Summary
Glycyrrhizin is an important bioactive compound that is used clinically to treat chronic hepatitis and is also used as a sweetener world‐wide. However, the key UDP‐dependent glucuronosyltransferases (UGATs) involved in the biosynthesis of glycyrrhizin remain unknown.
To discover unknown UGATs, we fully annotated potential UGATs from Glycyrrhiza uralensis using deep transcriptome sequencing. The catalytic functions of candidate UGATs were determined by an in vitro enzyme assay.
Systematically screening 434 potential UGATs, we unexpectedly found one unique GuUGAT that was able to catalyse the glucuronosylation of glycyrrhetinic acid to directly yield glycyrrhizin via continuous two‐step glucuronosylation. Expression analysis further confirmed the key role of GuUGAT in the biosynthesis of glycyrrhizin. Site‐directed mutagenesis revealed that Gln‐352 may be important for the initial step of glucuronosylation, and His‐22, Trp‐370, Glu‐375 and Gln‐392 may be important residues for the second step of glucuronosylation. Notably, the ability of GuUGAT to catalyse a continuous two‐step glucuronosylation reaction was determined to be unprecedented among known glycosyltransferases of bioactive plant natural products.
Our findings increase the understanding of traditional glycosyltransferases and pave the way for the complete biosynthesis of glycyrrhizin.
Keywords: biosynthesis, glucuronosylation, glucuronosyltransferase, glycosyltransferase, glycyrrhetinic acid, Glycyrrhiza uralensis, glycyrrhizin
Introduction
Triterpenoid saponins are an important class of natural plant products with a wide range of biological activities. These compounds can protect plants as a result of their antimicrobial, anti‐insect, and anti‐palatability functions (Tava & Odoardi, 1996; Osbourn, 2003; Xu et al., 2015), and they are crucial to human health. Triterpenoid saponins have useful pharmacological roles, including antimicrobial, antiviral, anti‐pathogen and anti‐cancer activities (Maes et al., 2004; Chan, 2007; Tang et al., 2015). In addition, triterpenoid saponins have been widely used in beverages, confectioneries and cosmetics (Uematsu et al., 2000; Sparg et al., 2004; Lee et al., 2008; Benchaar & Chouinard, 2009).
Glycyrrhizin, an important bioactive triterpenoid saponin in Glycyrrhiza plants, has various pharmacological anti‐inflammatory (Matsui et al., 2004), immunomodulatory (Jeong et al., 2002) and antiviral activities against different DNA and RNA viruses, including human immunodeficiency virus (HIV) and severe acute respiratory syndrome (SARS)‐associated coronavirus (Baba & Shigeta, 1987; Ito et al., 1987, 1988; Cinatl et al., 2003; Fiore et al., 2008; Wolkerstorfer et al., 2009). Clinically, glycyrrhizin has been widely used for the treatment of chronic hepatitis in Asian countries (van Rossum et al., 1998; Shibata, 2000). Sales of magnesium isoglycyrrhizinate injection reached CNY ¥1.6 billion in China in 2014. Glycyrrhizin is also commercially available world‐wide as a sweetener, as its sweetness is 150 times greater than that of sucrose (Kitagawa, 2002; Seki et al., 2011). The annual value of global trade in liquorice root from Glycyrrhiza plants was estimated at more than US $42.1 million in 2007 (Hayashi & Sudo, 2009; Kojoma et al., 2010). Therefore, the market demand for glycyrrhizin has increased (Seki et al., 2008, 2011). Currently, most of the key genes that are involved in the biosynthesis of glycyrrhizin have been successfully cloned and characterized, including β‐amyrin synthetase (bAS) (Shen et al., 2009), cytochrome P450 monooxygenase 88D6 (CYP88D6) (Seki et al., 2008) and CYP72A154 (Seki et al., 2011) (Fig. 1). UDP‐dependent glycosyltransferases (UGTs) have crucial roles in the last glucuronosylation reaction, which may greatly improve the sweetness and solubility of glycyrrhetinic acid.
Figure 1.
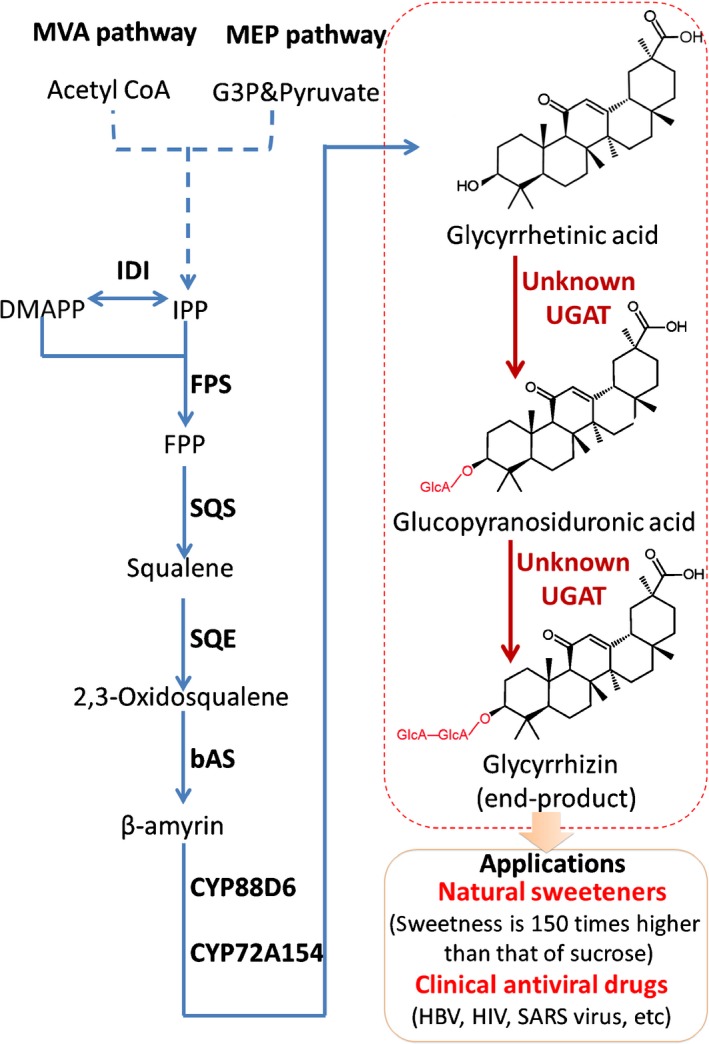
Glycyrrhizin biosynthesis pathway in Glycyrrhiza uralensis. One‐step catalytic reactions are indicated with solid arrows, and multi‐step catalytic reactions are indicated with dashed arrows. MVA pathway, mevalonate pathway; MEP pathway, plastid‐localized 2‐C‐methyl‐d‐erythritol 4‐phosphate pathway; IPP, isopentenyl diphosphate; DMAPP, dimethylallyl diphosphate; FPP, farnesyl pyrophosphate; IDI, isopentenyl diphosphate isomerase; FPS, farnesyl pyrophosphate synthase; SQS, squalene synthase; SQE, squalene monooxygenase or epoxidase; bAS, β‐amyrin synthetase; CYP88D6 and CYP72A154, cytochrome P450 monooxygenases; UGAT, glucuronosyltransferase; HBV, hepatitis B virus; HIV, human immunodeficiency virus; SARS virus, severe acute respiratory syndrome‐associated coronavirus.
The UDP‐dependent glucuronosyltransferase (UGAT) superfamily, members of which are a type of UGT, is one of the most important and prevalent superfamilies in plants. UGATs can greatly change the bioactivity, solubility or stability of metabolites (Kren & Martinkova, 2001), thereby playing important roles in plant growth, plant development and enzyme‐dependent modification of metabolites in metabolic engineering applications (Loos & Steinkellner, 2014; De Bruyn et al., 2015). In general, it is difficult to characterize the UGTs of natural products because natural product UGT families always have a wide variety of members with various functions in plants (Yonekura‐Sakakibara & Hanada, 2011). For instance, there are 115 UGTs in the Arabidopsis thaliana genome and 242 UGTs in the Glycine max genome (Yonekura‐Sakakibara & Hanada, 2011). In Panax ginseng, 512 contigs potentially encoding plant UGTs have been identified in expressed sequence tag (EST) data sets available from National Center for Biotechnology Information (NCBI) GenBank (Yan et al., 2014). However, only 13 triterpenoid UGTs have been characterized in plants (Supporting Information Table S1) (Seki et al., 2015). UGATs involved in the final catalytic steps of glycyrrhizin biosynthesis have been investigated ever since two key cytochrome P450 monooxygenases were cloned to produce glycyrrhetinic acid in 2011 (Seki et al., 2011). However, the key UGATs remain unknown (Fig. 1).
