Abstract
Severe acute respiratory syndrome (SARS) is a new human infectious disease with significant morbidity and mortality. The disease has been shown to be associated with a new coronavirus (SARS‐CoV). The clinical and epidemiological aspects of SARS have been described. Moreover, the viral genome of SARS‐CoV has been fully sequenced. However, much of the biological behaviour of the virus is not known and data on the tissue and cellular tropism of SARS‐CoV are limited. In this study, six fatal cases of SARS were investigated for the tissue and cellular tropism of SARS‐CoV using an in‐situ hybridization (ISH) technique. Among all the tissues studied, positive signals were seen in pneumocytes in the lungs and surface enterocytes in the small bowel. Infected pneumocytes were further confirmed by immunofluorescence–fluorescence in‐situ hybridization (FISH) analysis. These results provide important information concerning the tissue tropism of SARS‐CoV, which is distinct from previously identified human coronaviruses, and suggest the possible involvement of novel receptors in this infection. Whereas the lung pathology was dominated by diffuse alveolar damage, the gut was relatively intact. These findings indicated that tissue responses to SARS‐CoV infection are distinct in different organs. Copyright © 2004 John Wiley & Sons, Ltd.
Keywords: severe acute respiratory syndrome (SARS), coronavirus, viral tropism
Introduction
Severe acute respiratory syndrome (SARS) is an emerging human disease with significant morbidity and mortality. The worldwide outbreak affected more than 8000 patients from over 30 countries with an associated cumulative mortality of 9.2% 1. A new coronavirus (SARS‐CoV) has been aetiologically linked to SARS 2, 3, 4. The supporting evidence includes serological studies, detection of the viral genome by polymerase chain reaction (PCR)‐based molecular methods, and viral isolation from respiratory specimens 2, 3, 4, 5. Although a highly sensitive diagnostic test is not available and SARS‐CoV is not detected in every clinically‐defined case 5, monkey inoculation experiments reproduce a similar disease in the primate to that in the human and further strengthen the aetiological role of SARS‐CoV 6. Much has been documented on the clinical and epidemiological characteristics of SARS. Epidemiological modelling shows that SARS‐CoV is highly contagious. However, much of the biological behaviour of the virus is not known and data on the tissue and cellular tropism of SARS‐CoV are limited.
The known human coronaviruses, types 229E and OC43, generally cause the common cold. These viruses have only rarely been associated with more severe respiratory diseases, such as pneumonia in neonates, the elderly or immunocompromised patients 7, 8, 9. However, SARS is a lower respiratory tract disease. Upper respiratory tract symptoms are uncommon and mild 10. Moreover, the genomic sequence of SARS‐CoV is distinct from other coronaviruses, including the two known human pathogenic coronaviruses 229E and OC43 11, 12. On the basis of ultrastructural findings, SARS‐CoV appears to infect lung pneumocytes 13, 14. However, extra‐pulmonary manifestations including diarrhoea, lymphopaenia, impaired liver function, and elevation of non‐cardiac creatine kinase are common and suggest that the virus may have tropism for non‐pulmonary tissues 10. Thus, the tissue and cellular tropism of SARS‐CoV is likely to be distinct from that of the known human coronaviruses. At present, comprehensive documentation of the various cell types and organs infected by SARS‐CoV is lacking. In this report, we investigate the tissue and cellular tropism of SARS‐CoV in fatal SARS cases by in situ hybridization.
Materials and methods
Selection of cases
Local ethical guidelines were followed for this study, which, as part of our ongoing project on SARS, was approved by the local ethical committee. Six SARS autopsy cases in our hospital outbreak in March 2003 were included and the outbreak was also epidemiologically linked to outbreaks in other countries 10, 15. The diagnosis of SARS was based on the World Health Organization (WHO) criteria 16 and in these six cases, SARS‐CoV was isolated from post‐mortem tissues. All patients but one had received high‐dose steroids and ribarvirin treatment and all died with respiratory failure. The lungs and trachea, but not the other upper respiratory tract tissues, were studied. All other major organs and tissues, except the brain, were also available for investigation. The post‐mortem findings were dominated by diffuse alveolar damage 10. Virus culture results were also available. In brief, coronavirus isolation was performed using an African green monkey kidney (Vero) cell line. When a cytopathic effect was observed from Vero cells, the supernatants from these positive cell cultures were tested by reverse transcriptase (RT)‐PCR. Among these six cases, SARS‐CoV was isolated from lung tissues in three of six cases and small intestinal tissues in four of five cases. Virus was not isolated from other tissues, including the heart, liver, kidney, and spleen in all cases, and lymph nodes, bone marrow, and muscles in two cases (Table 1).
