Abstract
Recent advances in proteomics have been combined with traditional methods for isolation of nucleoli from mammalian and plant cells. This approach has confirmed the growing body of data showing a wide role for the nucleolus in eukaryotic cell biology beyond ribosome generation into many areas of cell function from regulation of the cell cycle, modulation of the cell stress response to innate immune responses. This has been reflected in the growing body of evidence that viruses specifically target the nucleolus by sequestering cellular nucleolar proteins or by targeting viral proteins to the nucleolus in order to maximise viral replication. This review covers those key areas and looks at the latest approaches using high‐throughput quantitative proteomics of the nucleolus in virus infected cells to gain an insight into the role of this fascinating compartment in viral infection.
Keywords: Microbiology, Nucleolus, Virology
1 Introduction
Although the nucleolus is traditionally the site of rRNA synthesis and ribosome assembly, over the last 20 years or so it has become clear that the nucleolus plays a role in a wide range of important cellular processes 1, 2, 3. For example, the nucleolus functions as a stress sensor; in UV irradiated cells only nucleolar damage induced p53 stabilisation and not DNA damage itself 4. The tumour suppressor protein, ARF, is stored in the nucleolus and is part of a complex pathway involving nucleolar antigen B23.1 and HDM2 to control p53 levels 5, 6, 7, 8, 9. In cell cycle control, the RENT complex helps govern cell cycle progression 10, 11, 12, 13. On a wider level of global control, SUMO specific proteases SENP3 and SENP5 are primarily nucleolar localised 14, 15. Even classical nucleolar antigens like nucleophosmin (B23.1), which have been shown to play a role in rRNA processing, have wider roles. For example, B23.1 helps to control centrosome duplication during mitosis 16, 17, 18, 19, 20. Thus, the nucleolus plays a more extensive role than that of ribosome biogenesis.
Given this information, it seems natural to attempt to understand the functions of the nucleolus in terms of resident proteins. This proteomic approach to understanding the role of the nucleolus in cell function has relied on efficient, robust purification of nucleoli from standard cell lines and the advent of high‐throughput quantitative proteomics.
2 Isolating nucleoli
Nucleoli can be isolated from cells in a relatively straightforward fractionation experiment developed some time ago 21, 22, 23. Cells are hypotonically swollen and the cytoplasm is sheared off using a dounce homogeniser. Next, the nuclei are enriched by centrifugation over a sucrose cushion. Purified nuclei are then gently sonicated to release the nucleoli, which are further separated from the nucleoplasm by centrifugation through another sucrose cushion. With care and practice, highly enriched nucleoli can be isolated in this manner. A key aspect of this protocol is the testing of the various fractions to ensure the high‐quality enrichment of nucleoli from the cytoplasm and nucleoplasm. This is normally monitored by Western blotting of the fractions with antiserum to classical components of each fraction, e.g. nuclear lamin should only be present in the nucleoplasmic fraction, nucleolin enriched in the nucleolar fraction.
Coupling this fractionation with the latest high‐throughput techniques has been pioneered by groups lead by A. Lamond, J. Diaz and M. Mann 24, 25, 26, 27. This has culminated in a comprehensive data set covering some 4000 members of the mammalian nucleolar proteome (http://www.lamondlab.com/NOPdb3.0). In parallel, the nucleolar proteome of the model plant system Arabidopsis has been analysed 28 in similar detail using isolation techniques specific for plant cells covering just over 200 proteins (http://bioinf.scri.sari.ac.uk/cgi-bin/atnopdb/home).
What these studies have revealed is that nucleoli contain many proteins whose primary functions are not related to ribosome synthesis or biogenesis. As stated earlier, there are a range of proteins present whose primary functions range from cell cycle control to p53 regulation 3. Comparison of the plant and mammalian nucleoli reveals intriguing differences that presumably reflect different evolutionary pathways. For example, very few proteins involved in mRNA transport are found in the mammalian cell nucleolus, whereas a number of mRNA transport factors are found in the plant cell nucleolus 28.
3 Dynamic proteomics of the nucleolus
The dynamic nature of the nucleolar proteome was illustrated by coupling high‐throughput proteomic analysis with stable isotope labelling with amino acids in cell culture (SILAC) 26. This technique compares two populations of cells: one labelled with normal 12C and 14N arginine and lysine; the other labelled with 13C and 15N labelled arginine and lysine for example. The cells grown in heavy labelled media are treated in this case with actinomycin D (ActD) which inhibits rRNA synthesis. The nucleoli are isolated from normal cells with normal amino acids and combined with nucleoli isolated from heavy labelled cells that have been exposed to ActD. These combined samples are separated by 1‐D electrophoresis and the gel divided into a number of slices. Normally, the more slices the lane is divided into the more sensitive and accurate the process of detection, identification and quantitation becomes because of reduction in sample complexity. Each slice is in‐gel digested with trypsin and the peptides are analysed by LC‐MS/MS. Since each peptide is chemically identical, both the light and heavy peptides are identified, detected and quantified together by MS/MS allowing changes in abundance to be inferred. After suitable analysis, the data returned represents a ratio or fold change in the quantity of each identified protein. That is to say this approach determines the relative enrichment or depletion of proteins in the nucleoli (or indeed any appropriately paired sample).
Using this technique has enabled a proteomic analysis of the changes in the nucleolar proteome over time in response to different treatments. For example, after ActD treatment, nearly a third of just over 500 nucleolar proteins analysed experienced at least a twofold change in their abundance. The types of proteins affected were as widespread as the nucleolar proteome itself, covering DEAD box helicases to snRNP proteins 26. Indeed, the authors conclude that the nucleolar proteome is not a fixed object but one that comprises overlapping sets of proteins that are present depending on the conditions of the cell at any one time.
A larger scale development of this approach uses triple labelling of cells to examine the content of the nucleolus, nucleoplasm and cytoplasm of cells in response to DNA damage. In this experiment, cells were grown in either light (12C14N l‐arginine and l‐lysine), or medium (l‐arginine‐13C6 14N4 and l‐lysine‐2H4) or heavy (l‐arginine‐13C6‐15N4 and l‐lysine‐13C6‐15N2) growth media. The cells were fractionated into nucleolus, nucleoplasm and cytoplasm and the fractions were recombined such that each subcellular component had a different isotopic label. In this way, proteins could be identified that were enriched in one compartment or present in two compartments or even distributed between all three. In the next step, cells were exposed to etoposide and the process repeated. In this manner, proteins that changed subcellular compartments in response to DNA damage could be tracked. In the case of nucleolar antigens, it seems that DNA damage increases the concentration of nucleolar antigens in the nucleolus relative to other compartments.