Transcriptome sequencing technologies are powerful tools for the characterization of genes that are involved in secondary metabolite biosynthesis in plants (Yonekura‐Sakakibara et al., 2007, 2008; Naoumkina et al., 2010), especially in plants without a sequenced genome. Here, we systematically screened 434 putative UGATs from Glycyrrhiza uralensis using deep transcriptome sequencing. Unexpectedly, we found one unique GuUGAT (c55437_g1) that was able to catalyse continuous two‐step glucuronosylation of glycyrrhetinic acid to directly yield glycyrrhizin; thus, the complete pathway of glycyrrhizin biosynthesis was determined. To our knowledge, this is the first description of a GuUGAT capable of catalysing a continuous two‐step glucuronosylation reaction; thus, this GuUGAT is unique among known glycosyltransferases of bioactive plant natural products. Knowledge regarding this gene may contribute to our understanding of traditional glycosyltransferases, pave the way for the complete biosynthesis of glycyrrhizin and be useful in the modification of natural products through genetic and metabolic engineering.
Materials and Methods
Plant material and stress treatment
Newly harvested seeds of Glycyrrhiza uralensis Fisch. were collected from the Ili cultivation base of Xinjiang Province in China and identified by Prof. Chunsheng Liu (Beijing University of Chinese Medicine, Beijing, China). The seeds were submerged in concentrated sulphuric acid for 1.5 h. The treated seeds were then washed with deionized water and soaked in it for 24 h at 25°C. The seeds were sown in vermiculite in an artificial climate box that was controlled at 25°C, with 16 h : 8 h, light : dark cycles. Drought or salt stress could lead to accumulation of glycyrrhizin in G. uralensis (Pan et al., 2006; Nasrollahi et al., 2014); thus, 21‐d‐old plants were subjected to treatment with 150 mM NaCl or 30% polyethylene glycol 6000 for 48 h. Treated plants were washed and frozen in liquid nitrogen and then stored at −80°C before RNA extraction.
Real‐time quantitative PCR
Total RNA was extracted from roots using the Plant RNA Extraction Kit (Biomed, Beijing, China). Real‐time quantitative (q)PCR was performed using the PrimeScript™ First‐Strand cDNA Synthesis Kit (Takara, Tokyo, Japan) and SYBR Master Mix (Takara) with gene‐specific primer pairs (Table S2). The relative amounts of the target genes were evaluated based on the relative expression index of mRNA using the 2−▵▵C(T) method, and β‐Actin (GenBank accession number EU190972.1) was used as the reference gene.
RNA extraction and cDNA library preparation
Total RNA was extracted from roots using the Plant RNA Extraction Kit (Biomed). The RNA was treated with RNase‐free DNase I and further purified using an RNA spin column (Biomed). RNA degradation and contamination were monitored on 1% agarose gels. RNA purity was checked using the NanoPhotometer® spectrophotometer (Implen, Carlsbad, CA, USA). The RNA concentration was measured using a Qubit® RNA Assay Kit and the Qubit® 2.0 Fluorometer (Life Technologies, Carlsbad, CA, USA). The RNA integrity was assessed using the RNA Nano 6000 Assay Kit and the Agilent Bioanalyzer 2100 system (Agilent Technologies, Carlsbad, CA, USA).
A total of 6 μg of RNA per sample was used as the input material for RNA sample preparation. Sequencing libraries were generated using the NEB Next® Ultra™ RNA Library Prep Kit for Illumina® (NEB (Beijing), Beijing, China) following the manufacturer's recommendations, and index codes were added to attribute sequences to each sample.
Deep Illumina sequencing, transcriptome assembly and gene functional annotation
After cluster generation, the library preparations were sequenced on an Illumina HiSeq 4000 platform, and paired‐end reads were generated. Raw data (raw reads) in the fastq format were first processed through in‐house Perl scripts. In this step, clean data (clean reads) were obtained by removing reads containing adapter sequences, reads containing poly‐N sequences and low‐quality reads from the raw data. All of the downstream analyses were based on clean data of high quality. Transcriptome assembly was accomplished based on Trinity (Grabherr et al., 2011). Gene function was annotated based on the following databases: Nr (NCBI nonredundant protein sequences), Nt (NCBI nonredundant nucleotide sequences), Pfam (protein families), KOG/COG (clusters of orthologous groups of proteins), Swiss‐Prot (a manually annotated and reviewed protein sequence database), KO (Kyoto encyclopedia of genes and genomes (KEGG) ortholog database) and GO (gene ontology).
Expression analysis of UGTs
The gene expression levels in each sample were estimated using rsem (Li & Dewey, 2011). Clean data were mapped back onto the assembled transcriptome. The read count for each gene was obtained from the mapping results. For each sequenced library, the read counts were adjusted using the edger program package through one scaling normalized factor. Differential expression analysis of two samples was performed using the DEGseq (2010) r package. P values were adjusted using the Q value. A Q value < 0.005&|log2(foldchange)|> 1 was set as the threshold for significant differential expression.
Heterologous expression of the GuUGAT protein
GuUGAT screened from comparative transcriptomic analysis was cloned and sequenced using gene‐specific primer pairs. Plasmids containing UGTs were digested using the restriction enzymes KpnI and XhoI and ligated into a pET‐32a(+) vector (Takara) that had been previously digested. The resultant plasmids were transformed into Escherichia coli BL21 (DE3). The transformants were precultured at 37°C for > 12 h on Luria−Bertani (LB) solid culture medium containing 50 μg ml−1 ampicillin. Single colonies were selected and transferred into liquid culture medium containing 50 μg ml−1 ampicillin. To screen colonies without mutagenesis, single positive colonies were amplified with universal primers designed to target the pET‐32a(+) vector and sequenced again. The screened colonies were then inoculated into 500 ml of liquid culture medium. After incubation at 37°C until the OD600 reached 0.5, isopropyl 1‐β‐d‐thiogalactoside (IPTG) was added to the medium at a final concentration of 0.1 mM, followed by further incubation at 15°C for 24 h. The recombinant E. coli cells were harvested by centrifugation (7000 g for 10 min at 4°C) and washed with distilled water. Recombinant proteins were purified and harvested using the Protein Purification Kit (CWBIO, Beijing, China). The purified proteins were then concentrated and desalted using VivaSpin 30000 MWCO (GE Healthcare, Buckinghamshire, UK). The protein concentration was determined using the Bradford method (Bradford, 1976). SDS‐PAGE was performed according to the method of Laemmli (Laemmli, 1970). Purified GuUGAT was verified by protein sequencing (Sangon, Shanghai, China).
High performance liquid chromatography‐electronic spray ion‐linear ion trap (HPLC‐ESI‐LTQ)‐Orbitrap MS analysis of the catalysed products
Reactions were performed in a volume of 50 μl. The reaction conditions were as follows: 50 mM Tris‐HCl (pH 8.0), 1 mM DTT, 1 mM UDP‐GlcA (Sigma‐Aldrich, Shanghai, China), 50 ng μl−1 purified proteins and 50 μM glycyrrhetinic acid or glucopyranosiduronic acid. The reactions were incubated for 2 h at 30°C and stopped by the addition of 200 μl of methanol. The precipitated proteins were removed by centrifugation (16 000 g at 30 min for 4°C), and the supernatants were concentrated by freeze‐drying and vacuum concentration. The residue was redissolved in 50 μl of 50% methanol and centrifuged at 12 000 g at 4°C for 30 min. A 5‐μl aliquot of the supernatant was injected into an HPLC‐ESI‐LTQ‐Orbitrap MS (Thermo Electron, Bremen, Germany) for analysis.
The chromatographic separations were performed on an SB‐C18 column (5 μm; 250 × 4.6 mm; Agilent Technologies) at room temperature with a flow rate of 1 ml min−1. A linear gradient elution was performed with water containing 0.1% formic acid (A) and methanol (B) as the mobile phases. The following programme was applied: 0–3.5 min, 79% B; 3.5–8.2 min, 79–98% B; 8.2–11 min, 98–79% B; and 11–20 min, 79% B. All of the chemical reference substances, including glycyrrhetinic acid (CAS: 471‐53‐4), glucopyranosiduronic acid (CAS: 34096‐83‐8), and glycyrrhizin (CAS: 1405‐86‐3), had > 98% purity and were commercially available (Weikeqi Biological Technology Co. Ltd, Sichuan, China).
