Abstract
Protein microarrays are a high‐throughput technology used increasingly in translational research, seeking to apply basic science findings to enhance human health. In addition to assessing protein levels, posttranslational modifications, and signaling pathways in patient samples, protein microarrays have aided in the identification of potential protein biomarkers of disease and infection. In this perspective, the different types of full‐length protein microarrays that are used in translational research are reviewed. Specific studies employing these microarrays are presented to highlight their potential in finding solutions to real clinical problems. Finally, the criteria that should be considered when developing next‐generation protein microarrays are provided.
Keywords: Biomarker, In vitro diagnostics, Protein microarray, Translational research
Abbreviations
- BLBC
basal‐like breast cancer
- CI
confidence interval
- DYRK2
dual‐specificity tyrosine phosphorylation‐regulated kinase 2
- GPCR
G‐coupled protein receptor
- HPV
human papillomavirus
- IVD
in vitro diagnostic
- MMTV
mouse mammary tumor virus
- Mtb
mycobacterium tuberculosis
- mTOR
mammalian Target of Rapamycin
- MV
membrane vesicle
- NAPPA
nucleic acid programmable protein array
- NPM
natural protein microarray
- Pf
Plasmodium falciparum
- Phe43
phenylalanine residue 43
- RCPA
reverse‐capture protein array
- RPPA
reverse‐phase protein array
- SLE
systemic lupus erythematosus
- T1D
type 1 diabetes
1. Introduction
Whereas the human genome has ∼20 000 protein‐coding genes, it is estimated that there are at least 70 000 different proteoforms as the result of polymorphisms, PTMs, and alternative splicing 1, 2. Proteins do not act alone; instead, they rely on complex interactions with other proteins and molecules to propagate signals leading to specific cellular responses. Given the breadth of protein species, high‐throughput methods are essential to gain a comprehensive understanding of the proteome. A popular, high‐throughput technology for studying the proteome is the protein microarray. Simply defined, protein microarrays are microscopic tools that display hundreds to thousands of different proteins on a fixed substrate in a high‐density array format, which can then be queried and analyzed simultaneously 3, 4, 5, 6. Protein microarrays can be distinguished from other protein display methods, such as yeast and phage display, because the identity of the protein and its associated array address are known in advance. Protein microarrays have helped tackle routine challenges in other proteomics technologies, including low protein abundance, complicated assay procedures, difficulty in modulating assay environment, low sensitivity, and low‐throughput analysis of proteins 6, 7, 8, 9, 10, 11, 12, 13, 14, 15.
In addition to basic research, protein microarrays have become useful in translational research, which aims to “translate” findings in fundamental research into clinical practice and meaningful health outcomes. Translational research often includes the use of clinical samples with the intent to discover or validate potential biomarkers that will aid in the diagnosis, prognosis, stratification, and treatment of patients. In order to achieve adequate statistical power during biomarker discovery and validation, the analyses of tens to hundreds of different clinical samples are often required due to high biological variation among patients. The expense associated with analyzing so many samples was, in part, responsible for the modest adoption of protein microarrays in translational research. For example, the production and purification of thousands of different proteins is much more complex than producing DNA oligonucleotides for a gene microarray. In addition, protein microarrays must achieve very tight tolerances and high reproducibility both within and between batches in order to discern subtle differences across clinical samples. New strategies and improved manufacturing techniques have led to a rapid increase in the application of protein microarrays in translational research 16, 17, 18, 19.
Various types of protein microarrays have been used in translational research, including protein fragment arrays 7 and peptide arrays that display random amino acid sequences 20, 21, known sequences, or sequences derived from predicted proteins 22. A recent hybrid system used bacteriophage to display barcoded peptides from human viruses to screen for antibodies that target these viral peptidomes and indicate the history of infection 23. Compared to standard protein microarrays that employ purified full‐length proteins, protein fragment and peptide microarrays are easier to produce due to their smaller sizes and simpler structures. Protein fragment arrays can also use antibody fragments lacking the Fc domain to probe for specific antigens in patient samples 6. The flexibility of the peptide microarray allows the incorporation of non‐natural amino acids and PTMs at specific sites or the use of unnatural peptide sequences 24. However, binding epitopes, especially conformational ones, may be lost with protein fragment and peptide microarrays. Given the diversity of protein microarrays and their applications, we have limited our scope in this perspective to the use of protein microarrays that display full‐length proteins.
In this article, we first outline the different full‐length protein microarrays that are available, as well as their advantages and disadvantages (Table 1). We then present translational research studies of cancer, autoimmune diseases, and infectious diseases that have employed full‐length protein microarrays. We discuss the role of protein microarrays in drug discovery and in vitro diagnostics (IVDs) tests, and last, we provide criteria that should be considered when designing next‐generation protein microarrays.
Table 1.
Comparison of the technologies used to prepare full‐length protein microarrays
| I. Purified | II. Natural | III. Cell‐free | |||||
|---|---|---|---|---|---|---|---|
| PPM | NPM | RPPA | RCPA | NAPPA | UPM | ||
| Expression system | E. coli | ✓ | ✓ | ✓ | |||
| Human | ✓ | ✓ | ✓ | ✓ | ✓ | ||
| Rabbit | ✓ | ✓ | |||||
| Wheat germ | ✓ | ✓ | |||||
| Insect | ✓ | ✓ | ✓ | ||||
| Phage | ✓ | ✓ | |||||
| Yeast | ✓ | ||||||
| Spotted molecules | Antibody | ✓ | |||||
| Cell/tissue lysate | ✓ | ||||||
| Plasmid cDNA | ✓ | ||||||
| Protein | ✓ | ||||||
| Proteins from cells/tissues | ✓ | ✓ | ✓ | ✓ | |||
| Displayed antigens | Denatured | ✓ | ✓ | ||||
| Folded | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |
| Functional | ? | ✓ | ✓ | ✓ | ? | ? | |
| In vivo folding | ? | ✓ | ✓ | ✓ | ? | ? | |
| PTMs | ✓ | ✓ | ✓ | ✓ | ✓ | ||
| Array antigens | Bacterial | ✓ 4, 55, 61, 62, 63, 65 | ✓ 66 | ✓ 9, 36, 37 | |||
| Human | ✓ 8, 17, 56, 58, 72, 73, 75 | ✓ 26 | ✓ 5, 54 | ✓ 10, 27 | ✓ 13, 31, 32, 38, 41, 42, 43, 51, 59 | ||
| Parasite | ✓ 44 | ✓ 34, 35, 68, 69 | |||||
| Virus | ✓ 53, 64 | ✓ 18, 41 | ✓ 19, 34 | ||||
| Applications | Antibody (immune) profiling | ✓ 6, 53, 55, 64, 65 | ✓ 18, 41, 44, 66 | ✓ 9, 19, 34, 35, 36, 37, 68, 69 | |||
| (Auto)antibody biomarker | ✓ 6, 8, 17, 48, 56, 58 | ✓ 26 | ✓ 10, 27 | ✓ 42, 43, 51, 59 | |||
| Protein biomarker | ✓ 5, 54 | ||||||
| Protein–DNA/RNA interactions | ✓ 4, 6, 61, 62 | ||||||
| Protein–lipid interactions | ✓ 6, 12, 15 | ||||||
| Protein–protein interactions | ✓ 6, 12, 63, 72, 73, 75 | ✓ 19, 31, 32, 38 | |||||
| PTMs | ✓ 6 | ✓ 5, 54 | ✓ 13, 39 | ||||
| Signaling pathway | ✓ 6 | ✓ 5, 54 | |||||
| Other | Concentration independent | ✓ | ✓ | ✓ | |||
| Cost‐effective | ✓ | ✓ | |||||
| Express large proteins | ✓ | ✓ | ✓ | ✓ | ✓ | ||
| Long shelf life | ✓ | ||||||
NAPPA: Nucleic Acid Programmable Protein Arrays; NPM: Natural protein microarray; PPM: purified protein microarray; RCPA: reverse capture protein array; UPM: unpurified protein microarray.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
2. Protein microarray technology
Planar protein microarrays displaying full‐length proteins that have been applied in translational research can be classified into three main types based on how the proteins are produced. These arrays have been referred to as purified, natural, and cell‐free expression protein microarrays (Fig. 1, Table 1). The most popular microarray format is the “purified protein microarray” in which proteins are expressed in heterologous systems, such as insect cells, Escherichia coli, or yeast. The proteins are purified in a 96‐well plate format via a fusion tag, and subsequently printed on the slide. The purified protein microarray was originally used to build the yeast proteome microarray containing 5800 ORFs, and has since been employed in various protein function and antibody biomarker discovery studies 12. Currently, the most comprehensive purified protein array is the HuProt™ human proteome microarray v2.0 manufactured by Arrayit Corporation and CDI Laboratories. The array has 19 394 proteins with 15 275 different genes, covering ∼75% of the human genome and representing different splicing isoforms. However, there are some disadvantages to the purified protein microarray approach; these include: (i) the expense and time requirement for the expression and purification of thousands of proteins, (ii) the challenge of expressing and purifying large proteins (>50 kDa) and membrane proteins, (iii) the short shelf life 25, and (iv) the possibility of incorrect protein folding and activity if the proteins are expressed in a different host milieu (e.g. human proteins expressed in E. coli) or dried following array production.