Proteomic analysis of the nucleolus confirms independent evidence that a wide range of proteins are present in this structure. In many cases, the nucleolus seems to act as a temporary storage or sequestration site that presumably facilitates a rapid response to stimuli.
4 Viruses and the nucleolus
Recently, we have become interested in the role of the nucleolus in viral infections. Over the last 10 years or so, it has become clear that the majority of viruses disrupt nucleolar function and/or they make proteins that are directed to the nucleolus. Perhaps, the defining viral nucleolar interaction is the observation that HIV uses the nucleolus to traffic its mRNA from the nucleus to the cytoplasm 29. Indeed, the interaction between HIV and the nucleolus has been shown to be critical since inhibiting the trafficking of HIV mRNA through the nucleolus effectively ablates viral replication 30, 31, 32, 33. From this body of work, it has emerged that modified T cell lines that inhibit the virus's ability to effectively use the nucleolus as a trafficking pathway are resistant to HIV infection.
This mRNA export strategy has recently been shown to be the case for a very distinct virus, Herpesvirus Saimiri (HVS) 34. In both cases, the reasons for using the nucleolus are clear – both viruses rely on mRNA that is not suited for efficient transport via normal mRNA export systems. In the case of HIV, incomplete splicing of viral transcripts make them a target for nonsense‐mediated decay and in the case of HVS most of the transcripts are intronless and would not therefore enter the normal mRNA export pathways, which are closely linked to splicing. What is intriguing is that both viruses have hijacked the nucleolus as a suitable (or even necessary) staging post for the export of their “aberrant” mRNA.
A number of reviews have highlighted that nucleolar interactions have important implications in the many distinct virus life cycles 35, 36, 37, 38. For example, a large number of virus‐encoded proteins traffic to and from the nucleolus. In addition, numerous host cell nucleolar proteins are redistributed to other cellular localisations during the virus replication cycle. However, the implications of these nucleolar modifications on the virus replication and host cell function have yet to be fully elucidated. Here, we highlight several examples that nucleolar proteins play essential roles in multiple steps of the virus replication cycle, from transcriptional regulation and RNA processing to virus entry and egress (summarised in Fig. 1).
Figure 1.
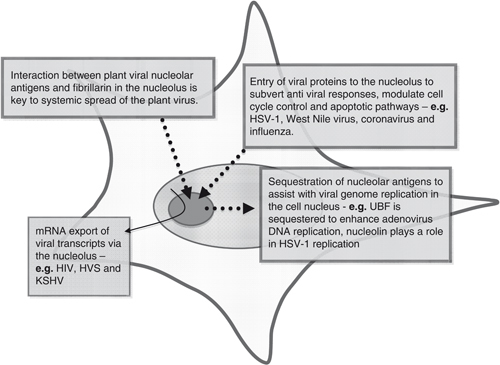
A schematic diagram of the cell with the nucleolus in dark green at the centre. This presents an overview of some of the role of the nucleolus in the life cycle of different viruses. Some viral proteins are directed to the nucleolus to aid viral spread either directly or by affecting apoptotic pathways for example. Some cellular nucleolar antigens are sequestered to aid viral replication and some viruses create a viral mRNA export pathway via the nucleolus.
5 DNA viruses
A large number of viruses with DNA genomes have been shown to interact with nucleolus, and this perhaps is not surprising as most DNA viruses replicate in the nucleus. For example, a genome‐wide screen of three distinct herpesviruses, herpes simplex virus 1 (HSV‐1), cytomegalovirus (CMV) and Epstein–Barr virus (EBV), has shown that at least 12 herpesvirus‐encoded proteins specifically localise to the nucleolus 39, which are implicated in many aspects of the herpesvirus life cycle. Therefore a number of proteomic studies are currently being undertaken to study changes, in a global context, within the nucleolar proteome during virus infections.
A significant area of virus biology currently being investigated is the role of viral proteins that traffic through the nucleolus. For example, a number of HIV proteins that traffic through the nucleolus have been implicated in virus mRNA processing 40, 41, 42, 43. Similar observations have also been made in herpesviruses. Initial studies utilising the prototype γ‐2 herpesvirus, HVS, demonstrated that the HVS nucleolar trafficking ORF57 protein induces nucleolar redistribution of the host cell human TREX proteins, which are involved in mRNA nuclear export 34. Intriguingly, ablating ORF57 nucleolar trafficking led to a failure of ORF57‐mediated viral mRNA nuclear export 34. The precise role of this nucleolar sequestration is yet to be determined, but possible effects on viral mRNA/protein processing and viral ribonucleoprotein particle assembly are currently being investigated. This property may also be conserved in other ORF57 homologues as recent analysis has shown that the ORF57 protein from Kaposi's sarcoma associated herpesvirus (KSHV) also dynamically traffics through the nucleolus 44. Moreover, upon the rapid disorganisation of the nucleolus a reduction is observed in virus mRNA nuclear export 45. The formation of an ORF57‐mediated export competent ribonucleoprotein particle within the nucleolus may also have implications for the translation of viral mRNAs. For example, it has recently been demonstrated that the cellular nucleo‐cytoplasmic shuttle protein, PYM, which is involved in translation enhancement, is redistributed to the nucleolus in the presence of the KSHV ORF57 protein 46. This interaction effectively enhances the translation of the predominantly intronless transcripts made by KSHV.
A second area of virus replication where nucleolar proteins are sequestered involves the replication of the virus DNA genome. For example, we and others have observed that nucleolar antigens upstream binding factor (UBF) and B23.1 are both sequestered into adenovirus DNA replication centres where they promote viral DNA replication 47, 48, 49, 50. Similarly, in HSV‐1 infected cells, a number of nucleolar proteins including nucleolin and UBF are recruited into viral DNA replication centres 51, 52. These are specific sites where replication and encapsidation of the HSV‐1 genome occurs. Evidence suggests that sequestration of UBF is essential for viral DNA replication as overexpression of tagged version of UBF act in a dominant‐negative fashion inhibiting virus DNA replication 52. Moreover, depletion of nucleolin results in reduced virus gene expression and infectious virion production 53, 54.