Table 1.
Viral isolation and ISH results in autopsy samples of fatal cases of SARS
| Tissue | Case 1 | Case 2 | Case 3 | Case 4 | Case 5 | Case 6 |
|---|---|---|---|---|---|---|
| Lung | +/+ | +/+ | –/– | –/– | –/– | +/+ |
| Small intestine | –/– | +/+ | +/+ | +/+ | +/+ | NA/– |
| Heart | –/– | –/– | –/– | –/– | –/– | –/– |
| Liver | –/– | –/– | –/– | –/– | –/– | –/– |
| Spleen | –/– | –/– | –/– | –/– | –/– | –/– |
| Kidney | –/– | –/– | –/– | –/– | –/– | –/– |
| Bone marrow | NA/– | NA/– | NA/– | NA/– | –/– | –/– |
| Lymph node | NA/– | NA/– | NA/– | NA/– | –/– | –/– |
+/ + indicates positive by viral isolation/positive by ISH; –/– indicates negative by viral isolation/negative by ISH. NA/– indicates not available for viral isolation/negative by ISH.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
In‐situ hybridization (ISH) studies
ISH was developed for the detection of SARS‐CoV. RNA from SARS‐CoV strain Su10 (Genbank Accession No AY282752) was used as a template to generate a 700‐bp RT‐PCR product. The RT‐PCR product corresponds to nucleotides 26 401–27 100, matching the putative membrane protein of SARS‐CoV. The nucleotide sequence of this probe does not show significant homology to the genomes of other coronaviruses, including the two known human pathogenic strains 229E and OC43. The probe was labelled with digoxigenin by nick translation (DIG‐Nick Translation Mix, Roche, Indianapolis, IN, USA). Two micrograms of the RT‐PCR products was incubated at 14 °C for 2 h. Completion of labelling was checked by testing an aliquot of the reaction mix on 2.5% agarose gel electrophoresis stained with ethidium bromide. Smearing and disappearance of the ∼700 bp band were used to judge the adequacy of labelling. Formalin‐fixed, paraffin wax‐embedded tissues were cut into 5 µm sections. After deparaffinization and rehydration, the sections were treated with microwave heating in an 800 W domestic microwave oven at medium power for 10 min in 0.01 m sodium citrate buffer. Post‐fixation was carried out in 4% paraformaldehyde for 5 min after washing and cooling in phosphate‐buffered saline (PBS). Hybridization was performed at high stringency with 2 ng/µl denatured probe at 42 °C for 16 h in 50 µl of hybridization mix [50% formamide, 10% dextran sulphate in 2× saline sodium citrate (SSC)] under a 22 × 50 mm coverslip in a moist chamber. Excess probe was removed by washing in 2× SSC for 5 min at room temperature. This was followed by three high stringency washes in 0.1× SSC for 10 min at 61 °C. Immunological detection was conducted using a DIG Nuclear Acid Detection Kit (Roche) following the manufacturer's instructions. Briefly, after incubation with 1% Blocking Reagent (bottle 5) for 30 min, the sections were incubated with sheep anti‐digoxigenin alkaline phosphatase conjugate (vial 3) diluted at 1 : 375 for 2 h at room temperature. After three separate washes with washing buffer [0.1 m maleic acid, 0.15 m NaCl (pH 7.5), 0.3% Tween 20], the signal was developed using 5‐bromo‐4‐chloro‐3‐indolyl phosphate (BCIP) and nitroblue tetrazolium salt (NBT) (vial 4), which produces an insoluble blue precipitate indicating hybrid molecules. Sections were counterstained with methyl green. Cell block preparations from Vero cell cultures with and without inoculation with SARS‐CoV were used as positive and negative controls, respectively. The signal specificity was also assessed by (1) digestion of the sections with RNase A (0.2 mg/ml, Sigma; RNase A was pretreated by boiling at 70 °C for 10 min to remove possible DNase contaminants) for 2 h at 37 °C before the hybridization and (2) omission of the probes in the hybridization mixture. In addition, tissue sections form three autopsy cases of non‐SARS patients collected during the SARS outbreak in our hospital were included as negative controls. All three of these cases were negative for viral isolation and serology for SARS. Tissues sections of various organs, including lung, intestine, heart, liver, kidney, spleen, lymph nodes, bone marrow and muscles, were studied.