Figure 1.
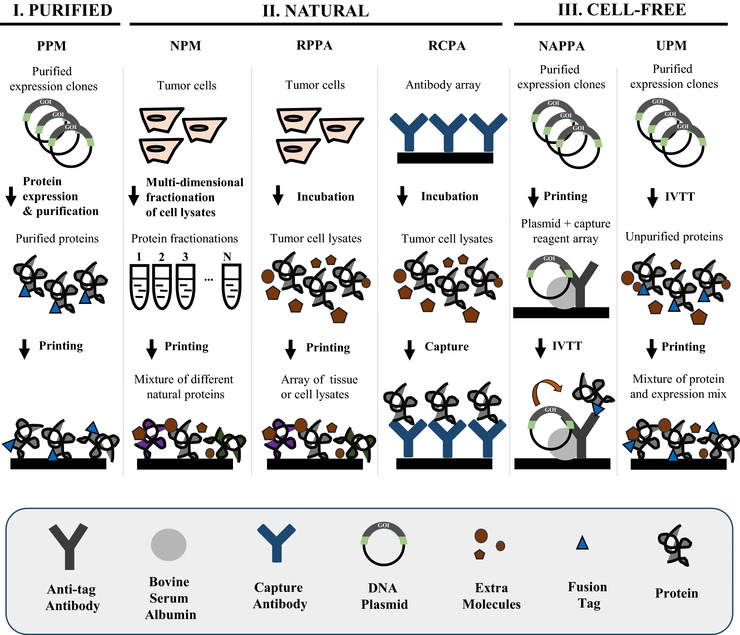
Comparison of the main fabrication technologies used to build functional full‐length protein microarrays. PPM: purified protein microarray; RCPA: reverse capture protein array; UPM: unpurified protein microarray.
Unlike the purified protein array that employs heterologous systems to produce protein, “natural protein microarrays” (NPMs) are printed using human cell lysate, tissue lysate, or bodily fluids (e.g. serum) containing naturally expressed proteins that likely have the same folding, activity, and PTMs as those in vivo at the time of sample collection. The proteins can be printed as a complete lysate, fractionated using multidimensional liquid‐based separation techniques, or captured to the array surface with specific antibodies. The translational applications differ depending on the fractionation state of the lysate and how the proteins are captured to slide. NPMs with fractionated samples are often probed with serum, and have been used to study the immune response to various cancers and their altered antigens (i.e. phosphorylated and glycosylated proteins) 26. The antibody target must then be subsequently identified, which is often done with mass spectrometry. Sample fractionation can also assist in the analyses of specific protein classes, for example, membrane or glycosylated proteins 26.
Unfractionated NPMs are also known as “reverse phase protein arrays” (RPPA), “lysate arrays,” or “reverse phase lysate arrays.” These lysate arrays are probed with antibodies that recognize key signaling proteins or their modified forms to help determine how specific signaling pathways are (dys) regulated during disease and infection, and may indicate potential therapeutic options. Moreover, numerous patients and control samples can be analyzed simultaneously with RPPAs 5. A primary advantage of RPPAs is that they are able to discern the dynamic posttranslational events (e.g. PTMs) that can greatly affect protein interactions within complex signaling networks, information of which cannot be determined by analyzing gene expression and protein levels alone.
Alternatively, “reverse‐capture protein arrays” (RCPAs) are coated with antibodies that will capture proteins expressed in vivo (e.g. tumor antigens), which are then used to probe for autoantibodies in serological samples. Unlike RPPAs that analyze one protein across many patient samples, RCPAs are generally used to study multiple proteins within one sample 10, 27. Disadvantages of fractionated, unfractionated (RPPAs), and captured (RCPAs) NPMs include: (i) their reliance on high‐quality antibodies specific to the pathway(s)‐of‐interest, which may not be available; (ii) low‐abundance target proteins may be difficult to detect; and (iii) proteins may lose their activity during purification, printing, and storage.
“Cell‐free protein microarrays” were developed to avoid challenges related to protein capture and purification for array production, as described in this section. Therefore, cell‐free protein microarrays use PCR‐generated DNA fragments or plasmid cDNA to express proteins in a cell‐free expression system that transcribes and translates the genes‐of‐interest 28. Various expression systems are commercially available, including lysate from E. coli, wheat germ, insect cells, rabbit reticulocyte, and human cells. Cell‐free protein microarrays include the protein in situ array 29, DNA array to protein array 30, nucleic acid programmable protein array (NAPPA) 13, 31, 32, in situ puromycin‐capture array 33, and the “unpurified” protein array developed by Davies et al. 34. Following in vitro transcription and translation that typically require 2–5 h, the expressed proteins are captured in situ by a capture antibody or molecule. The “unpurified” cell‐free microarray does not use any capturing agent; instead, the lysate containing the expressed protein‐of‐interest is spotted directly onto a nitrocellulose substrate, with the assumption that the coprinted lysate will have a similar background from feature to feature. Unpurified cell‐free protein microarrays have been successfully applied in immune profiling and antibody biomarker discovery for infectious diseases 9, 34, 35, 36, 37. In addition to the aforementioned applications, NAPPA has also been used to study protein–protein interactions and PTMs 13, 19, 38, 39.
There are many advantages of using the cell‐free expression approach, including the ability to express proteins in the appropriate expression milieu at the time of the experiment. The cell lysate, which includes chaperone proteins, also allows the proteins‐of‐interest to be expressed, folded, and maintained in physiologically compatible buffers from the time of translation through the experiment, thus significantly reducing the likelihood of unfolding, denaturing, or drying. Cell‐free expression arrays are cost‐effective because they do not require protein purification, and the printing of DNA (i.e. DNA array to protein array, NAPPA) results in a more stable array with a longer shelf life than protein‐based microarrays. These arrays can be easily made high throughput, with thousands of different proteins displayed on a standard‐sized microscope slide (i.e. 75 × 25 × 1.2 mm3). For example, NAPPA uses a routine density of ∼2300 proteins per slide, which has been increased to 8000 with the use of nanowells 40, 41. Finally, immune responses in disease may recognize both linear and conformation epitopes. Thus, biomarker screening using proteins that are both folded and denatured is likely to have the value of displaying both types of epitopes. NAPPA is amenable to studying both folded and denatured proteins, making it possible to identify autoantibody biomarkers to epitopes that would otherwise be buried during healthy homeostasis 42, 43, 44.