In addition to enhancing virus replication, nucleolar proteins are redistributed to alter cellular pathways during infection. For example, the nucleolar targeted HSV‐1 Us11 protein has been shown to interact with HIPK2, which plays a role in p53‐mediated cellular apoptosis and also participates in the regulation of the cell cycle. This interaction alters the subcellular localisation of HIPK2 and protects against HIPK2‐mediated cell cycle arrest 55. In contrast, the cellular protein, PICT‐1, can sequester the virally encoded apoptosis suppressor protein, KS‐Bcl‐2 protein, from the mitochondria into the nucleolus to downregulate its anti‐apoptotic activity 56. In animal models, nucleolar protein ARF has been show to play a protective role in innate immune responses to viral infection 57.
6 RNA viruses
The interaction of many of the RNA viruses with nucleolus on first inspection was unusual 58. In general, with the exception of influenza virus and several others, this group of viruses employs cytoplasmic replication strategies with all of the viral RNA synthesis and assembly occurring on membrane‐bound structures in the cytoplasm. Nevertheless, for many of these viruses examples can be found of viral proteins localising to the nucleolus with potential alterations in the nucleolar architecture and proteome.
One of the most well‐characterised examples in terms of localisation and functional relevance is with the nidovirus nucleocapsid (N) protein. The nidovirales are a group of viruses that incorporates the positive strand RNA viruses coronaviruses and arteriviruses and have similar genomic organisation and replication strategies. The N protein is a phosphoprotein that complexes and encapsidates the viral genomic RNA 59 and is therefore crucial to virus assembly. However, both the coronavirus 60 and arterivirus 61 N proteins localise to the cytoplasm and nucleolus during virus infection, and with the coronavirus N protein this may be cell cycle dependent 62. For both proteins, nucleolar localisation signals and nuclear export mechanisms have been characterised 63, 64, 65, 66.
Functionally, the nucleolar localisation of these proteins may be important for the virus life cycle. Blocking the nuclear export of the equine arteritis virus protein with the CRM1 dependent inhibitor leptomycin B resulted in the immediate retention of N protein in the nucleus and nucleolus, suggesting that the protein shuttles between the nucleus and the cytoplasm prior to its role in virus assembly 65. Live cell imaging analysis of fluorescently tagged N proteins coupled to FLIP and FRAP photo‐bleaching analysis supported these observations 67, 68. More definitively, in the context of a recombinant infectious virus, mutation of the arterivirus porcine reproductive and respiratory syndrome virus (PRRSV) N protein NoLS resulted in the retention of N protein in the cytoplasm, abrogation of nucleolar localisation and attenuation of virus replication 69, 70. In both infection in cell culture and in vivo, the genotypic alterations to the NoLS in the infectious recombinant virus were repaired and selected for through random mutation with the resulting phenotypic restoration of cytoplasmic and nucleolar localisation of the N protein and resulting of wild‐type pathogenicity 69. Another set of examples can be found in the filoviridae where there are numerous examples of West Nile virus and dengue virus interacting with the host cell nucleolus 71, 72, 73. Indeed, in the case of West Nile virus, there is evidence that the interaction with the nucleolus affects p53 stability through the ARF/HDM2/p53 pathway 73. Some RNA virus proteins that localise to the nucleolus have also been shown to interact with nucleolar proteins such as nucleolin 74 and fibrillarin 75. For the plant RNA umbraviruses, interaction with the nucleolus and nucleolar proteomes was shown to be crucial for systemic infection 76, 77. Together, these results clearly demonstrate the functional importance of the localisation of RNA virus proteins to the nucleolus during the virus life cycle and that abrogation of this interaction can be used in the generation of live‐attenuated recombinant vaccines. This has been most clearly demonstrated with the work on PRRSV by Pei et al. 70. From previous studies, mutant PRRSV that encoded N proteins with defective NLSs illustrated that wild‐type phenotypes were rapidly selected for and restored. Pei et al. generated NLS reversion resistant mutants. These progeny viruses were genetically stable for at least 20 passages in cell culture. More importantly, work in swine demonstrated that infection with the mutant viruses resulted in less viremia and of lower titre and shorter duration than wild‐type virus. More importantly, there was increased production of neutralising antibodies associated with the mutant viruses. This then is an ideal live attenuated vaccine candidate, in that the virus causes less or no disease but still stimulates the immune system. It would be interesting to determine whether viruses with N protein NoLS mutations also confer protection when animals are challenged with wild‐type virus.
The localisation of RNA virus proteins to the nucleolus and their interactions with nucleolar proteins may also have consequences on nucleolar function 78. Certainly, at a gross level, nucleolar morphology can change during RNA virus infection 79. As a first stage in investigating this, high‐throughput quantitative proteomics using SILAC have been applied by us to examine the nucleolar proteome in RNA virus infected cells focusing on the avian coronavirus 80 and influenza A virus (unpublished data). Strikingly, very similar to the findings recently reported by us for the DNA virus adenovirus 81, global changes to the nucleolar proteome do not occur and are restricted to selected nucleolar proteins. This suggests that the nucleolus may be broadly robust during virus infection despite the localisation of exogenous (viral) proteins and sequestration of cellular nucleolar proteins by viral infection.
7 Nucleolar proteomics and viral infection
Given the many roles of the nucleolus in the cell life cycle, including its role as a stress sensor, it would seem reasonable that comprehensive unbiased analysis of the nucleolar proteome would yield interesting data. We recently showed that SILAC based high‐throughput quantitative proteomics could be used to examine the nucleolus of human cells infected with adenovirus. Human adenoviruses are dsDNA viruses with a genome of approximately 360 000 bp condensed within an icosahedral protein capsid. Viral DNA and proteins enter the nucleus of the cell within an hour of attachment; indeed, we have shown that at least one viral capsid protein (protein V) is delivered to the nucleolus at this stage implying that adenovirus interaction with the nucleolus appears to be throughout the life cycle of the virus 82. Viral replication is primarily within the nucleus of the cell with the cell eventually succumbing 24–36 h later, releasing up to 10 000 infectious new virus particles. It has been known for some time that adenovirus infection has a profound effect on the processing and export of rRNA as well as affecting nucleolar antigens 83. More recently, we and others have reported on adenovirus proteins that are directed to the nucleolus and on the sequestration of nucleolar antigens B23.1, B23.2, nucleolin and UBF into viral replication centres 47, 48, 84, 85, 86, 87, 88, 89, 90, 91, 92. Most notably, we showed that UBF was functionally recruited into viral DNA replication centres apparently without affecting the function of RNA pol I. This finding was significant in that UBF is known to directly regulate the activity of RNA pol I, ultimately affecting the rate of rRNA synthesis 93, 94, 95, 96, 97. Moreover, the sequestration of UBF away from RNA pol I was unique to adenovirus infection – experiments in uninfected cells showing sequestration of UBF to non‐nucleolar locations also recruited RNA pol I 94. This provided us with a unique control when we examined the nucleolar proteome using high‐throughput quantitative MS 98.