Immunofluorescene–fluorescence in‐situ hybridization (FISH) studies
Cytokeratin (AE1/AE3, Dako, 1 : 50) was used as an epithelial marker and CD68 (Dako, 1 : 1000) was used as a macrophage marker. Double immunofluorescence–FISH was performed as for ISH, with the additional immunofluorescence performed by incubation of primary antibodies overnight before detection of signals with the appropriate fluorescent secondary antibodies. ISH signals were detected using anti‐digoxigenin‐rhodamine (Roche). The secondary antibody for immunofluorescence was generally anti‐mouse fluorescein thiocyanate (FITC). Nuclei were counterstained by 4,6‐diamidino‐2‐phenylindole (DAPI) in anti‐fade mountant (Vectorshield, Vector Laboratories). A hundred infected cells (FISH‐positive) in the alveolar spaces were counted in each immunofluorescence–FISH experiment. Epifluorescence microscopy was performed on a Zeiss Axoplan 2 microscope equipped with the appropriate sets of excitation and emission filters. Images were captured by a cooled charge‐coupled device as grey scale images.
Results
In Vero cell cultures infected by SARS‐CoV, ISH and FISH using the RT‐PCR product of the membrane protein gene (M) revealed strong signals in the cytoplasm (Figure 1A). This FISH signal is specific for infected cells. Similar cytoplasmic signals and autofluorescence were not detected in uninfected Vero cell cultures (Figure 1B). These results correlated with electron microscopic findings in which viral particles were mainly observed in the cytoplasm of infected cells 2, 3, 4, 13.
Figure 1.
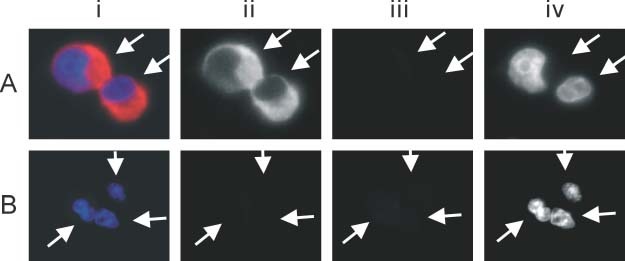
Fluorescence in‐situ hybridization (FISH) of SARS‐CoV in infected (A) and non‐infected (B) Vero cells. The Vero cells are indicated by arrows. The composite picture is shown in i. Images captured under the red, green, and blue filters are shown as grey scale images in ii, iii, and iv, respectively. FISH signals of SARS‐CoV were detected in the cytoplasm of infected, but not in uninfected, Vero cells (ii). No bleaching of filters or autofluorescence was detected (iii). The nuclei were counterstained with DAPI (iv)
We next performed ISH for SARS‐CoV on all available tissue sections obtained from autopsies of fatal cases of SARS and the control non‐SARS cases. The tissue sections from various organs of non‐SARS patients were all negative for ISH, indicating its specificity. Positive ISH signals were detected in those samples only when SARS‐CoV was isolated by virological cell culture in SARS patients. Lung sections from three cases and small intestinal sections from four cases were all positive for viral culture. These samples were all positive for ISH (Table 1). Other tissue samples, including the heart, liver, spleen, kidney, lymph nodes, bone marrow, and muscles, were all culture‐negative and they were all found to be ISH‐negative (Table 1). The trachea samples were also negative for ISH. The results of FISH in all cases were the same as those of ISH. Hence, the lung and the intestine were the only two organs where SARS‐CoV was explicitly identified.