There are also disadvantages to the cell‐free approach. For the unpurified protein microarrays: (i) protein functions may not be easily assayed as each spot is a mixture of different molecules; (ii) the lysate is allowed to dry on the nitrocellulose surface after printing, which may affect protein folding and activity; and (iii) serological antibodies will react to all of the printed lysate spots when E. coli lysate is used 34. For the other types of cell‐free microarrays, nonspecific binding of components in the cell‐free expression system to the array matrix may occur; however, these nonspecific interactions are largely removed during several washing steps, leaving behind lysate components that are at a level much lower than that of the protein‐of‐interest and are consistent across features. A bigger theoretical concern is that lysate proteins, which are also human, may interact specifically with some captured proteins‐of‐interest and be displayed as protein complexes. Therefore, all findings should be confirmed with subsequent studies. It is also worth noting that some degree of known PTM can occur to the protein‐of‐interest when expressed in some cell lysates, most notably phosphorylation with the human cell‐free expression system 45.
The above‐described methods typically employ a microscope slide as the substrate. However, bead‐based arrays offer an alternative approach, and have been frequently used in biomarker validation and the development of IVD tests. They are particularly useful when the number of proteins‐of‐interest is modest and the number of clinical specimens is high (Fig. 2) 14, 46; however, more recent advances in this approach are enabling the analyses of greater numbers of proteins 47. In bead‐based microarrays, the address of the target protein is embedded in the colors of their specifically bound beads. With Luminex xMAP® technology, each bead is barcoded using two fluorescent dyes that are mixed in an appropriate ratio; up to 500 different color beads are available. More colored beads (n = 1725) with a combination of four fluorescent dyes have also been developed for higher throughput analyses 47. As an example, antigens‐of‐interest on different color‐coated beads are incubated with serological samples. Captured antibodies from the serological samples are then bound by a fluorescently labeled secondary antibody. Two readings will take place during analyses: (i) the bead color will identify its associated antigen and (ii) the fluorescence from the secondary antibody will indicate the abundance of serological antibodies specifically bound to the antigen. The merits of the bead‐based assay include high sensitivity, wide dynamic range, excellent reproducibility, and ease of automation. Moreover, some bead‐based assays have been approved for clinical use, including the LIFECODES® LifeScreen Deluxe, an HLA antibody assay, (Immucor GTI Diagnostics, Inc.) 48 and BioPlex® 2200 HIV Ag‐Ab (Bio‐Rad Laboratories) 49. However, the production and binding of individual proteins to uniquely colored beads can be costly, especially for large numbers of proteins.
Figure 2.
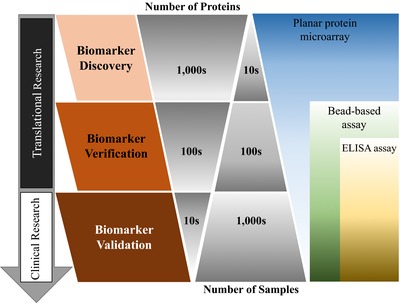
Proteomic biomarker discovery and validation pipeline.
3. Applications of protein microarrays in translational research
The proteomic biomarker discovery and validation pipeline is often divided into three consecutive phases, including identifying candidate biomarkers (i.e. discovery), verifying the biomarkers with (an) independent sample cohort(s), and finally validating the biomarkers for clinical applications (Fig. 2). The translational phase of this research refers to the first two steps of biomarker discovery and verification, where it is especially important to screen as many different candidate proteins as possible to avoid missing good markers. Thus, the planar protein microarray is an excellent tool for discovery because of its ability to display tens of thousands of different proteins in an easily addressable configuration. In later phases, where fewer candidate proteins need to be tested, the bead‐based protein microarrays and ELISA assays are ideal because they can analyze many patient samples quickly.
3.1. Immune profiling and (auto)antibody biomarker discovery
3.1.1. Cancer
Breast cancer is the second most common cause of cancer‐related mortality in women in the United States of America, with earlier cancer diagnosis improving patient prognosis. Currently, mammography is the standard screening method for the early detection of breast cancer; however, even though the test itself has higher sensitivity, the real‐world sensitivity of mammography has been reported to be as low as 30–48%, which reflects variations in radiologist assessments, tumor subtypes, patient age, patient compliance, and breast density 50. Therefore, the identification of circulating autoantibody biomarkers has the potential to fill a clinical niche for a noninvasive, inexpensive, and rapid approach for the early diagnosis of breast cancer that could complement imaging studies.
Anderson et al. 51 identified autoantibody biomarkers of breast cancer by screening the sera of early stage breast cancer patients (stages I–III) using the NAPPA protein microarray. Their array contained 5000 unique human proteins that included 1000 breast cancer proteins that were preselected from bioinformatics and data mining, 300 G‐coupled protein receptors (GPCRs), 500 kinases, and 700 transcription factors. Using three independent sample sets (155 total cases and 130 total age‐matched healthy controls), 28 potential autoantibody biomarkers were identified and verified with a sensitivity of 12–42% and a specificity of 66–100% under blinded conditions. These biomarkers could be used for the early detection of breast cancer in the future.
Ladd et al. 26 analyzed the dynamic interactions between tumor antigens and circulating autoantibodies during breast cancer development and progression. NPMs were prepared using lysates from an mouse mammary tumor virus (MMTV)‐neu breast cancer mouse model and an MCF7 human breast cancer cell line. The MMTV‐neu mouse model protein array was probed with the sera of a breast cancer mouse model, while the MCF7 human breast cancer cell line protein array was probed with sera from human patients recently diagnosed or soon‐to‐be diagnosed with breast cancer. Two glycolysis and splicesome autoantibody signatures were identified with an AUC of 0.68 (95% confidence interval [CI]: 0.59–0.78) and 0.73 (95% CI: 0.63–0.82), respectively. Although these signatures need to be validated in a larger patient population to determine whether they should be used to diagnose breast cancer in a clinical setting, the study provides compelling evidence that tumor autoantibodies are generated in the early development of breast cancer.
Basal‐like breast cancer (BLBC) is an aggressive type of breast cancer with relatively poor detection rates when screened with mammography. Using individually age‐matched plasma samples from the Polish Breast Cancer Study, Wang et al. 43 executed the largest unbiased discovery of autoantibodies associated with BLBC using NAPPA protein microarrays with 10 000 antigens (45 paired patients and controls). Seven hundred and forty‐eight antigens identified as potential targets of autoantibodies of disease in the discovery phase were printed on an independent array and screened using an independent sample set of 50 BLBC cases and 50 controls. Finally, 82 antigens were verified using ELISA (100 paired BLBC patients and controls). A panel of 13 verified autoantibodies (CTAG1B, CTAG2, TP53, RNF216, PPHLN1, PIP4K2C, ZBTB16, TAS2R8, WBP2NL, DOK2, PSRC1, MN1, and TRIM21) discriminated the BLBC samples from controls with 33% sensitivity and 98% specificity under blinded conditions. These biomarkers have great potential to be used in the early diagnoses of BLBC, thus improving patient prognosis.
Viral infection is closely associated with some types of cancer, including human papillomavirus (HPV) and cervical cancer. Therefore, the detection of antibodies to viral proteins in a clinical setting will be very useful in properly diagnosing and treating patients that have been infected with viruses that cause cancer 52. In 2005, Waterboer et al. 53 developed a bead‐based assay to measure serological antibodies to 27 HPV proteins by coupling glutathione S‐transferase fusion HPV proteins to glutathione‐coated beads. Seven hundred and fifty‐six sera of paired cervical cancer cases and controls were tested. The assay identified an association between cervical cancer and antibodies to the viral protein E6 from two HPV types, HPV52 and HPV58, with statistical significance (p‐value < 0.001) and a high correlation with ELISA (κ = 0.846). Their multiplexed serology assay required minimal sample volume (2 μL serum for <100 antigens) and showed greater sensitivity than GST‐captured ELISA. Moreover, the assay had a sensitivity of >1:1 000 000 serum dilution, dynamic range of 1.5 orders of magnitude, a coefficient of variation of 5.4%, and an assay reproducibility of R 2 = 0.97. Although it was previously known that cervical cancer patients could develop antibodies to the E6 protein, this study demonstrated the utility of the multiplexed bead‐based serology assay in biomarker verification.