For this experiment, approximately 108 HeLa cells that were grown in heavy labelled media were infected with adenovirus at a multiplicity of infection of 5. After 18 h, the cells were harvested and nucleoli isolated and combined with an equal amount of nucleoli from unlabelled uninfected cells. Our analysis of these samples using high‐throughput quantitative approaches quantified ratios for 351 proteins. Of those, just 24 (7%) showed a twofold or greater change in quantitation between uninfected and infected nucleoli. At the same time, we compared samples of isolated nucleoli from infected and uninfected cells using more traditional 2‐DE, which confirmed that in fact the samples are very similar overall.
That just 7% of proteins identified showed a two‐fold or greater change indicates that viral infection targets a specific subset of nucleolar antigens. By comparison, almost a third of nucleolar antigens show a greater than twofold change when cells are treated with ActD which inhibits rRNA synthesis (see Fig. 2A). What is notable is that direct comparison between the adenovirus data set and the ActD dataset shows no clear correlation (Fig. 2B) – further supporting the case that adenovirus induces effects on the nucleolus distinct from that of a generalised, non‐specific shut down of nucleolar function. This fits well with our previous observation that adenovirus infection does not affect rRNA synthesis even 36 h post infection 48.
Figure 2.
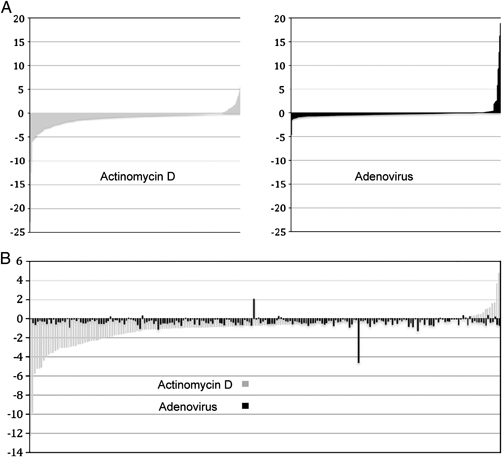
(A) Comparing overall profiles of changes in protein abundance in the nucleolus upon actinomycin D treatment or adenovirus infection. The left hand panel shows the ranges of observed fold changes in the nucleolus of actinomycin D treated cells at 180 minutes post treatment compared to untreated cells. On the right a similar graph shows the ranges of observed fold changes in proteins isolated from uninfected or adenovirus infected cells at 18 hours post infection. Both sets of data were collected in HeLa cells. (B) Comparing the effects of actinomycin D and adenovirus on the nucleolus by aligning proteins. Each pair of bars on the X axis represents an individual protein whose change in abundance in the nucleolus has a calculated ratio in both experiments which were in HeLa cells. In this example, black shows the ratio of change on adenovirus treatment and grey shows how the same protein changes abundance in the nucleolus on actinomycin D treatment.
One important feature of using the nucleolus as a source of proteomic information is that large numbers of proteins are primarily located elsewhere only having a low level presence in the nucleolus. In practice this means that alterations in the localisation of predominantly non‐nucleolar antigens is also highlighted, presumably since changes elsewhere impact on nucleolar levels as well.
Another useful observation is that examination of the nucleolar proteome gives a different, complementary view of the effects of viral infection on the cell compared to gene array studies. A very detailed analysis of the effect of viral infection on mRNA expression over time has been performed in HFFF cells. There are, of course, a number of difficulties in comparing the two sets of data, not least because they are in different cell types and the virus replicates with slightly slower kinetics in HFFF cells. In most cases, however, there is no real correlation between the two. Moreover, many proteins we observed to be depleted from the nucleolus are not being depleted as a result of mRNA levels dropping off (at least according to data generated in HFFF cells). Whilst we would ideally analyse proteomic and expression data side by side this comparison does help to illustrate the point that levels of mRNA and protein location studies can provide very different answers. This is more directly shown by our studies on nucleolar antigen UBF which regulates rRNA synthesis. UBF is depleted from the nucleolus as recorded by proteomic and microscopy approaches but overall levels of the protein in the cell are not affected as shown by Western blots 48, 98.
Building on this, in each of our laboratories, we have begun to examine the nucleolar proteome of a number of other viruses including KSHV, HSV‐1, infectious bronchitis virus 80, influenza and respiratory syncytial viruses. Whilst the data is relatively new, there are already encouraging signs that these types of analysis will prove highly informative. For example, there are intriguing similarities in the way the nucleolar proteome is affected by quite distinct viruses. Moreover, validation of findings by immunofluorescence (as was done for adenovirus) has again provided rapid reassurance that the MS/MS data are reflected in situ. Indeed, we feel that in situ approaches are essential to validate the proteomic data as it provides a rapid and wholly independent means of determining if there are changes in the nucleolar proteome on infection. Most recently, we have begun to examine the changes in the nucleolar proteome over time, which indicates there are distinct changes in the nucleolar proteome at different parts of the virus life cycle.
8 Perspectives
Thus, whilst studies of nucleolar proteome in uninfected cells are relatively well advanced, the application of these approaches to examine the role of the nucleolus in infection is just in its infancy. However, we anticipate that studies currently underway to examine the nucleolus of cells infected with different viruses and in cells expressing individual viral nucleolar antigens in isolation of infection will help us unravel the scope of this structure's role in the replication cycles of important pathogens. In turn, this should also prove highly informative in understanding the scope of influence of the nucleolus on the eukaryotic cell.