Determination of the enzyme kinetic parameters
For kinetic studies of GuUGAT, a typical assay contained 50 mM Tris‐HCl (pH 8.0), saturating UDP‐GlcA (1 mM) and varying concentrations of glycyrrhetinic acid or glucopyranosiduronic acid (0.3125–20 μM) at 30°C in a total volume of 20 μl. The reactions were incubated for 10 min at 30°C and stopped by the addition of 80 μl of methanol. The precipitated proteins were removed by centrifugation (16 000 g for 30 min at 4°C). The supernatants were analysed via ultra‐high‐performance liquid chromatography with electrospray ionization tandem mass spectrometry (UPLC‐ESI‐MS/MS) using a Waters Acquity UPLC system (Waters, Milford, MA, USA) coupled to a Xevo TQ‐S mass spectrometer (Waters, Etten‐Leur, the Netherlands) equipped with an ESI source. A Waters Acquity UPLC BEH C18 column (2.1 × 100 mm; 1.7 μm) was used for chromatographic separation with a column temperature of 30°C and a flow rate of 0.30 ml min−1. A linear gradient elution was performed with water containing 0.1% formic acid (A) and methanol (B) as mobile phases. The following programme was applied: 0–0.5 min, 1% B; 0.5–1.0 min, 1–80% B; 1.0–2.0 min, 80–90% B; and 2.0–3.5 min, 90–1% B. The injection volume for all the samples was 2 μl. The transitions were set at m/z 469.44→355.38 for glycyrrhetinic acid, m/z 645.53→113.02 for glucopyranosiduronic acid and m/z 821.56→351.16 for glycyrrhizin.
Phylogenetic analysis
The primary protein structures of characterized UGTs coupled with GuUGAT were aligned using clustalw and analysed using mega 6.0 (Tamura et al., 2013). A neighbour‐joining tree was constructed via the bootstrap method with 1000 replications.
Homology modelling and molecular docking
The protein−protein blast tool from NCBI was used to search for possible template structures from the Protein Data Bank (PDB). The Scoring Matrix was selected as PAM30. The academic version of modeler 9v11 was used for the homology modelling of the GuUGAT protein structure (Eswar et al., 2007). Here, the crystal structure of Medicago truncatula UGT71G1 (PDB ID 2ACW), determined at a resolution of 2.6 Å, showed the highest total score with maximum sequence homology and a low E‐value. Thus, this structure was selected as the template from which to establish the three‐dimensional (3D) structures of the GuUGAT protein.
After the addition of hydrogen atoms, the structures of the models were individually energy‐minimized using the staged minimization program of the sybyl x‐1.2 package in two steps (Eswar et al., 2007). First, the simple method was used for 20 cycles before switching to the AMBER FF99 force field for 1000 iterations with the steepest descent (SD) calculation. Then, the conjugated gradient (CG) calculation was implemented until the convergence on the gradient reached 0.05 kcal/(Å mol). After global energy minimization, the stereo chemical quality of the constructed models was assessed using various structure assessment tools.
Surflex‐Dock (Tri‐I, Shanghai, China) was used to perform a virtual screening and calculate the ligand−receptor interaction. The compounds, including UDP‐GlcA, glycyrrhetinic acid and glucopyranosiduronic acid, were prepared using the following procedure: the structures were checked, the hydrogen atoms were added, the atomic charges were added using the Gasteiger−Hückel method, and energy minimization was implemented using the Tripos force‐field for 1000 iterations. Then, the optimized compounds were docked one at a time into the active site of the GuUGAT protein using default settings. The best total score conformer with the best consensus score (CScore) was recorded in the docking results.
Mutagenesis and enzyme assay
Site‐directed mutants of GuUGAT, including H22A, D121A, W349A, Q352A, H367A, W370A, E375A, E391A and Q392A mutants, were constructed using a Site‐directed Mutagenesis Kit (Biomed). The primer pairs that were designed to construct the site‐directed mutants are listed in Table S2. The constructed site‐directed mutants were then verified by sequencing and were induced for protein expression. The mutant proteins were expressed, purified, and subsequently utilized in an enzyme assay as described earlier (see the section ‘Heterologous expression of the GuUGAT protein’ in Materials and Methods).
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL/DDBJ databases under the following accession numbers: GuUGAT, KT759000; transcriptome data set of GU_root1, SRS1141032; GU_root2, SRS1141168; and GU_root3, SRS1141169.
Results
Deep transcriptome sequencing of G. uralensis roots
Glycyrrhizin is mainly derived from the roots of Glycyrrhiza plants, and G. uralensis is the most commonly used species. Glycyrrhiza uralensis roots accumulate high concentrations of glycyrrhizin after exposure to suitable drought or salt stress (Pan et al., 2006; Nasrollahi et al., 2014), suggesting that the genes that are associated with glycyrrhizin biosynthesis may be induced when plants are stressed by drought or salt. An assessment of plants under these conditions may allow us to uncover the specific UGATs that are involved in glycyrrhizin biosynthesis. In our study, we subjected 21‐d‐old G. uralensis plants to suitable drought and salt stress conditions and chose GU_root1 (roots of plants that were treated with high salt), GU_root2 (roots of plants that were exposed to drought) and GU_root3 (control roots) for deep transcriptome sequencing with the Illumina HiSeq 4000 platform (Fig. S1). Empty reads, low‐quality reads and reads containing unknown bases were removed from the raw reads. The resulting clean reads were further assembled to identify high‐quality transcripts. In total, 12.25 G clean bases of GU_root1 (GenBank accession no. SRS1141032), 10.6 G clean bases of GU_root2 (GenBank accession no. SRS1141168) and 10.14 G clean bases of GU_root3 (GenBank accession no. SRS1141169) were examined (Table S3). A total of 152 246 transcripts were assembled with a mean length of 891 bp and an N50 of 1640 bp (Tables S3–S5). Compared with previously assembled G. uralensis transcriptome sequencing data (Ramilowski et al., 2013), the amount of sequencing data produced in this work was approximately five‐fold greater in each sample (Table S6) (Ramilowski et al., 2013). Thus, these data may provide a good foundation for the subsequent screening of putative UGTs.
Annotation of putative UGTs
Functional annotation based on sequence homology is usually the first step in studying the roles and biological functions of gene products. To avoid missing annotations, we used multiple databases to annotate the assembled data, including Nr (NCBI nonredundant protein sequence database), Nt (NCBI nonredundant nucleotide sequence database), Pfam (protein family database), KOG/COG (clusters of orthologous groups of protein database), Swiss‐Prot (manually annotated and reviewed protein sequence database), KO (KEGG orthologue database) and GO (gene ontology database). This analysis assigned significant matches to 112 307 unigenes with a mean length of 703 bp and an N50 of 1164 bp (Tables S4–S5, S7; Figs S2–S5). Among these unigenes, 434 putative UGTs were annotated (Notes S1).
Functional prediction of annotated UGTs
Transcriptome expression profiling provides a global and detailed picture of the activity of functional genes across various conditions. Nevertheless, the number of putative UGTs identified in this study was so large that it became difficult to identify UGATs involved in glycyrrhizin biosynthesis. Differential expression (DE) analysis and coexpression (CE) analysis are powerful tools to characterize functional genes in the transcriptome data of plants because enzymes that are involved in the same function are believed to be temporally and spatially induced and coexpressed in some cases (Jorgensen et al., 2005; Naoumkina et al., 2010). To further identify putative UGTs, they were first filtered by DE analysis (Figs S6–S8, 2a). Then, the resulting UGTs that were coupled with other DE unigenes (Notes S2, S3) were interrogated by hierarchical cluster analysis based on Pearson correlation (Figs S9, 2b,c). Putative UGTs that were up‐regulated under salt or drought stress or that were coexpressed with the bAS, CYP88D6 and CYP72A154 genes, which are involved in the glycyrrhizin biosynthetic pathway, were regarded as important candidates (Fig. S10). In this work, to avoid false‐positive and false‐negative errors, the samples were sequenced deeply with a high coverage of transcripts, precisely reflecting the expression levels of unigenes. Moreover, to fully screen the possible UGATs, putative UGTs that had high similarity with known plant triterpenoid UGTs were also considered key candidates (Fig. S11). In total, eight UGTs (Notes S4; Figs S10, S11) were screened as candidate UGATs for the subsequent in vitro catalytic experiments.
Figure 2.