On close examination of the lung, positive cytoplasmic signals were seen in pneumocytes lining the alveolar septa (Figure 2A) and within the alveolar spaces (Figure 2B). No signals were detected in the non‐SARS lung control (Figure 2C). The viral genome, as revealed by ISH, either was present diffusely throughout the cytoplasm of the infected cells or formed aggregates within the cytoplasm. In the lungs of fatal cases, multinucleate cells are one of the characteristic features of SARS. Detectable signals, however, were not seen in these cells. Detached cells in the alveoli might be detached pneumocytes or macrophages. The nature of the viral RNA‐positive detached cells in the alveolar spaces was ascertained by immunofluorescence–FISH with cytokeratin (AE1/AE3, Figures 3A and 3B) and macrophage (CD68) markers (Figure 3D). Among 100 infected cells in alveolar spaces counted in each immunofluorescence–FISH experiment, all were cytokeratin‐positive and none were CD68‐positive. Other cellular components in the lung, including bronchial epithelial cells, endothelial cells, fibroblasts, and lymphocytes, were all negative. No‐probe controls (with immunofluorescence for cytokeratin or macrophage markers, Figures 3C and 3E, respectively) indicated that the FISH signals seen were specific. Background fluorescence was limited to red blood cells and some connective tissue components. The cytoplasm of the detached pneumocytes and macrophages demonstrated minimal background signals.
Figure 2.
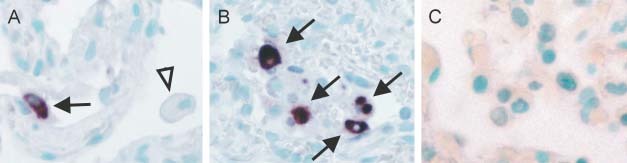
In situ hybridization (ISH) of SARS‐CoV in the lung. (A) Infected pneumocytes (arrow) with positive cytoplasmic signals are seen lining the alveolar septa. The other cellular components in the alveolar septa and histiocytes (arrow‐head) are negative (×400). (B) Scattered infected cells show a diffuse cytoplasmic distribution of the viral genome in infected cells (indicated by arrows; ×400). The infected cells, as suggested by the immunofluorescence–FISH study (Figure 3), were pneumocytes. (C) Control non‐SARS lung shows no specific ISH signals (×400)
Figure 3.
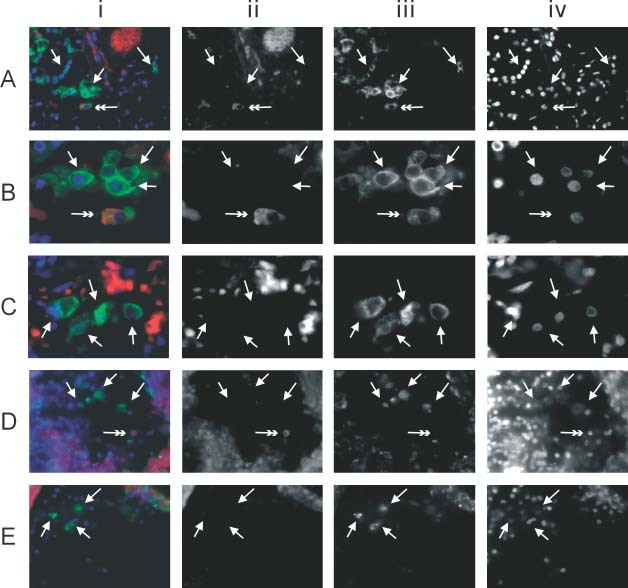
Immunofluorescence–FISH with cytokeratin (AE1/AE3) and macrophage (CD68) markers in lung tissue. The panels show (i) the composite picture; (ii) FISH signals of SARS‐CoV in the red channel; (iii) immunofluorescence in the green channel; and (iv) counterstaining of the nuclei in the blue channel. Panels ii–iv are shown as grey scale captured images. (A) Low‐power view of the lung showing a SARS‐CoV‐infected pneumocyte (double arrow) among other uninfected pneumocytes (arrows) (×400). Pneumocytes were highlighted by immunofluorescence using a cytokeratin marker (AE1/AE3). (B) High‐power view of A showing clearly the cytoplasmic signals (red and in ii) in the infected pneumocytes (double arrows) (×1000). Among 100 infected cells (double arrow) counted in each positive case, all were pneumocytes. (C) High‐power view of the no‐probe control in the same lung specimen. Arrows indicate some of the detached pneumocytes that stained positive for cytokeratin with minimal cytoplasmic signals when viewed under the