Acknowledgements
The authors thank the Wellcome Trust (DAM Grant No. 083604 and AW Grant No. 086168), Leverhulme Trust (JAH Grant No. RFG/11181) and BBSRC (AW Grant No. BB/G022836/1 and JAH DTG studentship) for funding. The authors also thank Sadie Young for assistance with the data analysis in Fig. 2.
The authors have declared no conflict of interest.
9 References
- 1. Pederson, T. , Tsai, R. Y. , In search of nonribosomal nucleolar protein function and regulation. J. Cell Biol. 2009, 184, 771–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pederson, T. , The plurifunctional nucleolus. Nucleic Acids Res. 1998, 26, 3871–3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Boisvert, F. M. , van Koningsbruggen, S. , Navascues, J. , Lamond, A. I. , The multifunctional nucleolus. Nat. Rev. Mol. Cell. Biol. 2007, 8, 574–585. [DOI] [PubMed] [Google Scholar]
- 4. Rubbi, C. P. , Milner, J. , Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. EMBO J. 2003, 22, 6068–6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gjerset, R. A. , DNA damage, p14ARF, nucleophosmin (NPM/B23), and cancer. J. Mol. Histol. 2006, 37, 239–251. [DOI] [PubMed] [Google Scholar]
- 6. Itahana, K. , Bhat, K. P. , Jin, A. , Itahana, Y. et al., Tumor suppressor ARF degrades B23, a nucleolar protein involved in ribosome biogenesis and cell proliferation. Mol. Cell 2003, 12, 1151–1164. [DOI] [PubMed] [Google Scholar]
- 7. Korgaonkar, C. , Hagen, J. , Tompkins, V. , Frazier, A. A. et al., Nucleophosmin (B23) targets ARF to nucleoli and inhibits its function. Mol. Cell. Biol. 2005, 25, 1258–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ivanchuk, S. M. , Mondal, S. , Rutka, J. T. , p14ARF interacts with DAXX: effects on HDM2 and p53. Cell Cycle 2008, 7, 1836–1850. [DOI] [PubMed] [Google Scholar]
- 9. Bothner, B. , Lewis, W. S. , DiGiammarino, E. L. , Weber, J. D. et al., Defining the molecular basis of Arf and Hdm2 interactions. J. Mol. Biol. 2001, 314, 263–277. [DOI] [PubMed] [Google Scholar]
- 10. Cockell, M. M. , Gasser, S. M. , The nucleolus: nucleolar space for RENT. Curr. Biol. 1999, 9, R575–576. [DOI] [PubMed] [Google Scholar]
- 11. Diaz‐Cuervo, H. , Bueno, A. , Cds1 controls the release of Cdc14‐like phosphatase Flp1 from the nucleolus to drive full activation of the checkpoint response to replication stress in fission yeast. Mol. Biol. Cell 2008, 19, 2488–2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hwang, W. W. , Madhani, H. D. , Nonredundant requirement for multiple histone modifications for the early anaphase release of the mitotic exit regulator Cdc14 from nucleolar chromatin. PLoS Genet. 2009, 5, e1000588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shou, W. , Sakamoto, K. M. , Keener, J. , Morimoto, K. W. et al., Net1 stimulates RNA polymerase I transcription and regulates nucleolar structure independently of controlling mitotic exit. Mol. Cell 2001, 8, 45–55. [DOI] [PubMed] [Google Scholar]
- 14. Yun, C. , Wang, Y. , Mukhopadhyay, D. , Backlund, P. et al., Nucleolar protein B23/nucleophosmin regulates the vertebrate SUMO pathway through SENP3 and SENP5 proteases. J. Cell Biol. 2008, 183, 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gong, L. , Yeh, E. T. , Characterization of a family of nucleolar SUMO‐specific proteases with preference for SUMO‐2 or SUMO‐3. J. Biol. Chem. 2006, 281, 15869–15877. [DOI] [PubMed] [Google Scholar]
- 16. Ferretti, R. , Palumbo, V. , Di Savino, A. , Velasco, S. et al., Morgana/chp‐1, a ROCK inhibitor involved in centrosome duplication and tumorigenesis. Dev. Cell 2010, 18, 486–495. [DOI] [PubMed] [Google Scholar]
- 17. Adon, A. M. , Zeng, X. , Harrison, M. K. , Sannem, S. et al., Cdk2 and Cdk4 regulate the centrosome cycle and are critical mediators of centrosome amplification in p53‐null cells. Mol. Cell. Biol. 2010, 30, 694–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ma, Z. , Kanai, M. , Kawamura, K. , Kaibuchi, K. et al., Interaction between ROCK II and nucleophosmin/B23 in the regulation of centrosome duplication. Mol. Cell. Biol 2006, 26, 9016–9034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang, W. , Budhu, A. , Forgues, M. , Wang, X. W. , Temporal and spatial control of nucleophosmin by the Ran‐Crm1 complex in centrosome duplication. Nat. Cell. Biol. 2005, 7, 823–830. [DOI] [PubMed] [Google Scholar]
- 20. Cha, H. , Hancock, C. , Dangi, S. , Maiguel, D. et al., Phosphorylation regulates nucleophosmin targeting to the centrosome during mitosis as detected by cross‐reactive phosphorylation‐specific MKK1/MKK2 antibodies. Biochem. J. 2004, 378, 857–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Busch, H. , Muramatsu, M. , Adams, H. , Steele, W. J. et al., Isolation of Nucleoli. Exp. Cell. Res. 1963, 24, 150–163. [PubMed] [Google Scholar]
- 22. Muramatsu, M. , Onishi, T. , Rapid isolation of nucleoli from detergent‐purified nuclei of tumor and tissue culture cells. Methods Cell Biol. 1977, 15, 221–234. [DOI] [PubMed] [Google Scholar]
- 23. Muramatsu, M. , Hayashi, Y. , Onishi, T. , Sakai, M. , Takai, K. , Rapid isolation of nucleoli from detergent purified nuclei of various tumor and tissue culture cells. Exp. Cell. Res. 1974, 88, 245–251. [DOI] [PubMed] [Google Scholar]
- 24. Scherl, A. , Coute, Y. , Deon, C. , Calle, A. et al., Functional proteomic analysis of human nucleolus. Mol. Biol. Cell 2002, 13, 4100–4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Andersen, J. S. , Lyon, C. E. , Fox, A. H. , Leung, A. K. et al., Directed proteomic analysis of the human nucleolus. Curr. Biol. 2002, 12, 1–11. [DOI] [PubMed] [Google Scholar]
- 26. Andersen, J. S. , Lam, Y. W. , Leung, A. K. , Ong, S. E. et al., Nucleolar proteome dynamics. Nature 2005, 433, 77–83. [DOI] [PubMed] [Google Scholar]