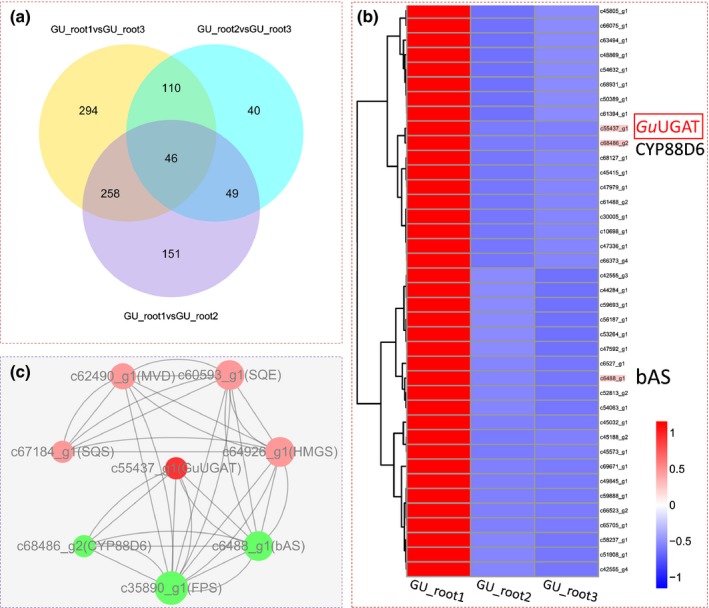
Transcriptome expression profile and analysis of differentially expressed unigenes from Glycyrrhiza uralensis. GU_root1, root sample treated with salt; GU_root2, root sample exposed to drought; GU_root3, control. (a) Venn diagram showing the number of differentially expressed genes in each of the two libraries. (b) Hierarchical clustering and corresponding heatmaps of the differentially expressed unigenes across all pairwise library comparisons (only the targeted subcluster is shown in Supporting Information Fig. S9). Glycyrrhiza uralensis UDP‐dependent glucuronosyltransferase (GuUGAT) is highlighted with a red box. (c) Network of differentially expressed unigenes involved in the biosynthesis of glycyrrhizin. Nodes with significant Pearson correlations (P < 0.05) are linked with lines.
Functional characterization of candidate UGATs in vitro
blast analysis revealed that all eight candidate UGATs contained the conserved plant secondary product glycosyltransferase (PSPG) domain (Notes S5). To determine the function of the candidate UGATs, they were homologously expressed in Escherichia coli. Crude enzymes were preliminarily used to characterize the activity in vitro, and only UGT3 (GuUGAT) was found to exhibit possible catalytic activity. GuUGAT was then purified using a protein purification kit (Fig. 3a), verified by protein sequencing (Fig. S12) and used in an in vitro catalytic reaction (Fig. 3b). Catalytic products were determined by high‐resolution HPLC‐ESI‐LTQ‐Orbitrap MS. To our surprise, compared with the control, the catalytic reaction containing the unique GuUGAT produced a new peak when glycyrrhetinic acid or glucopyranosiduronic acid was used as a substrate. The new peak was further identified as glycyrrhizin (Fig. 3d,e) by comparing the retention time of 4.48 min, accurate molecular ion at m/z 821.39865, and characteristic fragment ions (m/z 645.45285, m/z 469.34904 and m/z 351.03306) with an authentic reference (Fig. 3c). Analysis of the MS fragmentation pathway further verified its chemical structure (Fig. S13). Other UDP sugars, such as UDP‐glucose and UDP‐galactose, were also evaluated for comparison, indicating that GuUGAT is generally specific for the sugar moiety of UDP‐GlcA (Fig. S14). Interestingly, almost no glucopyranosiduronic acid or other byproducts were detected in the catalytic reaction of GuUGAT (Fig. 3d). These results suggest that GuUGAT may synthesize glycyrrhizin continuously via a two‐step glucuronosylation reaction with few by‐products.
Figure 3.
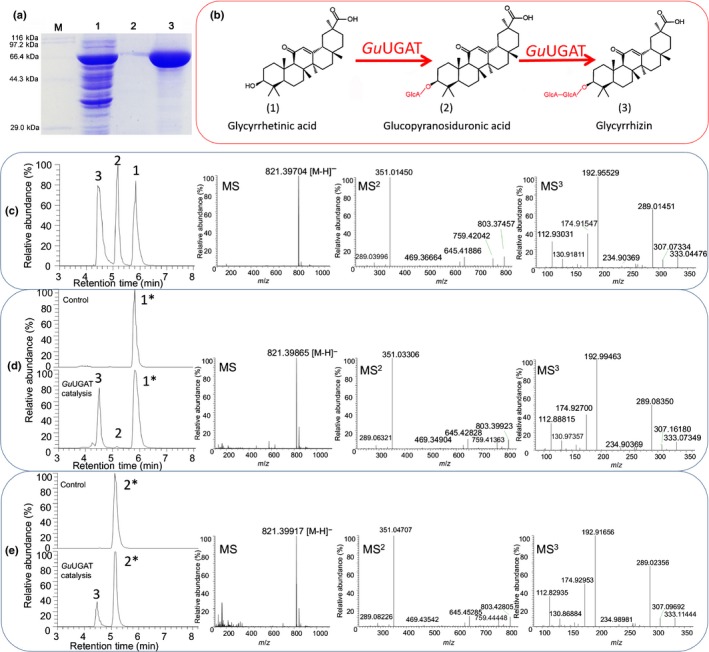
In vitro enzyme assays for determining activity of Glycyrrhiza uralensis UDP‐dependent glucuronosyltransferase (GuUGAT). (a) Sodium dodecyl sulfate−polyacrylamide gel electrophoresis (SDS‐PAGE) electropherogram of proteins expressed in Escherichia coli. M, standard protein markers; 1, crude protein after isopropyl 1‐β‐D‐thiogalactoside (IPTG) induction; 2, purified GuUGAT protein; 3, concentrated GuUGAT protein. (b) Continuous two‐step glycosylation reaction catalysed by GuUGAT. (c) Chemical reference substance determined by high performance liquid chromatography‐linear ion trap‐orbitrap mass spectrometry. 1, glycyrrhetinic acid (retention time: 5.84 min); 2, glucopyranosiduronic acid (retention time: 5.18 min); 3, glycyrrhizin (retention time: 4.46 min); MS fragments of glycyrrhizin are shown subsequently. (d, e) For each enzyme assay, (d) 50 μM glycyrrhetinic acid or (e) 50 μM glucopyranosiduronic acid was used as a substrate. Extracted chromatographic peaks for the substrates used are highlighted with asterisks (*). Enzymatic products are as indicated: 1, glycyrrhetinic acid (retention time: 5.83 min); 2, glucopyranosiduronic acid (retention time: 5.18 min); 3, glycyrrhizin (retention time: 4.48 min). MS fragments of peak 3 are shown subsequently.
Enzymes with the same function are believed to be temporally and spatially coexpressed across different conditions (Jorgensen et al., 2005), and bAS is regarded as a representative gene reflecting the expression profile of the glycyrrhizin biosynthesis pathway (Seki et al., 2008, 2011). Tissue expression analysis revealed that GuUGAT was generally expressed in the leaves, stems and roots and highly expressed in the roots under NaCl or drought stress (Fig. S15). To further demonstrate the key role of GuUGAT in the biosynthesis of glycyrrhizin, we analysed the change in the expression level of GuUGAT under drought and salt stress. Fig. 2 shows that GuUGAT was up‐regulated significantly under salt stress in particular and coexpressed with key genes involved in the biosynthesis of glycyrrhizin, including farnesyl pyrophosphate synthase (FPS), bAS and CYP88D6 (Notes S3). This result implies that GuUGAT may participate in the glycyrrhizin biosynthesis pathway, together with the bAS and CYP88D6 genes. Moreover, the real‐time qPCR of GuUGAT in vivo revealed that the expression profiles of GuUGAT and bAS are very similar across different conditions (Fig. 4). The content of glycyrrhizin in the roots increased when the expression level of the GuUGAT gene increased (Fig. S16), further confirming the key role of GuUGAT in the biosynthesis of glycyrrhizin.
Figure 4.
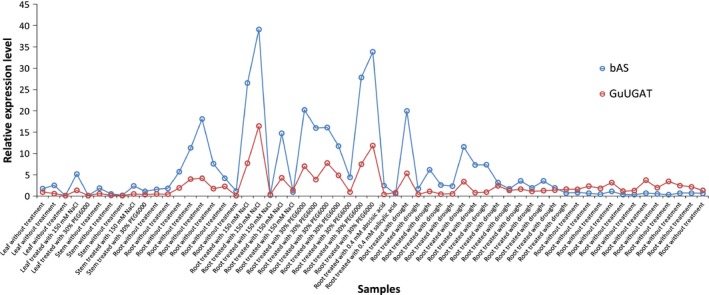
Real‐time quantitative PCR of the β‐amyrin synthetase (bAS) and UDP‐dependent glucuronosyltransferase (GuUGAT) genes from Glycyrrhiza uralensis.
Structural and biochemical characterization of the GuUGAT protein
Structures of UGTs are generally divided into two different groups: GT‐A and GT‐B. The structures of most plant UGTs contain a GT‐B fold with a highly conserved consensus signature sequence called the PSPG motif (Shao et al., 2005). The C terminus of plant UGTs is mainly involved in contact with the glycosyl donor, and the glycosyl acceptor primarily interacts with the N terminus. The carbohydrate‐active enzyme (CAZy) database provides a rich set of manually annotated UGTs that degrade, modify, or create glycosidic bonds. Using the CAZymes Analysis Toolkit in the CAZy database (Park et al., 2010; Lombard et al., 2014), GuUGAT was determined to have a GT‐B fold in inverted glycosylation mode.
