Summary
Transcriptomic methods are set to revolutionize the study of the immune system in naturally occurring nonmodel organisms. With this in mind, the present article focuses on ways in which the use of ‘nonmodel’ rodents (not the familiar laboratory species) can advance studies into the classical, but ever relevant, epidemiologic triad of immune defence, infectious disease and environment. For example, naturally occurring rodents are an interesting system in which to study the environmental stimuli that drive the development and homeostasis of the immune system and, by extension, to identify where these stimuli are altered in anthropogenic environments leading to the formation of immunopathological phenotypes. Measurement of immune expression may help define individual heterogeneity in infectious disease susceptibility and transmission and facilitate our understanding of infection dynamics and risk in the natural environment; furthermore, it may provide a means of surveillance that can filter individuals carrying previously unknown acute infections of potential ecological or zoonotic importance. Finally, the study of immunology in wild animals may reveal interactions within the immune system and between immunity and other organismal traits that are not observable under restricted laboratory conditions. Potentiating much of this is the possibility of combining gene expression profiles with analytical tools derived from ecology and systems biology to reverse engineer interaction networks between immune responses, other organismal traits and the environment (including symbiont exposures), revealing regulatory architecture. Such holistic studies promise to link ecology, epidemiology and immunology in natural systems in a unified approach that can illuminate important problems relevant to human health and animal welfare and production.
Keywords: disease, immunoepidemiology, immunoregulation, PRR, rodent, Th‐2
Background – Game‐Changing Measurements
Recent technological advances in the de novo sequencing and analysis of nucleic acids are revolutionizing the measurement of gene expression in nonmodel organisms, with promising applications in the study of the immune system 1. Although other phenotypic measurements of immunity remain relevant and useful, albeit limited in scope or technically difficult to apply in nonmodel organisms 2, these advances mean that studying the immunology of such organisms in the natural environment has become easier and can take on a genomewide perspective embodied, for example in techniques such as RNAseq. This can, in turn, be accompanied by powerful analytical approaches derived from systems biology 3 and statistical methodologies applied in ecology. When these elements are combined with the monitoring of natural fluctuation or experimental perturbation, it opens up the possibility of ‘reverse engineering’ the regulatory architecture of the immune system and its interaction with other organismal traits and with natural environmental pressures 3.
Such approaches, using natural systems, complement the strengths and weaknesses of modern immunology 4. Here, the great strengths are derived from the very refined use of inbred and genetically manipulated mice under controlled conditions that negate environmental variation. This is very successful for unpicking the structure of molecular pathways and workings of cellular populations, but relevance for natural environmental variation disappears where genetically unrepresentative individuals are studied under homogenous laboratory conditions and in the absence of a natural flora and fauna of symbionts 4, 5. (Here symbiont is defined as any organism involved in an intimate association with the host, including parasitic, commensal and mutualistic associations.) The present review will be concerned with how this ‘blind spot’ in modern immunology can be addressed by a focus on natural populations. It will scan the horizon for unique ways in which studies of nonmodel rodents can contribute to our wider understanding of the biology of the immune system and the way it interacts with the environment to determine health. Additionally, it will consider how immunological measurement, interpreted in the light of paradigms from laboratory mouse immunology, can define individual variation relevant to ecological and epidemiological studies of infectious disease in the natural environment 1, 6. Rather than produce an exhaustive list of possible interests, though, this review will concentrate on three broad reasons to study immunology in naturally occurring vertebrate hosts, reasons that seem particularly exciting because they could have major practical implications for human health and the welfare and productivity of domesticated animals.
Each of these themes will be considered in turn and then the reasons why nonmodel rodents (species excluding Mus musculus, M. domesticus and Rattus norvegicus) may be particularly useful models. Finally, selected case studies will be discussed that have begun to approach some of the issues raised. Whilst the focus in these case studies is on the measurement of expression in selected candidate genes, they also illustrate the potential for future work using broader transcriptomic approaches, such as RNAseq.