Rhabdomyosarcoma is the most frequent soft tissue tumor in children, and is often driven in part by dysregulation of the prosurvival PI3K/AKT/mammalian Target of Rapamycin (mTOR) pathway 3. Since 30 to 40% of children diagnosed with rhabdomyosarcoma will not respond to the standard chemotherapy treatment, Petricoin et al. 54 applied RPPAs to map the phosphoproteomic network within the PI3K/AKT/mTOR pathway with the objective of identifying better treatment options for nonresponders. Lysed microdissected tumor cells from 59 patients were printed directly onto a planar surface and then probed with antibodies specific to 16 proteins and 17 phosphoproteins in the PI3K/AKT/mTOR pathway. Higher levels of phosphorylated proteins that reflect PI3K/AKT/mTOR pathway activation were significantly associated with poor overall and disease‐free survival of childhood rhabdomyosarcoma. Further, mTOR inhibitors (i.e. rapamycin analogues) were able to profoundly reduce tumor growth in mouse xenografts, which suggests that administration of mTOR inhibitors to patients with high mTOR pathway activation may result in a more favorable chemotherapy response with improved cancer prognosis. The results from the studies described in this section illustrate the value of protein microarrays in finding circulating (auto)antibody biomarkers for early cancer detection as well as mapping protein signaling networks for improved cancer treatment, thus underscoring their importance in translational research.
3.1.2. Autoimmune disease
Protein microarrays have been employed to better understand autoimmune diseases, during which the immune system destroys healthy cells and tissues of the individual 55, 56. Multiple sclerosis is the most common autoimmune disorder affecting the CNS, causing symptoms that include fatigue, dizziness, pain, spasticity, depression, cognitive impairment, and bladder/bowel problems. As reported in 2008, 2–2.5 million people worldwide have multiple sclerosis 57. The development of new biomarkers would be helpful in diagnosing and predicting the onset of multiple sclerosis. To address this need, Ayoglu et al. 7 screened the sera of 90 multiple sclerosis patients using a purified protein microarray containing 11 520 protein fragments (∼7500 unique proteins) generated by the Human Protein Atlas project. Among the 2000 reactive autoantibodies that were initially identified, 384 priority targets were further verified in a large sample cohort (n = 376) using a bead‐based assay. Fifty one autoantibodies showed differential responses to eight subtypes of multiple sclerosis. This work demonstrates the value of protein microarrays in finding potential multiple sclerosis biomarkers and supports validating these biomarkers in a larger population. It also highlights the complementing roles of planar and bead‐based protein arrays in the proteomic biomarker discovery and validation pipeline (Fig. 2).
Autoantibodies to serological inflammatory factors (i.e. cytokines, chemokines, and growth factors) may inhibit immunity and be involved in inflammatory autoimmune disease. Price et al. 58 prepared a purified protein microarray containing 59 human cytokines and chemokines as well as 101 antigens related to autoimmune diseases. The authors then tested patients with systemic lupus erythematosus (SLE; 30 patients and 15 controls). In addition to confirming known autoantibodies, Price et al. 58 identified new SLE autoantibodies targeting TGF‐β1–β3, IL‐2, IL‐15, IL‐23, TNF, IFN‐α2B, and IFN‐γ. Furthermore, they observed that the autoantibody reactivity to B cell‐activating factor may be associated with SLE severity. Additional studies using an independent sample cohort(s) are warranted to validate these potential autoantibodies of SLE.
Type 1 diabetes (T1D) is a chronic autoimmune disease that significantly affects glucose metabolism and is characterized by the immune destruction of insulin‐producing β cells of the pancreatic islets, which is caused by complex and incompletely understood interactions between an individual's genetic background, environmental factors, and immune system. In the United States alone, over 1 million people are living with T1D, resulting in $14.9 billion of associated healthcare costs per year (CDC Diabetes Statistics Report, 2014; Economic Cost of Diabetes in the United States in 2012). Therefore, it is clear that biomarkers for the early prediction and diagnosis of T1D are urgently needed. Miersch et al. 59 performed a proteome‐wide screen for T1D serological autoantibody biomarkers using NAPPA protein microarrays containing ∼6000 human proteins. Ten potential autoantibody biomarkers identified in cohort 1 (50 cases and 20 controls) were validated in cohort 2 (75 cases and 74 controls) and cohort 3 (46 cases and 46 controls). Among them, the autoantibody to dual‐specificity tyrosine phosphorylation‐regulated kinase 2 (DYRK2) was further validated using the Luciferase Immuno Precipitation System assay with a sensitivity of 36% and a specificity of 98%. The combination of DYRK2A with the known T1D antigen, IA‐2A, generated a higher AUC of 0.90, compared to AUCs of 0.72 and 0.64 for DYRK2A and IA‐2A alone, respectively. As a recent follow‐up to better understand the environmental triggers of T1D, Bian et al. 18, 41 fabricated a viral proteome microarray containing 761 viral proteins from 25 different viruses using NAPPA technology and then analyzed the antibody responses of 42 patients with T1D and 42 age‐gender matched patients without T1D. This approach appeared to provide a reliable measure of historical infection because the antiviral antibody response to common viruses correlated well with known viral infection rates from epidemiological studies. Interestingly, the viral infection that was closely associated with T1D (OR 6.6; 95% CI: 2.0–25.7) was the Epstein–Barr virus, particularly in younger patients. This study demonstrates the potential value of immunoproteomics in better understanding the role of viral infections in chronic disease 18.
The above studies probed only a subset of the human proteome; despite this, the results are compelling. More comprehensive human proteome collections will no doubt expand the number of potential antibody biomarkers of autoimmune diseases that could be used to diagnose, stratify, and treat patients.
3.1.3. Infectious disease
Compared to traditional analytical technologies (e.g. ELISA), protein microarrays offer a comprehensive and systematic approach to profile the humoral immune response to numerous infectious pathogens with high sensitivity and specificity 18, 41, 60, 61, 62, 63. In 2006, Zhu et al. 64 prepared a protein microarray with 82 purified proteins from SARS coronavirus (SARS‐CoV) and five additional coronaviruses. They screened sera from ∼400 SARS and SARS‐CoV patients, respiratory illness patients, and healthcare professionals. Multiantibody signatures were then verified in two independent cohorts from Canada (181 SARS patients, 218 healthy; 76% sensitivity, 96% specificity) and China (35 SARS patients, 21 healthy; 100% sensitivity, 95% specificity). The results from the protein microarray and ELISA data exhibited good correlation (85%) in predicting the SARS infection in 147 patients with a fever during a SARS outbreak in China. These data demonstrate that a viral‐based protein microarray has the capacity to find specific antibodies associated with viral infection.
Mycobacterium tuberculosis (Mtb) is a bacterial pathogen that infects approximately one‐third of the world's population, with ∼10% of infected individuals eventually developing active tuberculosis. In addition to causing serious health complications (e.g. organ and joint damage), active tuberculosis can result in death. The percentage of active cases is significantly higher in developing countries, which have limited access and resources to medication. Thus, the comprehensive study of tuberculosis–host interactions will be invaluable in understanding the mechanism of infection and developing rapid diagnostics tests for Mtb. Kunnath‐Velayudhan et al. 37 evaluated the humoral response to the entire tuberculosis proteome across 500 patients with TB and non‐TB diseases using unpurified protein microarrays. The results revealed that ∼10% of the Mtb proteome can be recognized by the body's immune system, in which a high proportion of produced antibodies target membrane‐associated and extracellular proteins of the bacteria. In addition, active TB patients showed variable responses to 0.5% (13 proteins) of the Mtb proteome, which correlated with bacillary burden. Twelve of the proteins had been previously identified as antigens of B cells and T cells.