- 27. Ahmad, Y. , Boisvert, F. M. , Gregor, P. , Cobley, A. , Lamond, A. I. , NOPdb: Nucleolar Proteome Database – 2008 update. Nucleic Acids Res. 2009, 37, D181–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pendle, A. F. , Clark, G. P. , Boon, R. , Lewandowska, D. et al., Proteomic analysis of the Arabidopsis nucleolus suggests novel nucleolar functions. Mol. Biol. Cell 2005, 16, 260–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Michienzi, A. , Cagnon, L. , Bahner, I. , Rossi, J. J. , Ribozyme‐mediated inhibition of HIV 1 suggests nucleolar trafficking of HIV‐1 RNA. Proc. Natl. Acad. Sci. USA 2000, 97, 8955–8960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Michienzi, A. , Li, S. , Zaia, J. A. , Rossi, J. J. , A nucleolar TAR decoy inhibitor of HIV‐1 replication. Proc. Natl. Acad. Sci. USA 2002, 99, 14047–14052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Michienzi, A. , De Angelis, F. G. , Bozzoni, I. , Rossi, J. J. , A nucleolar localizing Rev binding element inhibits HIV replication. AIDS Res. Ther. 2006, 3, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Michienzi, A. , Castanotto, D. , Lee, N. , Li, S. et al., RNA‐mediated inhibition of HIV in a gene therapy setting. Ann. NY Acad. Sci. 2003, 1002, 63–71. [DOI] [PubMed] [Google Scholar]
- 33. Paul, C. P. , Good, P. D. , Li, S. X. , Kleihauer, A. et al., Localized expression of small RNA inhibitors in human cells. Mol. Ther. 2003, 7, 237–247. [DOI] [PubMed] [Google Scholar]
- 34. Boyne, J. R. , Whitehouse, A. , Nucleolar trafficking is essential for nuclear export of intronless herpesvirus mRNA. Proc. Natl. Acad. Sci. USA 2006, 103, 15190–15195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Olson, M. O. , Induction of apoptosis by viruses: what role does the nucleolus play? Cell Cycle 2009, 8, 3452–3453. [DOI] [PubMed] [Google Scholar]
- 36. Greco, A. , Involvement of the nucleolus in replication of human viruses. Rev. Med. Virol. 2009, 19, 201–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hiscox, J. A. , RNA viruses: hijacking the dynamic nucleolus. Nat. Rev. Microbiol. 2007, 5, 119–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hiscox, J. A. , The nucleolus – a gateway to viral infection? Arch. Virol. 2002, 147, 1077–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Salsman, J. , Zimmerman, N. , Chen, T. , Domagala, M. , Frappier, L. , Genome‐wide screen of three herpesviruses for protein subcellular localization and alteration of PML nuclear bodies. PLoS Pathog. 2008, 4, e1000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cao, Y. , Liu, X. , De Clercq, E. , Cessation of HIV‐1 transcription by inhibiting regulatory protein Rev‐mediated RNA transport. Curr. HIV Res. 2009, 7, 101–108. [DOI] [PubMed] [Google Scholar]
- 41. Ponti, D. , Troiano, M. , Bellenchi, G. C. , Battaglia, P. A. , Gigliani, F. , The HIV Tat protein affects processing of ribosomal RNA precursor. BMC Cell Biol. 2008, 9, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Peruzzi, F. , The multiple functions of HIV‐1 Tat: proliferation versus apoptosis. Front. Biosci. 2006, 11, 708–717. [DOI] [PubMed] [Google Scholar]
- 43. Dundr, M. , Leno, G. H. , Hammarskjold, M. L. , Rekosh, D. et al., The roles of nucleolar structure and function in the subcellular location of the HIV‐1 Rev protein. J. Cell. Sci. 1995, 108, 2811–2823. [DOI] [PubMed] [Google Scholar]
- 44. Boyne, J. R. , Colgan, K. J. , Whitehouse, A. , Recruitment of the complete hTREX complex is required for Kaposi's sarcoma‐associated herpesvirus intronless mRNA nuclear export and virus replication. PLoS Pathog. 2008, 4, e1000194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Boyne, J. R. , Whitehouse, A. , Nucleolar disruption impairs Kaposi's sarcoma‐associated herpesvirus ORF57‐mediated nuclear export of intronless viral mRNAs. FEBS Lett. 2009, 583, 3549–3556. [DOI] [PubMed] [Google Scholar]
- 46. Boyne, J. R. , Jackson, B. , Taylor, A. , Macnab, S. , Whitehouse, A. , KSHV ORF57 interacts with PYM to enhance the translation of viral intronless mRNAs. EMBO J. 2010, 29, 1851–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hindley, C. E. , Davidson, A. D. , Matthews, D. A. , Relationship between adenovirus DNA replication proteins and nucleolar proteins B23.1 and B23.2. J. Gen. Virol. 2007, 88, 3244–3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lawrence, F. J. , McStay, B. , Matthews, D. A. , Nucleolar protein upstream binding factor is sequestered into adenovirus DNA replication centres during infection without affecting RNA polymerase I location or ablating rRNA synthesis. J. Cell. Sci. 2006, 119, 2621–2631. [DOI] [PubMed] [Google Scholar]
- 49. Okuwaki, M. , Iwamatsu, A. , Tsujimoto, M. , Nagata, K. , Identification of nucleophosmin/B23, an acidic nucleolar protein, as a stimulatory factor for in vitro replication of adenovirus DNA complexed with viral basic core proteins. J. Mol. Biol. 2001, 311, 41–55. [DOI] [PubMed] [Google Scholar]
- 50. Samad, M. A. , Okuwaki, M. , Haruki, H. , Nagata, K. , Physical and functional interaction between a nucleolar protein nucleophosmin/B23 and adenovirus basic core proteins. FEBS Lett. 2007, 581, 3283–3288. [DOI] [PubMed] [Google Scholar]