To evaluate the affinity and catalytic efficiencies of GuUGAT, the kinetic parameters were investigated with glycyrrhetinic acid and glucopyranosiduronic acid as acceptors. It was found that the catalytic rate constant (Kcat) and Michaelis constant (Km) values of glycyrrhetinic acid were 2.85 s−1 and 36.2 μM, respectively, and the K cat and K m values of glucopyranosiduronic acid were 0.12 s−1 and 4.3 μM, respectively (Fig. S17). The K cat and K m values of GuUGAT are generally consistent with those of characterized triterpenoid UGTs, such as UGT73C11, UGT73C13, PgUGT74AE2 and PgUGT94Q2 (Augustin et al., 2012; Jung et al., 2014).
Phylogenetic analysis of the GuUGAT protein
GuUGAT was genetically and biochemically determined to encode a previously unidentified glycyrrhetinic acid glucuronosyltransferase. Phylogenetic analysis of different characterized UGTs indicated that GuUGAT was clustered in a group of UGT73 family members, including flavonoid glycosyltransferases, terpene glycosyltransferases, zeatin glycosyltransferases and phenylpropanoid glycosyltransferases (Fig. 5). This result implies that GuUGAT belongs to the UGT73 family. More importantly, in contrast to other triterpenoid UGTs (Fig. S18; Notes S6), GuUGAT was independently classified into a new subcluster that was closely related to flavonoid and phenylpropanoid glycosyltransferases. Phylogenetic analysis of the characterized triterpenoid UGTs also indicated that GuUGAT is in a new subclade within the UGT73 family (Fig. S19; Notes S7). These results suggest that GuUGAT may be a new representative of this subcluster.
Figure 5.

Phylogenetic tree of characterized plant glycosyltransferases. Only the key subclusters from Supporting Information Fig. S18 are shown. Amino acid sequences of these glycosyltransferases were aligned with clustalw using mega 6.0. The output was used to create a phylogenetic tree using mega 6.0 and the neighbour‐joining method, with Poisson correction as the model. The bootstrap confidence values were obtained based on 1000 replicates. The GuUGAT from Glycyrrhiza uralensis is highlighted with red text.
Site‐directed mutagenesis of GuUGAT proteins
GuUGAT, which catalyses a continuous two‐step glucuronosylation reaction, exhibits a different catalytic function from that of characterized triterpenoid UGTs. Aligned with characterized triterpenoid UGT protein sequences, all of the sequences showed conserved PSPG motifs, and 22 sites were highly conserved among them (Notes S8). To identify the crucial amino acids that are involved in the continuous glucuronosylation reaction, we performed homology modelling and optimized the 3D structure of GuUGAT (Fig. S20) according to the MtUGT71G1 crystal structure (Protein Data Bank ID code 2ACW) in view of their high protein sequence similarity and closely related biological functions (Shao et al., 2005), and we predicted key catalytic sites by molecular docking. Structural comparisons predicted that a few potential key sites, including the previously characterized important sites His‐22, Asp‐121 and Glu‐391 (Shao et al., 2005), may form hydrogen bonds with UDP‐glucuronic acid, glycyrrhetinic acid, and glucopyranosiduronic acid (Fig. 6a). These potential key sites probably contribute to the two‐step glucuronosylation reaction.
Figure 6.
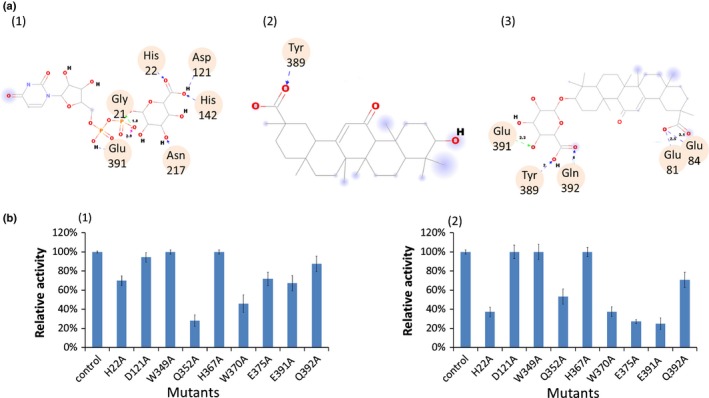
Molecular docking and mutagenesis assay of Glycyrrhiza uralensis UDP‐dependent glucuronosyltransferase (GuUGAT). (a) Molecular docking with UDP‐glucuronic acid (1), glycyrrhetinic acid (2) and glucopyranosiduronic acid (3). Hydrogen bonds are labelled with dashed arrows, and amino acid residues interacting with UDP‐glucuronic acid, glycyrrhetinic acid or glucopyranosiduronic acid via hydrogen bonding are highlighted with circles. (b) Relative catalytic activity of GuUGAT mutants when glycyrrhetinic acid (1) or glucopyranosiduronic acid (2) was used as a substrate. Error bars used in the figure indicate ± SDs.
To preliminarily determine the biochemical impact of these amino acids, we targeted a total of nine key sites for mutagenesis based on protein sequence conservation, mutagenesis analysis of other UGTs in previous studies (Lu et al., 2014), and predictions obtained from molecular docking simulations. Site‐directed mutagenesis and enzyme assays demonstrated that the Q352A mutation led to a c. 70% decrease in GuUGAT activity towards glycyrrhetinic acid, and the H22A, W370A, E375A and Q392A mutations resulted in a c. 60–70% decrease in GuUGAT activity towards glucopyranosiduronic acid (Fig. 6b). Our results suggest that Gln‐352 may contribute to the first step of glucuronosylation, and His‐22, Trp‐370, Glu‐375 and Gln‐391 may be important catalytic residues in the second step of glucuronosylation.
Discussion
Deep transcriptome sequencing for mining superfamily enzymes
UGTs, similar to cytochrome P450 monooxygenases (Chakrabarti et al., 2007; Li et al., 2013a; Weis et al., 2014; Guo et al., 2015), constitute one of the most important and prevalent superfamilies in plants. Glycosylation can greatly change the bioactivity, solubility or stability of metabolites (Kren & Martinkova, 2001) and confer a protective or adaptive function to plants (Tava & Odoardi, 1996; Osbourn, 2003). Plant‐derived glycosylated natural products also have several attractive characteristics and have a promising role in new drug development; thus, natural plant product biosynthesis and modification has become an important area of research in recent years (Liang et al., 2015). The characterization of specialized plant triterpenoid UGTs is quite difficult because of their rich diversity (Seki et al., 2015). The availability of inexpensive high‐throughput technologies has facilitated the identification and analysis of plant triterpenoid UGTs (Achnine et al., 2005; Naoumkina et al., 2010; Ruff et al., 2012). As a result, a few UGTs that are involved in triterpenoid saponin biosynthesis have been identified. One of the most noteworthy UGTs is UGT71A27 of Panax ginseng, which was successfully used to synthesize ginsenoside K, a bioactive compound used for treating arthritis (Yan et al., 2014). However, only 13 triterpenoid UGTs have been characterized thus far (Seki et al., 2015), and characterization of additional UGTs will a major challenge for future studies (Loos & Steinkellner, 2014).
Transcriptome sequencing is useful for the identification of functional genes that are involved in secondary metabolism, especially for plants without a sequenced genome. However, false‐positive or false‐negative errors in unigene expression profiles throughout HiSeq platforms can hinder further analysis. Here, we provide an effective deep transcriptome sequencing approach to increase transcript coverage, thereby decreasing false‐positive and false‐negative errors. DE and CE analyses in this work also demonstrated effectiveness in mining specialized GuUGATs. Thus, the deep transcriptome sequencing described in this work, which can be used to examine the expression profiles of unigenes in depth, may be a useful reference for the further characterization of UGTs or other superfamily enzymes.