Aberrant Inflammation in Anthropogenic Environments
Much of the ill‐health experienced in modern human populations is not caused directly by infectious agents but linked to aberrant inflammation 7. Thus, conditions including cancer 8, diabetes 9, asthma and allergies 10 and various neurological 11, 12 and psychiatric disorders 13 have, in part, immunological aetiologies. Trends in these immune‐based conditions have been, broadly, upward in ‘westernized’ human populations, the short time scale indicating the involvement of an environmental variable or variables 14 associated with ‘modern’ anthropogenic environments. In addition, the effects of environmental factors on individuals are likely to be modulated by genetic variation inherited from wild ancestral populations 15, 16 – amplifying individual variability in propensity to disease.
Identifying environmental factors
A major challenge facing biomedical science, then, is to pinpoint the environmental causes for this variance in health. In particular, given the observed epidemiological trends in immunologically based disease, the questions might be asked: What are the immunological changes that occur in the transition from natural to more anthropogenic environments, and what triggers these changes? Several causal mechanisms have been considered, but to parasite immunologists one particularly influential and intuitive (but unproven) line of thought is that increased dysregulation of the immune system is ultimately caused by the host's co‐adaptation to stimuli from co‐evolved symbionts. These symbionts might include macroparasites and other agents of chronic infection that tend to be lost in anthropogenic environments 17. This idea is embodied in what has been termed the ‘hygiene’ 5 or ‘old friends’ 13 hypothesis.
Surprisingly, modern immunology is not well placed to take up the challenge of identifying real‐world environmental drivers of the immune phenotype. This is because the remarkable laboratory models that have been established for revealing the molecular details of immunological pathways are unsuited to studying how these pathways interact with complex environments under natural conditions. It has often been noted that the genetics of laboratory mice does not reflect the natural situation 4. Typically, laboratory lineages are partly or completely inbred and often generated in a haphazard way that makes the range of allelic variation fixed in their genomes unrepresentative of natural variation 18. In the case of the fully inbred (isogenic) lines, their genomewide homozygosity is itself highly unnatural. Whilst these disadvantages would perhaps be overcome 19 by a range of carefully generated wild‐derived inbred and outbred lines 20, a much less tractable limitation lies in the inability to recreate natural environmental influences in the laboratory 5. Thus, wild animals experience a range of complex symbiont exposures and environmental stressors that cannot be sufficiently replicated in captivity 5. As such, wild mammals (and especially wild nonmodel rodents, due to some of the advantages discussed below) would seem a natural starting point to approach the problem of identifying environmental stimuli that drive immunological development and homeostasis in the wild. As previously considered in more detail by Friberg et al. 5, progress could be made either through in situ studies in natural populations tracking the effects of environmental variables using manipulative experimental or observational approaches (see for example, the wood mouse case study below), or through transplantation of naturally occurring lineages to (and monitoring of the changes occurring in) experimentally manipulated anthropogenic environments.
Identifying genetic loci under historical pathogen selection
Not all humans in modern environments develop immunologically based diseases (even though increasing numbers do), and those that succumb often have identifiable genetic predispositions. As noted above, causative environmental factors likely exert their effects upon a background of significant immunogenetic variability inherited from wild ancestral populations. The subject area of wild rodents as models for this immunogenetic variability was reviewed in detail by Turner and Paterson 15 and will only be considered here sufficiently to provide a general overview relevant to the present article. Briefly, a parallel challenge to the one of identifying environmental factors driving immunopathological phenotypes in anthropogenic environments described above, then, is the one of revealing genetic variation that places individuals at risk 15. In other words, finding genetic variation that natural selection has shaped in a way that, although adaptive in some natural settings, has the potential to be maladaptive in anthropogenic environments.
Such variation results from balancing selection, or from directional selection or genetic drift that has not proceeded to fixation, in ancestral populations. Here, selective agents (for example, naturally occurring pathogens) that were present in the past, or neutral processes, may have driven into wild populations alleles that are broadly deleterious in novel anthropogenic environments. Where potentially deleterious genes have been fixed, though, only variation due to environmental factors (see section above) or the wider genetic background is important.