- 51. Lymberopoulos, M. H. , Pearson, A. , Relocalization of upstream binding factor to viral replication compartments is UL24 independent and follows the onset of herpes simplex virus 1 DNA synthesis. J. Virol. 2010, 84, 4810–4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Stow, N. D. , Evans, V. C. , Matthews, D. A. , Upstream‐binding factor is sequestered into herpes simplex virus type 1 replication compartments. J. Gen. Virol. 2009, 90, 69–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Calle, A. , Ugrinova, I. , Epstein, A. L. , Bouvet, P. et al., Nucleolin is required for an efficient herpes simplex virus type 1 infection. J. Virol. 2008, 82, 4762–4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sagou, K. , Uema, M. , Kawaguchi, Y. , Nucleolin is required for efficient nuclear egress of herpes simplex virus type 1 nucleocapsids. J. Virol. 2010, 84, 2110–2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Giraud, S. , Diaz‐Latoud, C. , Hacot, S. , Textoris, J. et al., US11 of herpes simplex virus type 1 interacts with HIPK2 and antagonizes HIPK2‐induced cell growth arrest. J. Virol. 2004, 78, 2984–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kalt, I. , Borodianskiy‐Shteinberg, T. , Schachor, A. , Sarid, R. , GLTSCR2/PICT‐1, a putative tumor suppressor gene product, induces the nucleolar targeting of the Kaposi's sarcoma‐associated herpesvirus KS‐Bcl‐2 protein. J. Virol. 2010, 84, 2935–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Garcia, M. A. , Collado, M. , Munoz‐Fontela, C. , Matheu, A. et al., Antiviral action of the tumor suppressor ARF. EMBO J. 2006, 25, 4284–4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hiscox, J. A. , RNA viruses: hijacking the dynamic nucleolus. Nat. Rev. Microbiol. 2007, 5, 119–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chen, H. , Gill, A. , Dove, B. K. , Emmett, S. R. et al., Mass spectroscopic characterisation of the coronavirus infectious bronchitis virus nucleoprotein and elucidation of the role of phosphorylation in RNA binding using surface plasmon resonance. J. Virol. 2005, 79, 1164–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wurm, T. , Chen, H. , Britton, P. , Brooks, G. , Hiscox, J. A. , Localisation to the nucleolus is a common feature of coronavirus nucleoproteins and the protein may disrupt host cell division. J. Virol. 2001, 75, 9345–9356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Rowland, R. R. R. , Yoo, D. , Nucleolar‐cytoplasmic shuttling of PRRSV nucleocapsid protein: a simple case of molecular mimicry or the complex regulation by nuclear import, nucleolar localization and nuclear export signal sequences. Virus Res. 2003, 95, 23–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Cawood, R. , Harrison, S. M. , Dove, B. K. , Reed, M. L. , Hiscox, J. A. , Cell cycle dependent nucleolar localization of the coronavirus nucleocapsid protein. Cell Cycle 2007, 6, 863–867. [DOI] [PubMed] [Google Scholar]
- 63. Reed, M. L. , Dove, B. K. , Jackson, R. M. , Collins, R. et al., Delineation and modelling of a nucleolar retention signal in the coronavirus nucleocapsid protein. Traffic 2006, 7, 833–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Reed, M. L. , Howell, G. , Harrison, S. M. , Spencer, K. A. , Hiscox, J. A. , Characterization of the nuclear export signal in the coronavirus infectious bronchitis virus nucleocapsid protein. J. Virol. 2007, 81, 4298–4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Tijms, M. A. , van der Meer, Y. , Snijder, E. J. , Nuclear localization of non‐structural protein 1 and nucleocapsid protein of equine arteritis virus. J. Gen. Virol. 2002, 83, 795–800. [DOI] [PubMed] [Google Scholar]
- 66. Rowland, R. R. , Kerwin, R. , Kuckleburg, C. , Sperlich, A. , Benfield, D. A. , The localisation of porcine reproductive and respiratory syndrome virus nucleocapsid protein to the nucleolus of infected cells and identification of a potential nucleolar localization signal sequence. Virus Res. 1999, 64, 1–12. [DOI] [PubMed] [Google Scholar]
- 67. Emmott, E. , Dove, B. K. , Howell, G. , Chappell, L. A. et al., Viral nucleolar localisation signals determine dynamic trafficking within the nucleolus. Virology 2008, 380, 191–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. You, J. H. , Howell, G. , Pattnaik, A. K. , Osorio, F. A. , Hiscox, J. A. , A model for the dynamic nuclear/nucleolar/cytoplasmic trafficking of the porcine reproductive and respiratory syndrome virus (PRRSV) nucleocapsid protein based on live cell imaging. Virology 2008, 378, 34–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Lee, C. , Hodgins, D. , Calvert, J. G. , Welch, S. K. et al., Mutations within the nuclear localization signal of the porcine reproductive and respiratory syndrome virus nucleocapsid protein attenuate virus replication. Virology 2006, 346, 238–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Pei, Y. , Hodgins, D. C. , Lee, C. , Calvert, J. G. et al., Functional mapping of the porcine reproductive and respiratory syndrome virus capsid protein nuclear localization signal and its pathogenic association. Virus Res. 2008, 135, 107–114. [DOI] [PubMed] [Google Scholar]
- 71. Wang, S. H. , Syu, W. J. , Huang, K. J. , Lei, H. Y. et al., Intracellular localization and determination of a nuclear localization signal of the core protein of dengue virus. J. Gen. Virol. 2002, 83, 3093–3102. [DOI] [PubMed] [Google Scholar]
- 72. Sangiambut, S. , Keelapang, P. , Aaskov, J. , Puttikhunt, C. et al., Multiple regions in dengue virus capsid protein contribute to nuclear localization during virus infection. J. Gen. Virol. 2008, 89, 1254–1264. [DOI] [PubMed] [Google Scholar]