Deng et al. 65 also evaluated the humoral antibody response to the Mtb proteome; however, they analyzed the immune response in 21 patients with active Mtb and 20 “recovered” patients following antibiotic treatment using purified protein microarrays. Fourteen antibodies to Mtb antigens that discriminated active from recovered patients were identified and subsequently verified in two independent cohorts (cohort 2: 64 active TB and 43 “recovered” TB; cohort 3: 104 active TB and 87 “recovered” TB) with a p‐value of 0.05.
Prados‐Rosales et al. 66 assessed the vaccine potential of the Mtb membrane vesicle (MV), which has been shown to be secreted by Mtb in a mouse model. They applied sera obtained from mice immunized with MV or challenged with the Bacillus Calmette‐Guérin vaccine to a cell‐free expression Mtb proteome microarray (i.e. NAPPA). The Bacillus Calmette‐Guérin vaccine served as a negative control in this experiment since it, too, has an MV. The results indicated that a dozen humoral antibodies were specifically induced in the mouse immunized with MV from Mtb, with target antigens primarily from membrane and cell wall proteins. This study generated evidence that Mtb MV could be used to develop a vaccine against Mtb infection.
Despite large variations in study aims, sample origins, study designs, and microarray platforms (purified vs. NAPPA vs. unpurified), as well as the geographical locations of sample acquisition, it is interesting to see that Rv0934c (pstS1/38kD) and Rv3616c (espA) were shown as the top hits in the Kunnath‐Velayudhan and Prados‐Rosales studies, and Rv3881c (espB) was found in the Kunnath‐Velayudhan and Deng studies. Rv0934c, Rv3616c, and Rv3881c are all functionally annotated as cell‐wall and cell‐processes proteins (TubercuList database, http://tuberculist.epfl.ch/). Moreover, Rv0934c is a diagnostics biomarker for TB with a previously shown sensitivity of 64.21% and specificity of 80.74% in 594 Chinese patients (312 patients with active pulmonary tuberculosis and 282 control subjects) 67. In view of these studies, Rv0934c and Rv3616c might be investigated as targets for vaccine development, whereas Rv3881c may be a good predictive marker for therapeutic treatment of TB patients.
Plasmodium falciparum (Pf) is a protozoan parasite that is transmitted by the female Anopheles mosquito and causes the most fatal form of malaria in human with high complications and mortality. The immune system can protect the host during repeated malaria infections and has the potential to be exploited for therapeutic treatment of malaria patients. Crompton et al. 69 prepared an unpurified protein microarray containing 23% of the 5400 Pf proteins, and analyzed the humoral antibody response of 220 individuals in Mali before and after the 6‐month malaria season. Four hundred and ninety‐one immunogenic proteins were recognized by the immune system, in which 26 were confirmed from a previous study with Pf‐exposed Kenyan adults using unpurified cell‐free expression protein microarrays 68. The results further revealed that the antibody response to Pf proteins was significantly increased in children during the 6‐month malaria season, after which the level of some antibodies decreased; the authors hypothesized that the decreased levels were likely due to the antibody half‐life 69.
To find biomarkers that indicate malaria infection, Helb et al. 35 screened 186 Ugandan children for antibodies targeting 856 Pf antigens with unpurified protein microarrays. The results revealed a three‐antibody biomarker signature (hyp2, GEXP18, a putative exonuclease) that accurately classifies individuals 30, 90, or 365 days postinfection with an AUC of 0.86–0.93. These data have not been verified yet. However, the ability to estimate Pf exposure is extremely useful in effectively controlling and eliminating malaria. Unfortunately, the biomarker candidates from Crompton et al. 69 and Helb et al. 35 are not easily comparable due to the different gene annotations that they used. These studies demonstrate that high‐density protein microarrays can be applied in profiling the dynamic host immune response in regards to viral infection, the cycle of parasitic infection, identifying novel diagnostic antibody biomarkers, and identifying potential vaccine targets for host protection 16.
3.2. Drug discovery
Fifty to 70% of prescribed drugs target membrane proteins due to their crucial roles in molecular binding, signal transportation, and ion regulation. Thus, a high‐throughput membrane protein array platform will significantly facilitate the drug screening process. In 2002, Fang et al. 70 demonstrated the feasibility of such arrays by printing three extracted GPCR proteins (adrenergic receptor, neurotensin receptor, and dopamine receptor) on γ‐aminopropylsilane modified slides. The binding of fluorescein‐conjugated antagonists (CGP 12177, ICI 118551, neurotensin) to their corresponding receptors (beta‐adrenergic receptor, neurotensin receptor) were then detected 71. Further membrane protein array development by Hong et al. 72, 73 illustrated the screening ability of these arrays by using a mixture of fluorescein‐labeled ligands with an array displaying ten GPCR receptor proteins and sandwich‐based competition assay. Although the throughput of these arrays is still quite low, the feasibility of the approach shows promise.
Until recently, the high‐throughput capability of membrane protein arrays has been limited by detergent‐based protein extraction from mammalian cells and subsequent array fabrication. In 2013, Hu et al. 74 developed a novel high‐throughput strategy by exploiting the infection machinery utilized by herpes viruses. Following molecular cloning and transfection, mammalian cells were infected with herpes virus, and the membrane proteins‐of‐interest were displayed on large, secreted spherical herpesvirus virions. The virions were then purified with a simple sucrose gradient and printed onto a nitrocellulose membrane slide. The activities of both a single‐pass (CD4) and a multipass (GPR77) transmembrane proteins on the Virion Display Array were confirmed using antibody staining and ligand interaction assays.
Protein microarrays can also be used to screen the effect of small molecules on protein–protein interactions. For example, the glycoprotein of gp120 plays a critical function in the entry of the HIV virus into host cells by binding to the T‐cell CD4 receptor through a conserved binding pocket, in which the phenylalanine residue 43 (Phe43) has been suggested to play an important role. Alterations in gp120‐CD4 receptor binding were characterized with a gp120 purified protein array queried with seven GFP‐tagged CD4 analogs containing Phe43 derivatives of different sizes and steric conformations. The binding of each CD4 analog was assessed using a rabbit anti‐GFP antibody and an Alexa555‐labeled antirabbit secondary antibody. Compared to wild‐type Phe43, the gp120–CD4 receptor interaction became stronger when Phe43 was modified with hydrophobic groups. The data point toward a new direction for the development of gp120 inhibitors and assay reagents in HIV treatment 75. All of these results demonstrate that protein microarrays have the capability to screen antibodies, membrane proteins, and small compounds for drug discovery in a high‐throughput way.
3.3. Protein microarray for IVDs
The majority of IVD tests that implement protein arrays are used to diagnose autoimmune and infectious diseases. The most advanced IVD tests using bead‐based arrays include the AtheNA Multi‐Lyte® Test System and the BioPlex 2200® multiplexed platform. The IVD tests for autoimmune diseases include systemic rheumatoid diseases, vasculitis, thyroid diseases, celiac disease, and Antiphospholipid Syndrome. The IVD tests for infectious diseases include Epstein–Barr virus, herpes simplex virus, TORCH, and Treponemapallidum.
Randox Laboratories has developed a fully automated planar‐based Evidence‐Biochip Analyzer system that is capable of analyzing 1200 tests per hour 76. A variety of kits for cardiac, cerebral, cytokine, endocrine, metabolic syndrome, and tumor markers are commercially available. Moreover, an immunoassay array testing for 13 common drugs of abuse has a CE Mark and FDA approval for use as an IVD device.
Despite the tremendous effort that has been spent on the discovery of biomarkers for various cancers, only a few cancer biomarkers have been FDA‐approved for IVD tests. One of these tests is the bead‐based OVA1 test (ASPiRA LABS™), which combines five biomarkers (i.e. beta‐2 microglobulin, CA 125II, apolipoprotein A1, prealbumin, and transferrin) to assess the likelihood that an ovarian mass is malignant prior to surgery. The OVA1 test employs a score between 0 and 10 corresponding to the likelihood of malignancy 77. With the completion of the human genome project in 2003 and an average of 15 years for a newly discovered biomarker to become part of an IVD test (Market Trends for Biomarker‐Based IVT Tests (2003–2014). Amplion Inc. [www.amplion.com] 2015), it is anticipated that more biomarkers will be approved for IVD tests within the next 10 years.