Elucidation of the complete biosynthetic pathway of glycyrrhizin
Elucidation of the biosynthetic pathway of bioactive natural plant products is crucial for the future genetic and metabolic engineering of plants. The biosynthetic pathway of glycyrrhizin involves > 20 enzymes. The upstream biosynthetic pathway of glycyrrhizin, similar to that of other triterpenoid saponins, includes mevalonate (MVA) and plastid‐localized 2‐C‐methyl‐d‐erythritol 4‐phosphate (MEP) pathways, which provide important isopentenyl diphosphates (IPPs). Then, IPPs and dimethylallyl diphosphates (DMAPPs) are catalysed by FPS, squalene synthase (SQS), squalene monooxygenase or epoxidase (SQE) and bAS to yield β‐amyrin. Various P450s and UGTs are involved in the downstream biosynthetic pathway of glycyrrhizin (Fig. 1). To elucidate the downstream biosynthetic pathway of glycyrrhizin, Seki et al. successfully cloned beta‐amyrin 11‐oxidase (CYP88D6) in 2008 and demonstrated that it is the cytochrome P450 involved in the biosynthesis of glycyrrhizin (Seki et al., 2008). More recently, CYP72A154, another cytochrome P450 monooxygenase, was also found to be capable of producing glycyrrhetinic acid (Seki et al., 2011). UGATs are crucial UGTs for the completion of the last glucuronosylation reaction, which may greatly improve the sweetness and solubility of glycyrrhetinic acid. Mining of UGATs that are involved in glycyrrhizin biosynthesis has been performed ever since cytochrome P450 monooxygenases were cloned in 2011 (Seki et al., 2011); however, these UGATs still remain unknown. Here, we systematically screened candidate UGTs from 434 putative UGTs through deep transcriptome sequencing, and we successfully identified one unique GuUGAT that catalyses the glucuronosylation of the C‐3 hydroxyl group of glycyrrhetinic acid, thus uncovering the complete pathway of glycyrrhizin biosynthesis. Unexpectedly, this UGAT produced glycyrrhizin directly via two‐step continuous glucuronosylation (Fig. 3). In fact, glycosylation of bioactive triterpenoids and other natural plant products was generally believed to occur through the addition of only one sugar moiety by specialized UGTs (Thimmappa et al., 2014; Liang et al., 2015; Seki et al., 2015), although this was not the case for a few UGTs of Streptomyces species (Luzhetskyy et al., 2005; Li et al., 2013b). The GuUGAT that we identified in this study can catalyse two‐step glucuronosylation reactions continuously and is therefore a novel UGT of bioactive natural plant products. Furthermore, previously reported plant triterpenoid UGTs can only use four UDP sugars as glycosyl donors, namely UDP‐galactose, UDP‐glucose, UDP‐rhamnose and UDP‐xylose; however, the glycosyl donor used by UDP‐glucuronic acid (GlcA) remains unclear (Table S1) (Seki et al., 2015). GuUGAT is also the first plant triterpenoid UGT that can use UDP‐GlcA as a glycosyl donor. Phylogenetic analysis of characterized UGTs also indicated that GuUGAT is in a new subclade in the plant UGT73 family (Fig. 5), further demonstrating the novelty of GuUGAT in the plant UGT73 family. The discovery of GuUGAT in this work helps to elucidate the complete pathway of glycyrrhizin biosynthesis, providing new insight into the function of glycosyltransferases. Additionally, GuUGAT may be useful for the modification of important natural products.
Complete biosynthesis of glycyrrhizin in the future
Glycyrrhizin, the catalytic product of GuUGAT, is an important bioactive natural product with high economic value. This compound is mainly derived from the belowground portion of the Glycyrrhiza plant. Glycyrrhizin is an important low‐calorie sweetener that is available world‐wide and is usually used as a flavouring agent in food, beverages and confectioneries; its sweetness is c. 150 times greater than that of sucrose (Kitagawa, 2002; Seki et al., 2011). Glycyrrhizin also has useful pharmacological roles, including anti‐cancer, antivirus and anti‐inflammation activities (Baba & Shigeta, 1987; Ito et al., 1987, 1988; Cinatl et al., 2003; Fiore et al., 2008; Wolkerstorfer et al., 2009). Glycyrrhizin preparations for clinical treatment of chronic hepatitis have been widely used in Asian countries, and sales of these preparations have reached approximately CNY ¥2 billion yr−1 (van Rossum et al., 1998; Shibata, 2000). In addition, glycyrrhizin is now being used as a cosmetic ingredient as a consequence of its beneficial effect on skin (Hayashi & Sudo, 2009). Consequently, the market demand for glycyrrhizin has increased in recent years as a result of its useful properties (Hayashi & Sudo, 2009). Nevertheless, the acquisition of glycyrrhizin depends wholly on artificial extraction from roots of Glycyrrhiza plants, especially from wild G. uralensis, which greatly accelerates the consumption of natural resources and may damage the ecosystem. The complete biosynthesis of important natural products using synthetic biological techniques has been carried out in recent years, with the goals of generating new chemicals, improving human health, and addressing environmental issues (Way et al., 2014; Galanie et al., 2015; Winzer et al., 2015). GuUGAT, which can catalyse the formation of glycyrrhizin via glycyrrhetinic acid with few by‐products, may be suitable for genetic modification, thereby allowing the complete biosynthesis of glycyrrhizin and its synthesis using synthetic biology. GuUGAT and similar genes are likely to have important roles in future industrial applications involving genetic and metabolic engineering.
Author contributions
G.X., C.L. and W.G. planned and designed the research. G.X. performed experiments, conducted fieldwork and wrote the manuscript. W.C. analysed data.
Supporting information
Please note: Wiley Blackwell are not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Quality assessment of total RNA used for deep transcriptome sequencing.
Fig. S2 Whole distribution of annotated unigenes.
Fig. S3 Classification of annotated unigenes by gene function.
Fig. S4 KOG classification of annotated unigenes.
Fig. S5 KEGG classification of annotated unigenes.
Fig. S6 Gene expression distribution among three RNA‐seq samples.
Fig. S7 Pearson correlation analysis among samples.
Fig. S8 Volcano plots of differentially expressed genes.
Fig. S9 Detailed hierarchical clustering and corresponding heat maps of the unigenes across all of the pairwise library comparisons.
Fig. S10 Expression levels of eight candidate UGTs, bAS and two CYPs.
Fig. S11 Phylogenetic tree of eight candidate UGTs and characterized plant triterpenoid UGTs.
Fig. S12 Protein sequencing of GuUGAT.
Fig. S13 Analysis of the MS fragmentation pathway of the catalytic product.
Fig. S14 Catalytic specificity of GuUGAT towards UDP sugars.
Fig. S15 Expression pattern of GuUGAT based on real‐time PCR.
Fig. S16 Expression level of the GuUGAT gene and content of glycyrrhizin in roots under stress conditions.
Fig. S17 Determination of kinetic parameters for recombinant GuUGAT.
Fig. S18 Phylogenetic tree of characterized plant glycosyltransferases.
Fig. S19 Phylogenetic tree of characterized plant triterpenoid UGTs.
Fig. S20 Optimized 3D structure of GuUGAT for molecular docking.
Table S1 Characterized triterpenoid glycosyltransferases collected in the GenBank database
Table S2 Primers that were used in this study
Table S3 Overall quality assessment of raw data from RNA‐seq
Table S4 Overall transcript length of assembled data from RNA‐seq
Table S5 Length distribution of assembled data from RNA‐seq
Table S6 Data sets for the construction of a genus Glycyrrhiza cDNA database
Table S7 Number of unigenes annotated in databases
Notes S1 The 434 putative UGTs that were annotated in this study.
Notes S2 Differential expression analysis results.
Notes S3 Annotation of unigenes in the targeted subcluster shown in Fig. S9.
Notes S4 Eight screened candidate UGATs.
Notes S5 Alignment of the protein sequences of the eight screened candidate UGATs.
Notes S6 Amino acid sequences of the UGTs that were used for phylogenetic analysis as shown in Fig. 5.
Notes S7 Amino acid sequences of triterpenoid UGTs that were used for phylogenetic analysis as shown in Fig. S12.
Notes S8 Alignment of the protein sequences of plant triterpenoid UGTs.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (grant nos. 81373909 and 81422053). The authors also acknowledge Fei Li, Huihui Duan, Xing Wang, Xue Zhang and Chunguo Wang for supporting this project.