red filter. The oval background red signals represent autofluorescence from red blood cells. (D) Macrophages were seen among the floating cells and were recognized by their CD68 immunoreactivity (arrows). Among 100 infected cells (double arrow) counted in each positive case, none were macrophages (×400). (E) High‐power view of the no‐probe control in the same lung specimen. Arrows indicate macrophages in the alveolar space. No autofluorescence signal is seen in these cells
In the small intestine, positive cytoplasmic signals of SARS‐CoV ISH were detected in the surface enterocytes but not in the other cells (Figures 4Ai and 4Aii). The signals appeared concentrated at the apical regions of the enterocytes. No detectable ISH signals were seen in non‐SARS small intestinal control (Figures 4Bi and 4Bii). FISH showed the same findings as ISH in the small intestinal sections. Double immunofluorescence–FISH with cytokeratin (AE1/AE3) further confirmed that the SARS‐CoV‐positive cells were enterocytes (Figure 5A, arrows). The no‐probe control (Figure 5B) again indicated that autofluorescence signals were minimal in the detached surface enterocytes (arrows). It is important to note that the tissue responses in the intestines were quite different from those in the lungs. Apart from autolytic changes, no gross or histological abnormality was apparent in the small intestine of any of these fatal cases of SARS. In particular, inflammatory processes were not detected.
Figure 4.
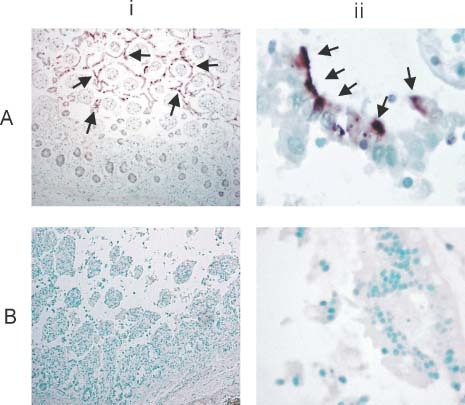
In situ hybridization (ISH) of SARS‐CoV in the small intestine. The panels show (i) a low‐power view (×40) and (ii) a high‐power view (×400). (A) Despite the autolytic changes, positive signals were seen and mainly detected in the surface enterocytes (arrows). High‐power view showing the positive cytoplasmic signals detected in the partially detached surface enterocytes. The positive signals appear polarized towards the apical part of the enterocytes. (B) Control non‐SARS intestine section reveals no specific ISH signals. High‐power view shows no cytoplasmic signals in the detached surface enterocytes
Figure 5.
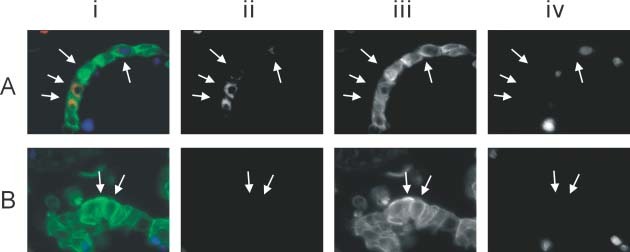
Immunofluorescence–FISH with cytokeratin (AE1/AE3) in the small intestine. The panels show (i) the composite picture; (ii) FISH signals of SARS‐CoV in the red channel; (iii) immunofluorescence in the green channel; and (iv) counterstaining of the nuclei in the blue channel. Panels ii–iv are shown as grey scale captured images. (A) High‐power view of the small intestine showing SARS‐CoV‐infected surface enterocytes (arrows) that have become detached (×1000). Surface enterocytes are highlighted by immunofluorescence using the cytokeratin marker (AE1/AE3). (B) High‐power view of the no‐probe control showing minimal autofluorescence signals in the detached enterocytes (arrows)
Discussion
We have studied the distribution of SARS‐CoV in fatal cases of SARS using in situ hybridization. Among all of the tissues available, only the lungs and the small intestines were positive. Cell types infected were limited to pneumocytes and enterocytes in these two organs, respectively.