4. Perspectives
Protein microarrays have increased our understanding of protein function, protein signaling, and the immune response during infection and disease. Future studies will employ both well‐established and novel platforms, which will address current limitations of protein microarrays. For example, protein microarrays have been used to study a handful of the most common and well‐studied PTMs (e.g. phosphorylation, glycosylation, AMPylation). However, it is likely that future platforms will be adapted to analyze additional modifications should they be shown to affect protein activity, location, degradation, stability, or homeostatic interactions. Protein microarrays have also been unable to provide the kinetics and affinities of protein–protein interactions in a high‐throughput manner. The comprehensive characterization of transient and stable protein–protein interactions may help identify critical decision proteins within complex interaction networks, which could be used as therapeutic targets of disease. These next generation of protein microarrays should take the following into consideration: (i) protein expression and activity, (ii) cost and ease‐of‐use or flexibility, (iii) sensitivity and dynamic range of the assay, and (iv) reproducibility.
Critical factors for full‐length protein microarrays include protein expression and activity. As described in “Protein microarray technology,” the use of heterologous systems to express human proteins may not result in full‐length, functional products. Mammalian‐based expression systems offer an excellent alternative. In a previous study, the human HeLa cell lysate in vitro transcription translation system had a successful expression rate of 87% using 31 full‐length human proteins of various sizes and function when compared to 73% with E. coli 45. These HeLa cell‐produced proteins showed functionality in protein–protein interactions and PTM (i.e. AMPylation) assays, and were recognized by serological antibodies 13, 38. In addition to HeLa cell lysate, lysates from human erythroleukemic cells (K562) and Chinese hamster ovary cells have also been able to produce human proteins 78.
The high cost of full‐length protein microarrays has been a major limitation in making the platform a standard laboratory tool, especially for high‐density proteome microarrays. Protein microarray affordability is particularly relevant in translational research due to the requirement for tens to hundreds of different samples. The use of different cell‐free methods to produce protein microarrays offer some advantages, including: (i) low cost of DNA production and quality control; (ii) fresh protein production in a few hours; (iii) DNA arrays are stable (>6 months) at room temperature; and (iv) custom arrays of any protein can be made if the cDNAs are available. Access to a wide variety of cDNAs is becoming easier and cheaper with plasmid repositories generated from programs such as the Protein Structure Initiative 79.
Other important aspects of protein microarrays for consideration are sensitivity and dynamic range, which are relevant in acquiring information about low‐abundance biomarker proteins in biological samples and accurately quantifying the changes of these proteins in different conditions, respectively. For example, Zhang et al. 17 developed a plasmonic gold chip spotted with purified proteins to detect autoantibodies during T1D using near‐infrared fluorescence‐enhanced detection, which has two to three orders higher sensitivity than glass slides (<0.1 U/mL). With the plasmonic array, autoantibodies to three antigens (insulin, GAD65, and IA2) distinguished T1D from type 2 diabetes.
Finally, protein microarrays should be highly reproducible and robust, particularly when used in translational research and clinical assays. Reproducibility can be significantly improved with the automation of array processing, sample incubation, and signal readout. The screening of clinical samples with high‐density protein microarrays using an automated station resulted in an intra‐ and interreproducibility of R = 0.99 43.
Protein microarrays have increased our understanding of proteome networks in regards to normal homeostasis, disease, and infection. Technological advancements, particularly in the areas listed above, will further the contribution of protein microarrays in drug screening and biomarker discovery for improved patient care.
The authors have declared no conflict of interest.
Acknowledgment
This project is funded by the Early Detection Research Network (5U01CA117374) and the start‐up grant from the National Center for Protein Sciences (The PHOENIX Center, Beijing).
Colour Online: See the article online to view Figs. 1 and 2 in colour.
5 References
- 1. Ezkurdia, I. , Juan, D. , Rodriguez, J. M. , Frankish, A. et al., Multiple evidence strands suggest that there may be as few as 19,000 human protein‐coding genes. Hum. Mol. Gen. 2014, 23, 5866–5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jost, J. , Scherrer, K. , Information theory, gene expression, and combinatorial regulation: a quantitative analysis. Theory Biosci. 2014, 133, 1–21. [DOI] [PubMed] [Google Scholar]
- 3. Beck, J. T. , Ismail, A. , Tolomeo, C. , Targeting the phosphatidylinositol 3‐kinase (PI3K)/AKT/mammalian target of rapamycin (mTOR) pathway: an emerging treatment strategy for squamous cell lung carcinoma. Cancer Treat. Rev. 2014, 40, 980–989. [DOI] [PubMed] [Google Scholar]
- 4. Chen, C. S. , Korobkova, E. , Chen, H. , Zhu, J. et al., A proteome chip approach reveals new DNA damage recognition activities in Escherichia coli . Nat. Methods 2008, 5, 69–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pierobon, M. , Wulfkuhle, J. , Liotta, L. , Petricoin, E. , Application of molecular technologies for phosphoproteomic analysis of clinical samples. Oncogene 2015, 34, 805–814. [DOI] [PubMed] [Google Scholar]
- 6. Sutandy, F. X. , Qian, J. , Chen, C. S. , Zhu, H. , Overview of protein microarrays. Current Protocols in Protein Science/Editorial Board, John E. Coligan … [et al.] 2013, Chapter 27, Unit 27 21. [DOI] [PMC free article] [PubMed]
- 7. Ayoglu, B. , Haggmark, A. , Khademi, M. , Olsson, T. et al., Autoantibody profiling in multiple sclerosis using arrays of human protein fragments. Mol. Cell. Proteomics 2013, 12, 2657–2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hu, C. J. , Song, G. , Huang, W. , Liu, G. Z. et al., Identification of new autoantigens for primary biliary cirrhosis using human proteome microarrays. Mol. Cell. Proteomics 2012, 11, 669–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Felgner, P. L. , Kayala, M. A. , Vigil, A. , Burk, C. et al., A Burkholderia pseudomallei protein microarray reveals serodiagnostic and cross‐reactive antigens. Proc. Natl. Acad. Sci. USA 2009, 106, 13499–13504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ehrlich, J. R. , Qin, S. , Liu, B. C. , The ‘reverse capture’ autoantibody microarray: a native antigen‐based platform for autoantibody profiling. Nat. Protoc. 2006, 1, 452–460. [DOI] [PubMed] [Google Scholar]
- 11. Phizicky, E. , Bastiaens, P. I. , Zhu, H. , Snyder, M. , Fields, S. , Protein analysis on a proteomic scale. Nature 2003, 422, 208–215. [DOI] [PubMed] [Google Scholar]
- 12. Zhu, H. , Bilgin, M. , Bangham, R. , Hall, D. et al., Global analysis of protein activities using proteome chips. Science 2001, 293, 2101–2105. [DOI] [PubMed] [Google Scholar]