- 73. Yang, M. R. , Lee, S. R. , Oh, W. , Lee, E. W. et al., West Nile virus capsid protein induces p53‐mediated apoptosis via the sequestration of HDM2 to the nucleolus. Cell Microbiol. 2008, 10, 165–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Chen, H. , Wurm, T. , Britton, P. , Brooks, G. , Hiscox, J. A. , Interaction of the coronavirus nucleoprotein with nucleolar antigens and the host cell. J. Virol. 2002, 76, 5233–5250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Yoo, D. , Wootton, S. K. , Li, G. , Song, C. , Rowland, R. R. , Colocalization and interaction of the porcine arterivirus nucleocapsid protein with the small nucleolar RNA‐associated protein fibrillarin. J. Virol. 2003, 77, 12173–12183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Kim, S. H. , Macfarlane, S. , Kalinina, N. O. , Rakitina, D. V. et al., Interaction of a plant virus‐encoded protein with the major nucleolar protein fibrillarin is required for systemic virus infection. Proc. Natl. Acad. Sci. USA 2007, 104, 11115–11120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Kim, S. H. , Ryabov, E. V. , Kalinina, N. O. , Rakitina, D. V. et al., Cajal bodies and the nucleolus are required for a plant virus systemic infection. EMBO J. 2007, 26, 2169–2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Emmott, E. , Hiscox, J. A. , Nucleolar targetting: the hub of the matter. EMBO Rep. 2009, 10, 231–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Dove, B. K. , You, J. H. , Reed, M. L. , Emmett, S. R. et al., Changes in nucleolar morphology and proteins during infection with the coronavirus infectious bronchitis virus. Cell. Microbiol. 2006, 8, 1147–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Emmott, E. , Rodgers, M. , Macdonald, A. , McCrory, S. et al., Quantitative proteomics using stable isotope labeling with amino acids in cell culture (SILAC) reveals changes in the cytoplasmic, nuclear and nucleolar proteomes in Vero cells infected with the coronavirus infectious bronchitis virus. Mol. Cell. Proteomics doi: 10.1074/mcp.M900345-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Lam, Y. W. , Evans, V. C. , Heesom, K. J. , Lamond, A. I. , Matthews, D. A. , Proteomics analysis of the nucleolus in adenovirus‐infected cells. Mol. Cell. Proteomics 2009, 9, 117–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Hindley, C. E. , Lawrence, F. J. , Matthews, D. A. , A role for transportin in the nuclear import of adenovirus core proteins and DNA. Traffic 2007, 8, 1313–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Castiglia, C. L. , Flint, S. J. , Effects of adenovirus infection on rRNA synthesis and maturation in HeLa cells. Mol. Cell. Biol. 1983, 3, 662–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Miron, M. J. , Gallouzi, I. E. , Lavoie, J. N. , Branton, P. E. , Nuclear localization of the adenovirus E4orf4 protein is mediated through an arginine‐rich motif and correlates with cell death. Oncogene 2004, 23, 7458–7468. [DOI] [PubMed] [Google Scholar]
- 85. Lee, T. W. , Blair, G. E. , Matthews, D. A. , Adenovirus core protein VII contains distinct sequences that mediate targeting to the nucleus and nucleolus, and colocalization with human chromosomes. J. Gen. Virol. 2003, 84, 3423–3428. [DOI] [PubMed] [Google Scholar]
- 86. Lee, T. W. , Lawrence, F. J. , Dauksaite, V. , Akusjarvi, G. et al., Precursor of human adenovirus core polypeptide Mu targets the nucleolus and modulates the expression of E2 proteins. J. Gen. Virol. 2004, 85, 185–196. [DOI] [PubMed] [Google Scholar]
- 87. Matthews, D. A. , Adenovirus protein V induces redistribution of nucleolin and B23 from nucleolus to cytoplasm. J. Virol. 2001, 75, 1031–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Lutz, P. , Puvion‐Dutilleul, F. , Lutz, Y. , Kedinger, C. , Nucleoplasmic and nucleolar distribution of the adenovirus IVa2 gene product. J. Virol. 1996, 70, 3449–3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Miron, M. J. , Blanchette, P. , Groitl, P. , Dallaire, F. et al., Localization and importance of the adenovirus E4orf4 protein during lytic infection. J. Virol. 2009, 83, 1689–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Puvion‐Dutilleul, F. , Christensen, M. E. , Alterations of fibrillarin distribution and nucleolar ultrastructure induced by adenovirus infection. Eur. J. Cell Biol. 1993, 61, 168–176. [PubMed] [Google Scholar]
- 91. Rodrigues, S. H. , Silva, N. P. , Delicio, L. R. , Granato, C. , Andrade, L. E. , The behavior of the coiled body in cells infected with adenovirus in vitro. Mol. Biol. Rep. 1996, 23, 183–189. [DOI] [PubMed] [Google Scholar]
- 92. Tollefson, A. E. , Ying, B. , Doronin, K. , Sidor, P. D. , Wold, W. S. , Identification of a new human adenovirus protein encoded by a novel late l‐strand transcription unit. J. Virol. 2007, 81, 12918–12926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Copenhaver, G. P. , Putnam, C. D. , Denton, M. L. , Pikaard, C. S. , The RNA polymerase I transcription factor UBF is a sequence‐tolerant HMG‐box protein that can recognize structured nucleic acids. Nucleic Acids Res. 1994, 22, 2651–2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Mais, C. , Wright, J. E. , Prieto, J. L. , Raggett, S. L. , McStay, B. , UBF‐binding site arrays form pseudo‐NORs and sequester the RNA polymerase I transcription machinery. Genes Dev. 2005, 19, 50–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Meraner, J. , Lechner, M. , Loidl, A. , Goralik‐Schramel, M. et al., Acetylation of UBF changes during the cell cycle and regulates the interaction of UBF with RNA polymerase I. Nucleic Acids Res. 2006, 34, 1798–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Prieto, J. L. , McStay, B. , Recruitment of factors linking transcription and processing of pre‐rRNA to NOR chromatin is UBF‐dependent and occurs independent of transcription in human cells. Genes Dev. 2007, 21, 2041–2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Voit, R. , Grummt, I. , Phosphorylation of UBF at serine 388 is required for interaction with RNA polymerase I and activation of rDNA transcription. Proc. Natl. Acad. Sci. USA 2001, 98, 13631–13636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Lam, Y. W. , Evans, V. C. , Heesom, K. J. , Lamond, A. I. , Matthews, D. A. , Proteomic analysis of the nucleolus in adenovirus‐infected cells. Mol. Cell. Proteomics 2009, 9, 117–130. [DOI] [PMC free article] [PubMed] [Google Scholar]