References
- Achnine L, Huhman DV, Farag MA, Sumner LW, Blount JW, Dixon RA. 2005. Genomics‐based selection and functional characterization of triterpene glycosyltransferases from the model legume Medicago truncatula . Plant Journal 41: 875–887. [DOI] [PubMed] [Google Scholar]
- Augustin JM, Drok S, Shinoda T, Sanmiya K, Nielsen JK, Khakimov B, Olsen CE, Hansen EH, Kuzina V, Ekstrom CT et al 2012. UDP‐glycosyltransferases from the UGT73C subfamily in Barbarea vulgaris catalyze sapogenin 3‐O‐glucosylation in saponin‐mediated insect resistance. Plant Physiology 160: 1881–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba M, Shigeta S. 1987. Antiviral activity of glycyrrhizin against varicella‐zoster virus in vitro . Antiviral Research 7: 99–107. [DOI] [PubMed] [Google Scholar]
- Benchaar C, Chouinard PY. 2009. Short communication: assessment of the potential of cinnamaldehyde, condensed tannins, and saponins to modify milk fatty acid composition of dairy cows. Journal of Dairy Science 92: 3392–3396. [DOI] [PubMed] [Google Scholar]
- Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein‐dye binding. Analytical Biochemistry 72: 248–254. [DOI] [PubMed] [Google Scholar]
- Chakrabarti M, Meekins KM, Gavilano LB, Siminszky B. 2007. Inactivation of the cytochrome P450 gene CYP82E2 by degenerative mutations was a key event in the evolution of the alkaloid profile of modern tobacco. New Phytologist 175: 565–574. [DOI] [PubMed] [Google Scholar]
- Chan PK. 2007. Acylation with diangeloyl groups at C21‐22 positions in triterpenoid saponins is essential for cytotoxicity towards tumor cells. Biochemical Pharmacology 73: 341–350. [DOI] [PubMed] [Google Scholar]
- Cinatl J, Morgenstern B, Bauer G, Chandra P, Rabenau H, Doerr HW. 2003. Glycyrrhizin, an active component of liquorice roots, and replication of SARS‐associated coronavirus. Lancet 361: 2045–2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bruyn F, Maertens J, Beauprez J, Soetaert W, De Mey M. 2015. Biotechnological advances in UDP‐sugar based glycosylation of small molecules. Biotechnology Advances 33: 288–302. [DOI] [PubMed] [Google Scholar]
- Eswar N, Webb B, Marti‐Renom MA, Madhusudhan MS, Eramian D, Shen MY, Pieper U, Sali A. 2007. Comparative protein structure modeling using MODELLER. Current Protocols in Protein Science Chapter 2: Unit 2.9. [DOI] [PubMed] [Google Scholar]
- Fiore C, Eisenhut M, Krausse R, Ragazzi E, Pellati D, Armanini D, Bielenberg J. 2008. Antiviral effects of Glycyrrhiza species. Phytotherapy Research 22: 141–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galanie S, Thodey K, Trenchard IJ, Filsinger Interrante M, Smolke CD. 2015. Complete biosynthesis of opioids in yeast. Science 349: 1095–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q et al 2011. Full‐length transcriptome assembly from RNA‐Seq data without a reference genome. Nature Biotechnology 29: 644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Ma X, Cai Y, Ma Y, Zhan Z, Zhou YJ, Liu W, Guan M, Yang J, Cui G et al 2015. Cytochrome P450 promiscuity leads to a bifurcating biosynthetic pathway for tanshinones. New Phytologist 210: 525–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi H, Sudo H. 2009. Economic importance of licorice. Plant Biotechnology 26: 101–104. [Google Scholar]
- Ito M, Nakashima H, Baba M, Pauwels R, De Clercq E, Shigeta S, Yamamoto N. 1987. Inhibitory effect of glycyrrhizin on the in vitro infectivity and cytopathic activity of the human immunodeficiency virus [HIV (HTLV‐III/LAV)]. Antiviral Research 7: 127–137. [DOI] [PubMed] [Google Scholar]
- Ito M, Sato A, Hirabayashi K, Tanabe F, Shigeta S, Baba M, De Clercq E, Nakashima H, Yamamoto N. 1988. Mechanism of inhibitory effect of glycyrrhizin on replication of human immunodeficiency virus (HIV). Antiviral Research 10: 289–298. [DOI] [PubMed] [Google Scholar]
- Jeong HG, You HJ, Park SJ, Moon AR, Chung YC, Kang SK, Chun HK. 2002. Hepatoprotective effects of 18beta‐glycyrrhetinic acid on carbon tetrachloride‐induced liver injury: inhibition of cytochrome P450 2E1 expression. Pharmacological Research 46: 221–227. [DOI] [PubMed] [Google Scholar]
- Jorgensen K, Rasmussen AV, Morant M, Nielsen AH, Bjarnholt N, Zagrobelny M, Bak S, Moller BL. 2005. Metabolon formation and metabolic channeling in the biosynthesis of plant natural products. Current Opinion in Plant Biology 8: 280–291. [DOI] [PubMed] [Google Scholar]
- Jung SC, Kim W, Park SC, Jeong J, Park MK, Lim S, Lee Y, Im WT, Lee JH, Choi G et al 2014. Two ginseng UDP‐glycosyltransferases synthesize ginsenoside Rg3 and Rd. Plant and Cell Physiology 55: 2177–2188. [DOI] [PubMed] [Google Scholar]
- Kitagawa I. 2002. Licorice root. A natural sweetener and an important ingredient in Chinese medicine. Pure & Applied Chemistry 74: 1189–1198. [Google Scholar]
- Kojoma M, Ohyama K, Seki H, Hiraoka Y, Asazu SN, Sawa S, Sekizaki H, Yoshida S, Muranaka T. 2010. In vitro proliferation and triterpenoid characteristics of licorice (Glycyrrhiza uralensis Fischer, Leguminosae) stolons. Plant Biotechnology 27: 59–66. [Google Scholar]
- Kren V, Martinkova L. 2001. Glycosides in medicine: “The role of glycosidic residue in biological activity”. Current Medicinal Chemistry 8: 1303–1328. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685. [DOI] [PubMed] [Google Scholar]
- Lee SH, Kim SY, Kim DW, Jang SH, Lim SS, Kwon HJ, Kang TC, Won MH, Kang IJ, Lee KS et al 2008. Active component of Fatsia japonica enhances the transduction efficiency of Tat‐SOD fusion protein both in vitro and in vivo . Journal of Microbiology and Biotechnology 18: 1613–1619. [PubMed] [Google Scholar]
- Li B, Dewey CN. 2011. RSEM: accurate transcript quantification from RNA‐Seq data with or without a reference genome. BMC Bioinformatics 12: 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Jiang L, Youn JH, Sun W, Cheng Z, Jin T, Ma X, Guo X, Wang J, Zhang X et al 2013a. A comprehensive genetic study reveals a crucial role of CYP90D2/D2 in regulating plant architecture in rice (Oryza sativa). New Phytologist 200: 1076–1088. [DOI] [PubMed] [Google Scholar]
- Li S, Xiao J, Zhu Y, Zhang G, Yang C, Zhang H, Ma L, Zhang C. 2013b. Dissecting glycosylation steps in lobophorin biosynthesis implies an iterative glycosyltransferase. Organic Letters 15: 1374–1377. [DOI] [PubMed] [Google Scholar]
- Liang DM, Liu JH, Wu H, Wang BB, Zhu HJ, Qiao JJ. 2015. Glycosyltransferases: mechanisms and applications in natural product development. Chemical Society Reviews 44: 8350–8374. [DOI] [PubMed] [Google Scholar]
- Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. 2014. The carbohydrate‐active enzymes database (CAZy) in 2013. Nucleic Acids Research 42: D490–D495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loos A, Steinkellner H. 2014. Plant glyco‐biotechnology on the way to synthetic biology. Frontiers in Plant Science 5: 523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Xue F, Liu C, Yang M, Ma L. 2014. Crystal structures of plant uridine diphosphate‐dependent glycosyltransferases. Sheng Wu Gong Cheng Xue Bao 30: 838–847. [PubMed] [Google Scholar]
- Luzhetskyy A, Fedoryshyn M, Durr C, Taguchi T, Novikov V, Bechthold A. 2005. Iteratively acting glycosyltransferases involved in the hexasaccharide biosynthesis of landomycin A. Chemistry & Biology 12: 725–729. [DOI] [PubMed] [Google Scholar]
- Maes L, Vanden Berghe D, Germonprez N, Quirijnen L, Cos P, De Kimpe N, Van Puyvelde L. 2004. In vitro and in vivo activities of a triterpenoid saponin extract (PX‐6518) from the plant Maesa balansae against visceral leishmania species. Antimicrobial Agents and Chemotherapy 48: 130–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui S, Matsumoto H, Sonoda Y, Ando K, Aizu‐Yokota E, Sato T, Kasahara T. 2004. Glycyrrhizin and related compounds down‐regulate production of inflammatory chemokines IL‐8 and eotaxin 1 in a human lung fibroblast cell line. International Immunopharmacology 4: 1633–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naoumkina MA, Modolo LV, Huhman DV, Urbanczyk‐Wochniak E, Tang Y, Sumner LW, Dixon RA. 2010. Genomic and coexpression analyses predict multiple genes involved in triterpene saponin biosynthesis in Medicago truncatula . Plant Cell 22: 850–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasrollahi V, Mirzaie‐asl A, Piri K, Nazeri S, Mehrabi R. 2014. The effect of drought stress on expression of key genes involved in biosynthesis of triterpenoid saponins in licorice (Glycyrrhiza glabra). Phytochemistry 103: 32–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osbourn AE. 2003. Saponins in cereals. Phytochemistry 62: 1–4. [DOI] [PubMed] [Google Scholar]