In this study, the results of the ISH hybridization study were concordant with viral isolation results in our autopsy series. Despite anti‐viral treatment in most of these cases, persistent infection was detected in the lungs and small intestines in these fatal cases. The identification of SARS‐CoV RNA in the pneumocytes is consistent with the reported ultrastructural findings 13, 14. As SARS is a lower respiratory tract disease, one may expect to see SARS‐CoV infecting pneumocytes. This is in contrast to the other two known pathogenic coronaviruses, types 229E and OC43. These two viruses produce the common cold and are expected to affect the upper respiratory tract. However, apart from trachea samples, other upper respiratory tract tissues were not available from these SARS victims in our series for further study.
Macrophages are commonly seen in lung tissues from SARS patients and it is reasonable to suggest that they might play an important role in the pathogenesis of pulmonary injury. However, it is not known whether these macrophages are infected by the SARS‐CoV. Our study clearly demonstrates that, at least in fatal cases, persistent SARS‐CoV infection is demonstrable in pneumocytes but not in macrophages or other cell types in lung tissues. This result is consistent with the report of the haematological manifestations in SARS patients in which direct infection of SARS‐CoV in haemato‐lymphoid tissues has not been documented 17.
The presence of viral RNA in the surface enterocytes is intriguing. Data from recent outbreaks indicate that concurrent gastrointestinal symptoms, especially diarrhoea, are common 10, 18, 19. Human coronavirus or coronavirus‐like particles (CVLPs) have been suggested to be the aetiological agent of diarrhoea in susceptible subjects, including patients with acquired immunodeficiency syndrome and in children from developing countries 20, 21. However, enteric tissue tropism of SARS‐CoV has not been documented. Our results indicate that the surface enterocytes are infected by the SARS‐CoV, but in contrast to the lungs, no evidence of tissue destruction or inflammatory processes was seen in association with viral infection in the intestines. The mechanism underlying diarrhoea in SARS remains unclear. However, our observations suggest that the underlying pathology might well be in the small intestine and that active viral replication within intestinal cells rather than an inflammatory lesion might be responsible for the pathophysiology of the SARS‐related diarrhoea. Nevertheless, our study indicates that tissue responses to SARS‐CoV infection are distinct in different organs.
Although brain tissues and samples during the early phase of SARS were not available to assess comprehensively the temporal tissue tropism and neurotropism of the virus, our study does not support persistent viral infection in other organs in these fatal cases. Thus, it is possible that the extra‐pulmonary manifestations may be related to systemic effects of the abnormal inflammatory or immune reactions towards SARS‐CoV.
Multiple factors contribute to tissues and cellular tropism. On the basis of genomic data, SARS‐CoV is distinct from other coronaviruses, including the two known human pathogenic coronaviruses 229E and OC43 11, 12. Emerging data suggest that differences in the spike protein of the virus may be related to tissue tropism of bovine coronavirus 22. The host cell surface proteins are also important. The membrane protein aminopeptidase N (CD13) acts as a receptor for human coronavirus 229E and mediates its tissue tropism 23. CD13 is expressed on various polarized epithelial cells, including surface enterocytes, but not pneumocytes. It is also interesting to note that the macrophages in the lungs in these cases also expressed CD13 (KFT, unpublished data). However, even in areas with virus‐positive pneumocytes, none of the adjacent macrophages were infected. The surface receptor for SARS‐CoV is currently unknown, but our data suggest that CD13 is probably not the receptor for SARS‐CoV.
In conclusion, we have demonstrated SARS‐CoV tropism towards lung pneumocytes and intestinal surface enterocytes in fatal cases of SARS. The results contribute to our understanding of the pathology of this new human infectious disease. The distinct tissues and cellular tropism highlight the novel biology of SARS‐CoV, which may require a distinct set of receptor proteins for its tropism. Further studies of these receptors may have an important impact on the development of drugs for the treatment of this new human disease.
References
- 1. World Health Organization . Severe acute respiratory syndrome (SARS). Multi‐countries outbreak update 73. http://www.who.int/csr/don/2003_06_04/en/. Access date 4 June 2003.
