. Leong, W. F. , Chow, V. T. , Transcriptomic and proteomic analyses of rhabdomyosarcoma cells reveal differential cellular gene expression in response to enterovirus 71 infection. Cell Microbiol. 2006, 8, 565–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zheng, X. , Hong, L. , Shi, L. , Guo, J. et al., Proteomics analysis of host cells infected with infectious bursal disease virus. Mol. Cell. Proteomics 2008, 7, 612–625. [DOI] [PubMed] [Google Scholar]
- 40. Go, E. P. , Wikoff, W. R. , Shen, Z. , O'Maille, G. et al., Mass spectrometry reveals specific and global molecular transformations during viral infection. J. Proteome Res. 2006, 5, 2405–2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zantema, A. , de Jong, E. , Lardenoije, R. , van der Eb, A. J. , The expression of heat shock protein hsp27 and a complexed 22‐kilodalton protein is inversely correlated with oncogenicity of adenovirus‐transformed cells. J. Virol. 1989, 63, 3368–3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sun, J. , Jiang, Y. , Shi, Z. , Yan, Y. et al., Proteomic alteration of PK‐15 cells after infection by classical swine fever virus. J. Proteome Res. 2008, 7, 5263–5269. [DOI] [PubMed] [Google Scholar]
- 43. Tong, S. W. , Yang, Y. X. , Hu, H. D. , An, X. et al., HSPB1 is an intracellular antiviral factor against hepatitis B virus. J. Cell Biochem. 2013, 114, 162–173. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary Data 1
Figure s1
Table 1. Information for each significantly altered protein (P < 0.05) identified in PEDV‐infected cells
Table 2. Functional characterization of up‐ and down‐regulated proteins in PEDV‐infected cells