- 13. Yu, X. , LaBaer, J. , High‐throughput identification of proteins with AMPylation using self‐assembled human protein (NAPPA) microarrays. Nat. Protoc. 2015, 10, 756–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yu, X. , Schneiderhan‐Marra, N. , Joos, T. O. , Protein microarrays for personalized medicine. Clin. Chem. 2010, 56, 376–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lu, K. Y. , Tao, S. C. , Yang, T. C. , Ho, Y. H. et al., Profiling lipid‐protein interactions using nonquenched fluorescent liposomal nanovesicles and proteome microarrays. Mol. Cell. Proteomics 2012, 11, 1177–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chung, A. W. , Kumar, M. P. , Arnold, K. B. , Yu, W. H. et al., Dissecting polyclonal vaccine‐induced humoral immunity against HIV using systems serology. Cell 2015, 163, 988–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang, B. , Kumar, R. B. , Dai, H. , Feldman, B. J. , A plasmonic chip for biomarker discovery and diagnosis of type 1 diabetes. Nat. Med. 2014, 20, 948–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bian, X. , Wallstrom, G. , Davis, A. , Wang, J. et al., Immunoproteomic profiling of anti‐viral antibodies in new‐onset Type 1 diabetes using protein arrays. Diabetes 2015, 65, 285–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yu, X. , Bian, X. , Throop, A. , Song, L. et al., Exploration of panviral proteome: high‐throughput cloning and functional implications in virus‐host interactions. Theranostics 2014, 4, 808–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Stafford, P. , Cichacz, Z. , Woodbury, N. W. , Johnston, S. A. , Immunosignature system for diagnosis of cancer. Proc. Natl. Acad. Sci. USA 2014, 111, E3072–E3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Legutki, J. B. , Zhao, Z. G. , Greving, M. , Woodbury, N. et al., Scalable high‐density peptide arrays for comprehensive health monitoring. Nat. Commun. 2014, 5, 4785. [DOI] [PubMed] [Google Scholar]
- 22. Mock, A. , Warta, R. , Geisenberger, C. , Bischoff, R. et al., Printed peptide arrays identify prognostic TNC serumantibodies in glioblastoma patients. Oncotarget 2015, 6, 13579–13590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xu, G. J. , Kula, T. , Xu, Q. , Li, M. Z. et al., Viral immunology. Comprehensive serological profiling of human populations using a synthetic human virome. Science 2015, 348, aaa0698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Katz, C. , Levy‐Beladev, L. , Rotem‐Bamberger, S. , Rito, T. et al., Studying protein‐protein interactions using peptide arrays. Chem. Soc. Rev. 2011, 40, 2131–2145. [DOI] [PubMed] [Google Scholar]
- 25. Chandra, H. , Reddy, P. J. , Srivastava, S. , Protein microarrays and novel detection platforms. Expert Rev. Proteomics 2011, 8, 61–79. [DOI] [PubMed] [Google Scholar]
- 26. Ladd, J. J. , Chao, T. , Johnson, M. M. , Qiu, J. et al., Autoantibody signatures involving glycolysis and splicesome proteins precede a diagnosis of breast cancer among postmenopausal women. Cancer Res. 2013, 73, 1502–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ehrlich, J. R. , Tang, L. , Caiazzo, R. J., Jr. , Cramer, D. W. et al., The "reverse capture" autoantibody microarray : an innovative approach to profiling the autoantibody response to tissue‐derived native antigens. Methods Mol. Biol. 2008, 441, 175–192. [DOI] [PubMed] [Google Scholar]
- 28. Gonzalez‐Gonzalez, M. , Jara‐Acevedo, R. , Matarraz, S. , Jara‐Acevedo, M. et al., Nanotechniques in proteomics: protein microarrays and novel detection platforms. Eur. J. Pharm. Sci. 2012, 45, 499–506. [DOI] [PubMed] [Google Scholar]
- 29. He, M. , Taussig, M. J. , Single step generation of protein arrays from DNA by cell‐free expression and in situ immobilisation (PISA method). Nucleic Acids Res. 2001, 29, E73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. He, M. , Stoevesandt, O. , Palmer, E. A. , Khan, F. et al., Printing protein arrays from DNA arrays. Nat. Methods 2008, 5, 175–177. [DOI] [PubMed] [Google Scholar]
- 31. Ramachandran, N. , Raphael, J. V. , Hainsworth, E. , Demirkan, G. et al., Next‐generation high‐density self‐assembling functional protein arrays. Nat. Methods 2008, 5, 535–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ramachandran, N. , Hainsworth, E. , Bhullar, B. , Eisenstein, S. et al., Self‐assembling protein microarrays. Science 2004, 305, 86–90. [DOI] [PubMed] [Google Scholar]
- 33. Tao, S. C. , Zhu, H. , Protein chip fabrication by capture of nascent polypeptides. Nat. Biotechnol. 2006, 24, 1253–1254. [DOI] [PubMed] [Google Scholar]
- 34. Davies, D. H. , Liang, X. , Hernandez, J. E. , Randall, A. et al., Profiling the humoral immune response to infection by using proteome microarrays: high‐throughput vaccine and diagnostic antigen discovery. Proc. Natl. Acad. Sci. USA 2005, 102, 547–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Helb, D. A. , Tetteh, K. K. , Felgner, P. L. , Skinner, J. et al., Novel serologic biomarkers provide accurate estimates of recent Plasmodium falciparum exposure for individuals and communities. Proc. Natl. Acad. Sci. USA 2015, 112, E4438–E4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lee, S. J. , Liang, L. , Juarez, S. , Nanton, M. R. et al., Identification of a common immune signature in murine and human systemic Salmonellosis. Proc. Natl. Acad. Sci. USA 2012, 109, 4998–5003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kunnath‐Velayudhan, S. , Salamon, H. , Wang, H. Y. , Davidow, A. L. et al., Dynamic antibody responses to the Mycobacterium tuberculosis proteome. Proc. Natl. Acad. Sci. USA 2010, 107, 14703–14708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yu, X. , Decker, K. B. , Barker, K. , Neunuebel, M. R. et al., Host‐pathogen interaction profiling using self‐assembling human protein arrays. J. Proteome Res. 2015, 14, 1920–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yu, X. , Woolery, A. R. , Luong, P. , Hao, Y. H. et al., Copper‐catalyzed azide‐alkyne cycloaddition (click chemistry)‐based detection of global pathogen‐host AMPylation on self‐assembled human protein microarrays. Mol. Cell. Proteomics 2014, 13, 3164–3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Takulapalli, B. R. , Qiu, J. , Magee, D. M. , Kahn, P. et al., High density diffusion‐free nanowell arrays. J. Proteome Res. 2012, 11, 4382–4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bian, X. , Wiktor, P. , Kahn, P. , Brunner, A. et al., Antiviral antibody profiling by high‐density protein arrays. Proteomics 2015, 15, 2136–2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang, J. , Barker, K. , Steel, J. , Park, J. et al., A versatile protein microarray platform enabling antibody profiling against denatured proteins. Proteomics Clin. Appl. 2013, 7, 378–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang, J. , Figueroa, J. D. , Wallstrom, G. , Barker, K. et al., Plasma autoantibodies associated with basal‐like breast cancers. Cancer Epidemiol. Biomarkers Prev. 2015, 24, 1332–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Manzano‐Roman, R. , Diaz‐Martin, V. , Gonzalez‐Gonzalez, M. , Matarraz, S. et al., Self‐assembled protein arrays from an Ornithodoros moubata salivary gland expression library. J Proteome Res 2012, 11, 5972–5982. [DOI] [PubMed] [Google Scholar]
- 45. Saul, J. , Petritis, B. , Sau, S. , Rauf, F. et al., Development of a full‐length human protein production pipeline. Protein Sci. 2014, 23, 1123–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yu, X. , Schneiderhan‐Marra, N. , Hsu, H. Y. , Bachmann, J. , Joos, T. O. , Protein microarrays: effective tools for the study of inflammatory diseases. Methods Mol. Biol. 2009, 577, 199–214. [DOI] [PubMed] [Google Scholar]
- 47. Slaastad, H. , Wu, W. , Goullart, L. , Kanderova, V. et al., Multiplexed immuno‐precipitation with 1725 commercially available antibodies to cellular proteins. Proteomics 2011, 11, 4578–4582. [DOI] [PubMed] [Google Scholar]
