Presence of SARS coronavirus in macrophages
We read with great interest the excellent article by To et al, which was published in the February issue of The Journal of Pathology 1. Using in situ hybridization (ISH), To et al demonstrated the presence of severe acute respiratory syndrome‐associated coronavirus (SARS‐CoV) in pneumocytes and in the surface enterocytes of the small intestine. Their findings are compatible with those of another study conducted in Taiwan 2. This observation indicates the cellular tropism of the infectious agent and thus has an impact on our understanding of the disease. Supplementing their findings, we report here our recent study, which suggests that the SARS‐CoV may not be exclusively located in pneumocytes, but also in pulmonary macrophages.
The lung tissue that we studied was a necropsy specimen provided by one of us (CHH). The patient was a 36‐year‐old woman who died of SARS. Histological examination of the lungs revealed diffuse alveolar damage. The SARS viral genome had already been verified in the tissue by nested RT‐PCR 2. The full‐length spike protein (S) cDNA, membrane protein (M) cDNA, and nucleoprotein (N) cDNA clones were kindly provided by the National Research Program for Genomic Medicine and the Genome Research Center, National Yang‐Ming University, Taiwan. These inserts were amplified by PCR and labelled with digoxigenin using the DIG‐High Prime kit (Roche, Germany). The tissue sections were hybridized with the mixed probes (S, M, and N), 3 ng/µl, at 42°C overnight. BCIP/NBT was employed as a chromogen 3.
Under low magnification, scattered cells containing dark bluish signals, which represented the SARS‐CoV, were visualized (Figure 1A, nuclear fast red counterstain) within the damaged alveolar spaces, small bronchioles, and even the vascular lumina. High magnification revealed that the virus‐carrying cells harbouring the viral particles were frequently multinucleated and laden with anthracotic pigment (Figures 1B and 1C: empty arrows indicate anthracotic pigments; arrows indicate virus‐carrying multinucleated cells; nuclear fast red counterstain).
Figure 1.
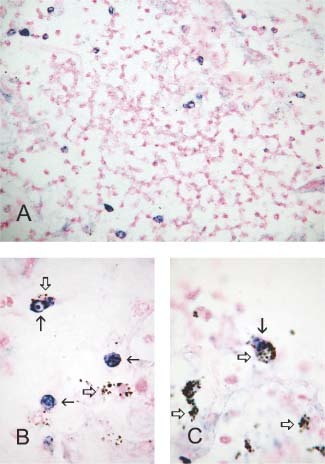
(A) Virus‐carrying cells are identified by dark bluish signals in lung (200×). (B and C) Viral signals (arrows) and anthrotic pigments (empty arrows) sometimes coexist in the same cells, indicating that these macrophages also harbour SARS‐CoV (1000×)
On the basis of our findings, we believe that some, if not all, positive cells are of the histiophagocytic lineage. First, the co‐existence of viral signals and anthracotic pigment in the same cells indicates that those cells are bona‐fide macrophages. Second, we observed that some virus‐harbouring cells were in the vascular lumina. It is unlikely that pneumocytes can be seen in vascular spaces even in a lung undergoing an extensively destructive process. Third, the virus‐harbouring cells were predominantly arranged as single discrete cells interspersed within the parenchyma, a finding also observed in previous reports. We did not identify any adherent virus‐positive cells, nor did we discern positive signals in the residual intact pneumocytes attached to the alveolar septa. This feature partly argues for a haematological, rather than an epithelial, origin for the virus‐carrying cells.
Monocytes/histiocytes/macrophages play important roles in the immune response. They produce a plethora of cytokines that have potent regulatory activities in haematopoiesis, inflammation, and diverse immune reactions. In addition to their involvement in cytokine production, they are actively motile cells that respond to chemotactic stimuli and they can phagocytose particulate material and kill micro‐organisms 4. The mortality of patients with SARS has been attributed to a ‘cytokine storm’ triggered by the host immune response to SARS‐CoV that results in diffuse alveolar damage 5. Although the underlying mechanism of the ‘cytokine storm’ has not been clarified, such a hypothesis fits our observation that cells of the histiophagocytic lineage are another carrier of SARS‐CoV. The presence of SARS‐CoV in these cells may induce catastrophic cytokine release.
On the other hand, the localization of SARS‐CoV in the cytoplasm of monocytes/histiocytes/macrophages does not justify their role as a host. These cells can phagocytose viral particles, with or without surface virus‐specific receptors. Also, the presence of many copies of the viral genome does not necessarily indicate viral propagation in these cells.
Due to the limited availability of SARS‐CoV‐infected tissue, an ISH study on a larger scale would be difficult to perform. Nonetheless, this piece of information is critical for future SARS investigation–for example, in the search for SARS‐CoV‐specific receptors–and also has clinical relevance, such as developing therapeutic agents that target histiocytes.
References
- 1. To KF, Tong JH, Chan PK, et al. Tissue and cellular tropism of the coronavirus associated with severe acute respiratory syndrome: an in‐situ hybridization study of fatal cases. J Pathol 2004; 202: 157–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chow KC, Hsiao CH, Lin TY, Chen CL, Chiou SH. Detection of severe acute respiratory syndrome‐associated coronavirus in pneumocytes of the lung. Am J Clin Pathol 2004; 121: 574–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chen PC, Pan CC, Yang AH, Wang LS, Chiang H. Detection of Epstein–Barr virus genome within thymic epithelial tumours in Taiwanese patients by nested PCR, PCR in situ hybridization, and RNA in situ hybridization. J Pathol 2002; 197: 684–688. [DOI] [PubMed] [Google Scholar]
- 4. Wickramasinghe SN. Normal blood cell In Blood and Bone Marrow Pathology, Wickramasinghe SN, McCullough J. (eds). Churchill Livingstone: London, 2003; 3–22. [Google Scholar]
- 5. Lee N, Sung J. The use of corticosteroids in SARS. The authors reply. N Engl J Med 2003; 348: 2034–2035. [DOI] [PubMed] [Google Scholar]
Paul Chih‐Hsueh Chen*, Cheng‐Hsiang Hsiao, * Department of Pathology, Taipei Veterans General Hospital and National Yang‐Ming University, Taipei, Taiwan, Department of Pathology, National Taiwan University Hospital, Taipei, Taiwan
Acknowledgements
This work was supported by grants SARS393‐2 from Taipei Veterans General Hospital and CSTVGH92‐02 from the Medical Research & Advancement Foundation in Memory of Dr Chi‐Shuen Tsou, Taiwan. The research use of the specimen and the procedures followed the ethical standards of the human experimentation committee of the institute.