- 2. Peiris JS, Lai ST, Poon LL, et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 2003; 361: 1319–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Drosten C, Gunther S, Preiser W, et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med 2003; 348: 1967–1976. [DOI] [PubMed] [Google Scholar]
- 4. Ksiazek TG, Erdman D, Goldsmith CS, et al. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med 2003; 348: 1953–1966. [DOI] [PubMed] [Google Scholar]
- 5. World Health Organization . Recommendations for laboratories testing for SARS. http://www.who.int/csr/sars/labmethods/en/#lab. Access date 21 July 2003.
- 6. Fouchier RA, Kuiken T, Schutten M, et al. Aetiology: Koch's postulates fulfilled for SARS virus. Nature 2003; 423: 240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Holmes KV. Coronavirus In Fields Virology (4th edn), Knipe DM, Howley PM. (eds). Lippincott Williams and Wilkins: Philadelphia, 2001; 1187–1203. [Google Scholar]
- 8. El‐Sahly HM, Atmar RL, Glezen WP, Greenberg SB. Spectrum of clinical illness in hospitalized patients with ‘common cold’ virus infections. Clin Infect Dis 2000; 31: 96–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fotz RJ, Elkordy MA. Coronavirus pneumonia following autologous bone marrow transplantation for breast cancer. Chest 1999; 115: 901–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lee N, Hui D, Wu A, et al. A major outbreak of severe acute respiratory syndrome in Hong Kong. N Engl J Med 2003; 348: 1986–1994. [DOI] [PubMed] [Google Scholar]
- 11. Marra MA, Jones SJ, Astell CR, et al. The genome sequence of the SARS‐associated coronavirus. Science 2003; 300: 1399–1404. [DOI] [PubMed] [Google Scholar]
- 12. Rota PA, Oberste MS, Monroe SS, et al. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 2003; 300: 1394–1399. [DOI] [PubMed] [Google Scholar]
- 13. Nicholls JM, Poon LL, Lee KC, et al. Lung pathology of fatal severe acute respiratory syndrome. Lancet 2003; 361: 1773–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ding Y, Wang H, Shen H, et al. The clinical pathology of severe acute respiratory syndrome (SARS): a report from China. J Pathol 2003; 200: 282–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Update: Outbreak of severe acute respiratory syndrome—worldwide, 2003. MMWR Morb Mortal Wkly Rep 2003; 52: 269–272. [PubMed] [Google Scholar]
- 16. World Health Organization . Case definitions for surveillance of severe acute respiratory syndrome (SARS). http://www.who.int/csr/sars/casedefinition/en/. Access date 21 July 2003.
- 17. Wong RSM, Wu AKL, To KF, et al. Haematological manifestations in patients with severe acute respiratory syndrome: retrospective analysis. Br Med J 2003; 326: 1358–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Peiris JSM, Chu CM, Cheng VCC, et al. Clinical progression and viral load in a community outbreak of coronavirus‐associated SARS pneumonia: a prospective study. Lancet 2003; 361: 1767–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Booth CM, Matukas LM, Tomlinson GA, et al. Clinical features and short‐term outcomes of 144 patients with SARS in the greater Toronto area. J Am Med Assoc 2003; 289: 2801–2809. [DOI] [PubMed] [Google Scholar]
- 20. Gerna G, Passarani N, Battaglia M, et al. Human enteric coronaviruses: antigenic relatedness to human coronavirus OC43 and possible etiologic role in viral gastroenteritis. J Infect Dis 1985; 151: 796–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Simhon A, Mata L. Fecal rotaviruses, adenoviruses, coronavirus‐like particles, and small round viruses in a cohort of rural Costa Rican children. Am J Trop Med Hyg 1985; 34: 931–936. [DOI] [PubMed] [Google Scholar]
- 22. Sanchez CM, Izeta A, Sanchez‐Morgado JM, et al. Targeted recombination demonstrates that the spike gene of transmissible gastroenteritis coronavirus is a determinant of its enteric tropism and virulence. J Virol 1999; 73: 7607–7618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yeager CL, Ashmun RA, Williams RK, et al. Human aminopeptidase N is a receptor for human coronavirus 229E. Nature 1992; 357: 420–422. [DOI] [PMC free article] [PubMed] [Google Scholar]