- 48. Middelburg, R. A. , Porcelijn, L. , Lardy, N. , Briet, E. , Vrielink, H. , Prevalence of leucocyte antibodies in the Dutch donor population. Vox Sanguinis 2011, 100, 327–335. [DOI] [PubMed] [Google Scholar]
- 49. Salmona, M. , Delarue, S. , Delaugerre, C. , Simon, F. , Maylin, S. , Clinical evaluation of BioPlex 2200 HIV Ag‐Ab, an automated screening method providing discrete detection of HIV‐1 p24 antigen, HIV‐1 antibody, and HIV‐2 antibody. J. Clin. Microbiol. 2014, 52, 103–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Madjar, H. , Role of breast ultrasound for the detection and differentiation of breast lesions. Breast Care 2010, 5, 109–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Anderson, K. S. , Sibani, S. , Wallstrom, G. , Qiu, J. et al., Protein microarray signature of autoantibody biomarkers for the early detection of breast cancer. J. Proteome Res. 2011, 10, 85–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. De Paoli, P. , Carbone, A. , Carcinogenic viruses and solid cancers without sufficient evidence of causal association. Int. J. Cancer 2013, 133, 1517–1529. [DOI] [PubMed] [Google Scholar]
- 53. Waterboer, T. , Sehr, P. , Michael, K. M. , Franceschi, S. et al., Multiplex human papillomavirus serology based on in situ‐purified glutathione s‐transferase fusion proteins. Clin. Chem. 2005, 51, 1845–1853. [DOI] [PubMed] [Google Scholar]
- 54. Petricoin, E. F., 3rd. , Espina, V. , Araujo, R. P. , Midura, B. et al., Phosphoprotein pathway mapping: Akt/mammalian target of rapamycin activation is negatively associated with childhood rhabdomyosarcoma survival. Cancer Res. 2007, 67, 3431–3440. [DOI] [PubMed] [Google Scholar]
- 55. Chen, P. C. , Syu, G. D. , Chung, K. H. , Ho, Y. H. et al., Antibody profiling of bipolar disorder using Escherichia coli proteome microarrays. Mol. Cell. Proteomics 2015, 14, 510–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sjoberg, R. , Mattsson, C. , Andersson, E. , Hellstrom, C. et al., Exploration of high‐density protein microarrays for antibody validation and autoimmunity profiling. N. Biotechnol. 2015, pii: S1871–6784(15)00154‐5. [DOI] [PubMed] [Google Scholar]
- 57. Mortality, G. B. D. , Causes of death, C., global, regional, and national age‐sex specific all‐cause and cause‐specific mortality for 240 causes of death, 1990–2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet 2015, 385, 117–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Price, J. V. , Haddon, D. J. , Kemmer, D. , Delepine, G. et al., Protein microarray analysis reveals BAFF‐binding autoantibodies in systemic lupus erythematosus. J. Clin. Invest. 2013, 123, 5135–5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Miersch, S. , Bian, X. , Wallstrom, G. , Sibani, S. et al., Serological autoantibody profiling of type 1 diabetes by protein arrays. J. Proteomics 2013, 94, 486–496. [DOI] [PubMed] [Google Scholar]
- 60. Lu, D. D. , Chen, S. H. , Zhang, S. M. , Zhang, M. L. et al., Screening of specific antigens for SARS clinical diagnosis using a protein microarray. Analyst 2005, 130, 474–482. [DOI] [PubMed] [Google Scholar]
- 61. Chen, Y. W. , Teng, C. H. , Ho, Y. H. , Jessica Ho, T. Y. et al., Identification of bacterial factors involved in type 1 fimbria expression using an Escherichia coli K12 proteome chip. Mol. Cell. Proteomics 2014, 13, 1485–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Fan, B. , Lu, K. Y. , Reymond Sutandy, F. X. , Chen, Y. W. et al., A human proteome microarray identifies that the heterogeneous nuclear ribonucleoprotein K (hnRNP K) recognizes the 5’ terminal sequence of the hepatitis C virus RNA. Mol. Cell. Proteomics 2014, 13, 84–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ho, Y. H. , Sung, T. C. , Chen, C. S. , Lactoferricin B inhibits the phosphorylation of the two‐component system response regulators BasR and CreB. Mol. Cell. Proteomics 2012, 11, M111 014720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zhu, H. , Hu, S. , Jona, G. , Zhu, X. et al., Severe acute respiratory syndrome diagnostics using a coronavirus protein microarray. Proc. Natl. Acad. Sci. USA 2006, 103, 4011–4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Deng, J. , Bi, L. , Zhou, L. , Guo, S. J. et al., Mycobacterium tuberculosis proteome microarray for global studies of protein function and immunogenicity. Cell Rep. 2014, 9, 2317–2329. [DOI] [PubMed] [Google Scholar]
- 66. Prados‐Rosales, R. , Carreno, L. J. , Batista‐Gonzalez, A. , Baena, A. et al., Mycobacterial membrane vesicles administered systemically in mice induce a protective immune response to surface compartments of Mycobacterium tuberculosis. mBio 2014, 5, e01921–e01914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Chiang, I. H. , Suo, J. , Bai, K. J. , Lin, T. P. et al., Serodiagnosis of tuberculosis. A study comparing three specific mycobacterial antigens. Am. J. Respir. Crit. Care Med. 1997, 156, 906–911. [DOI] [PubMed] [Google Scholar]
- 68. Doolan, D. L. , Mu, Y. , Unal, B. , Sundaresh, S. et al., Profiling humoral immune responses to P. falciparum infection with protein microarrays. Proteomics 2008, 8, 4680–4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Crompton, P. D. , Kayala, M. A. , Traore, B. , Kayentao, K. et al., A prospective analysis of the Ab response to Plasmodium falciparum before and after a malaria season by protein microarray. Proc. Natl. Acad. Sci. USA 2010, 107, 6958–6963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Fang, Y. , Frutos, A. G. , Lahiri, J. , Membrane protein microarrays. J. Am. Chem. Soc. 2002, 124, 2394–2395. [DOI] [PubMed] [Google Scholar]
- 71. List, C. , Qi, W. , Maag, E. , Gottstein, B. et al., Serodiagnosis of Echinococcus spp. infection: explorative selection of diagnostic antigens by peptide microarray. PLoS Negl. Trop. Dis. 2010, 4, e771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Hong, Y. , Webb, B. L. , Su, H. , Mozdy, E. J. et al., Functional GPCR microarrays. J. Am. Chem. Soc. 2005, 127, 15350–15351. [DOI] [PubMed] [Google Scholar]
- 73. Hong, Y. , Webb, B. L. , Pai, S. , Ferrie, A. et al., G‐protein‐coupled receptor microarrays for multiplexed compound screening. J. Biomol. Screen 2006, 11, 435–438. [DOI] [PubMed] [Google Scholar]
- 74. Hu, S. , Feng, Y. , Henson, B. , Wang, B. et al., VirD: a virion display array for profiling functional membrane proteins. Anal. Chem. 2013, 85, 8046–8054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Yu, X. , Talukder, P. , Bhattacharya, C. , Fahmi, N. E. et al., Probing of CD4 binding pocket of HIV‐1 gp120 glycoprotein using unnatural phenylalanine analogues. Bioorg. Med. Chem. Lett. 2014, 24, 5699–5703. [DOI] [PubMed] [Google Scholar]
- 76. Fitzgerald, S. P. , Lamont, J. V. , McConnell, R. I. , Benchikh el, O. , Development of a high‐throughput automated analyzer using biochip array technology. Clin. Chem. 2005, 51, 1165–1176. [DOI] [PubMed] [Google Scholar]
- 77. Fung, E. T. , A recipe for proteomics diagnostic test development: the OVA1 test, from biomarker discovery to FDA clearance. Clin. Chem. 2010, 56, 327–329. [DOI] [PubMed] [Google Scholar]
- 78. Brodel, A. K. , Wustenhagen, D. A. , Kubick, S. , Cell‐free protein synthesis systems derived from cultured mammalian cells. Methods Mol. Biol. 2015, 1261, 129–140. [DOI] [PubMed] [Google Scholar]
- 79. Seiler, C. Y. , Park, J. G. , Sharma, A. , Hunter, P. et al., DNASU plasmid and PSI:Biology‐Materials repositories: resources to accelerate biological research. Nucleic Acids Res. 2014, 42, D1253–D1260. [DOI] [PMC free article] [PubMed] [Google Scholar]