- Pan Y, Wu LJ, Yu ZL. 2006. Effect of salt and drought stress on antioxidant enzymes activities and SOD isoenzymes of liquorice (Glycyrrhiza uralensis Fisch). Plant Growth Regulation 49: 157–165. [Google Scholar]
- Park BH, Karpinets TV, Syed MH, Leuze MR, Uberbacher EC. 2010. CAZymes Analysis Toolkit (CAT): web service for searching and analyzing carbohydrate‐active enzymes in a newly sequenced organism using CAZy database. Glycobiology 20: 1574–1584. [DOI] [PubMed] [Google Scholar]
- Ramilowski JA, Sawai S, Seki H, Mochida K, Yoshida T, Sakurai T, Muranaka T, Saito K, Daub CO. 2013. Glycyrrhiza uralensis transcriptome landscape and study of phytochemicals. Plant and Cell Physiology 54: 697–710. [DOI] [PubMed] [Google Scholar]
- van Rossum TG, Vulto AG, de Man RA, Brouwer JT, Schalm SW. 1998. Review article: glycyrrhizin as a potential treatment for chronic hepatitis C. Alimentary Pharmacology & Therapeutics 12: 199–205. [DOI] [PubMed] [Google Scholar]
- Ruff AJ, Dennig A, Wirtz G, Blanusa M, Schwaneberg U. 2012. Flow cytometer‐based high‐throughput screening system for accelerated directed evolution of P450 monooxygenases. Acs Catalysis 2: 2724–2728. [Google Scholar]
- Seki H, Ohyama K, Sawai S, Mizutani M, Ohnishi T, Sudo H, Akashi T, Aoki T, Saito K, Muranaka T. 2008. Licorice beta‐amyrin 11‐oxidase, a cytochrome P450 with a key role in the biosynthesis of the triterpene sweetener glycyrrhizin. Proceedings of the National Academy of Sciences, USA 105: 14204–14209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki H, Sawai S, Ohyama K, Mizutani M, Ohnishi T, Sudo H, Fukushima EO, Akashi T, Aoki T, Saito K et al 2011. triterpene functional genomics in licorice for identification of CYP72A154 involved in the biosynthesis of glycyrrhizin. Plant Cell 23: 4112–4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki H, Tamura K, Muranaka T. 2015. P450s and UGTs: key players in the structural diversity of triterpenoid saponins. Plant and Cell Physiology 56: 1463–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao H, He X, Achnine L, Blount JW, Dixon RA, Wang X. 2005. Crystal structures of a multifunctional triterpene/flavonoid glycosyltransferase from Medicago truncatula . Plant Cell 17: 3141–3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z, Liu C, Wang X. 2009. Cloning and characterization of open reading frame encoding beta‐amyrin synthase in Glycyrrhiza uralensis . Zhongguo Zhong Yao Za Zhi 34: 2438–2440. [PubMed] [Google Scholar]
- Shibata S. 2000. A drug over the millennia: pharmacognosy, chemistry, and pharmacology of licorice. Yakugaku Zasshi 120: 849–862. [DOI] [PubMed] [Google Scholar]
- Sparg SG, Light ME, van Staden J. 2004. Biological activities and distribution of plant saponins. Journal of Ethnopharmacology 94: 219–243. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Molecular Biology and Evolution 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Li W, Cao J, Zhao Y. 2015. Bioassay‐guided isolation and identification of cytotoxic compounds from Bolbostemma paniculatum . Journal of Ethnopharmacology 169: 18–23. [DOI] [PubMed] [Google Scholar]
- Tava A, Odoardi M. 1996. Saponins from Medicago Spp.: chemical characterization and biological activity against insects. Advances in Experimental Medicine and Biology 405: 97–109. [DOI] [PubMed] [Google Scholar]
- Thimmappa R, Geisler K, Louveau T, O'Maille P, Osbourn A. 2014. Triterpene biosynthesis in plants. Annual Review of Plant Biology 65: 225–257. [DOI] [PubMed] [Google Scholar]
- Uematsu Y, Hirata K, Saito K, Kudo I. 2000. Spectrophotometric determination of saponin in Yucca extract used as food additive. Journal of AOAC International 83: 1451–1454. [PubMed] [Google Scholar]
- Way JC, Collins JJ, Keasling JD, Silver PA. 2014. Integrating biological redesign: where synthetic biology came from and where it needs to go. Cell 157: 151–161. [DOI] [PubMed] [Google Scholar]
- Weis C, Hildebrandt U, Hoffmann T, Hemetsberger C, Pfeilmeier S, Konig C, Schwab W, Eichmann R, Huckelhoven R. 2014. CYP83A1 is required for metabolic compatibility of Arabidopsis with the adapted powdery mildew fungus Erysiphe cruciferarum . New Phytologist 202: 1310–1319. [DOI] [PubMed] [Google Scholar]
- Winzer T, Kern M, King AJ, Larson TR, Teodor RI, Donninger SL, Li Y, Dowle AA, Cartwright J, Bates R et al 2015. Plant science. Morphinan biosynthesis in opium poppy requires a P450‐oxidoreductase fusion protein. Science 349: 309–312. [DOI] [PubMed] [Google Scholar]
- Wolkerstorfer A, Kurz H, Bachhofner N, Szolar OH. 2009. Glycyrrhizin inhibits influenza A virus uptake into the cell. Antiviral Research 83: 171–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Liberatore KL, MacAlister CA, Huang Z, Chu YH, Jiang K, Brooks C, Ogawa‐Ohnishi M, Xiong G, Pauly M et al 2015. A cascade of arabinosyltransferases controls shoot meristem size in tomato. Nature Genetics 47: 784–792. [DOI] [PubMed] [Google Scholar]
- Yan X, Fan Y, Wei W, Wang P, Liu Q, Wei Y, Zhang L, Zhao G, Yue J, Zhou Z. 2014. Production of bioactive ginsenoside compound K in metabolically engineered yeast. Cell Research 24: 770–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekura‐Sakakibara K, Hanada K. 2011. An evolutionary view of functional diversity in family 1 glycosyltransferases. Plant Journal 66: 182–193. [DOI] [PubMed] [Google Scholar]
- Yonekura‐Sakakibara K, Tohge T, Matsuda F, Nakabayashi R, Takayama H, Niida R, Watanabe‐Takahashi A, Inoue E, Saito K. 2008. Comprehensive flavonol profiling and transcriptome coexpression analysis leading to decoding gene‐metabolite correlations in Arabidopsis. Plant Cell 20: 2160–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekura‐Sakakibara K, Tohge T, Niida R, Saito K. 2007. Identification of a flavonol 7‐O‐rhamnosyltransferase gene determining flavonoid pattern in Arabidopsis by transcriptome coexpression analysis and reverse genetics. Journal of Biological Chemistry 282: 14932–14941. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: Wiley Blackwell are not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Quality assessment of total RNA used for deep transcriptome sequencing.
Fig. S2 Whole distribution of annotated unigenes.
Fig. S3 Classification of annotated unigenes by gene function.
Fig. S4 KOG classification of annotated unigenes.
Fig. S5 KEGG classification of annotated unigenes.
Fig. S6 Gene expression distribution among three RNA‐seq samples.
Fig. S7 Pearson correlation analysis among samples.
Fig. S8 Volcano plots of differentially expressed genes.
Fig. S9 Detailed hierarchical clustering and corresponding heat maps of the unigenes across all of the pairwise library comparisons.
Fig. S10 Expression levels of eight candidate UGTs, bAS and two CYPs.
Fig. S11 Phylogenetic tree of eight candidate UGTs and characterized plant triterpenoid UGTs.
Fig. S12 Protein sequencing of GuUGAT.
Fig. S13 Analysis of the MS fragmentation pathway of the catalytic product.
Fig. S14 Catalytic specificity of GuUGAT towards UDP sugars.
Fig. S15 Expression pattern of GuUGAT based on real‐time PCR.
Fig. S16 Expression level of the GuUGAT gene and content of glycyrrhizin in roots under stress conditions.
Fig. S17 Determination of kinetic parameters for recombinant GuUGAT.
Fig. S18 Phylogenetic tree of characterized plant glycosyltransferases.
Fig. S19 Phylogenetic tree of characterized plant triterpenoid UGTs.
Fig. S20 Optimized 3D structure of GuUGAT for molecular docking.
Table S1 Characterized triterpenoid glycosyltransferases collected in the GenBank database
Table S2 Primers that were used in this study
Table S3 Overall quality assessment of raw data from RNA‐seq
Table S4 Overall transcript length of assembled data from RNA‐seq
Table S5 Length distribution of assembled data from RNA‐seq
Table S6 Data sets for the construction of a genus Glycyrrhiza cDNA database
Table S7 Number of unigenes annotated in databases
Notes S1 The 434 putative UGTs that were annotated in this study.
Notes S2 Differential expression analysis results.
Notes S3 Annotation of unigenes in the targeted subcluster shown in Fig. S9.
Notes S4 Eight screened candidate UGATs.
Notes S5 Alignment of the protein sequences of the eight screened candidate UGATs.
Notes S6 Amino acid sequences of the UGTs that were used for phylogenetic analysis as shown in Fig. 5.
Notes S7 Amino acid sequences of triterpenoid UGTs that were used for phylogenetic analysis as shown in Fig. S12.
Notes S8 Alignment of the protein sequences of plant triterpenoid UGTs.