Immunogenetic polymorphism driven by balancing or directional selection is thought to cause at least some of the variation underlying immunopathological conditions in modern humans 21, 22, 23, 24, 25, 26 and could, equally, be responsible for a great deal of natural variation in wild animals. Although it is increasingly recognized that adaptive evolution has structured the polymorphism in genes controlling the vertebrate immune system 26, 27, 28, our knowledge of the dynamics of this selection is still rudimentary. Crucially, the types of genes and pathways that tend to undergo balancing selection, or frequent intermittent directional selection, in the natural environment are poorly known. Also, the nature of the selection involved and the phenotypic manifestations of the polymorphisms are not well understood. Studies of wild populations 29, 30, 31, 32, 33 are pivotal in this regard, as the wild is the only place in which natural selection, and the phenotypes upon which it operates, can be measured. Furthermore, given the conservation of the mammalian immune system, it seems likely that similar genes and pathways may be the target of pathogen‐mediated balancing selection across taxa. Future studies into the causes of immunogenetic variation (and the characterization of the associated phenotypes) in natural populations are thus likely to feed insights into the biomedical and veterinary fields through focussing attention on the types of gene predisposed to drive immunopathology due to ancestral adaptive evolution.
The Dynamics of Infectious Disease in the Environment
Infectious disease risk
Another goal of immunological studies in wildlife is to provide an improved understanding of the dynamics of infection in natural populations 6, 34. And rodents are of particular interest in this regard, given their ecological importance and role as reservoirs for many zoonotic infectious agents 35, 36. In general, the dynamics of zoonotic and other ecologically important infectious diseases are likely to be influenced by heterogeneities amongst individual hosts 37, which will affect disease severity and transmission. As the immune system is the host's defence against infectious agents, variation within it is likely to generate much of this heterogeneity 32, 38. The measurement of immunological variation in real ecological contexts may, then, allow us to better define individual heterogeneity, and moreover, to address its environmental causes and epidemiological consequences 1, 39. This could help anticipate infectious disease risks in the environment. For example, such information may help to identify variations in immune defence that occur in time [e.g. seasonal 40, 41, 42] or space [e.g. habitat–specific or along invasion fronts 43, 44] and that alter susceptibility to infectious agents; or it may help identify species 45, 46, or subsets within populations 47, whose immunological profiles indicate an increased likelihood to serve as permissive reservoir hosts for certain pathogens. For example, studies on variability in the expression of tnfα and mx2 genes in bank voles (Myodes glareolus) have suggested the possibility of environmentally driven landscape‐level patterns that may affect the epidemiology of zoonotic Puumala hantavirus 47.
Infectious disease surveillance
As the most numerous group of mammals (~1500 species) the rodents represent, through sheer weight of numbers and their wide distributions, very significant potential reservoirs for emerging zoonotic infections. Much emphasis has recently been placed on emerging and re‐emerging infectious diseases from wildlife reservoirs [for example, the One Health initiative 48] and the importance of undertaking pro‐active monitoring programmes. In addition to infection risk for humans and domesticated animals, it is likely that epidemic and endemic infectious disease may also contribute significantly to the dynamics of wildlife themselves and indirectly to higher level ecosystem functioning. Discovery of infectious agents of ecological importance, or that present a risk of zoonotic emergence is, however, limited by the fact that diagnostic techniques are specific to individual pathogens, or groups of pathogens. Whilst next‐generation sequencing (NGS) approaches are becoming available that allow very broad nonspecific surveys for pathogen sequences, these are costly and many individuals would have to be processed were a population to be sampled randomly. Given this, the detection of potential emergent infectious agents in wildlife could, in some cases, be facilitated by the monitoring of immunological expression 49. This would narrowly focus attention on individuals with aberrant immunological profiles that might be indicative of infection states. For example, these individuals could then be selected for further NGS studies 50, 51 in order to detect pathogen‐specific sequences correlating with the aberrant immune expression profiles.
Whilst endemic reservoirs may be highly co‐adapted with their pathogens and be relatively asymptomatic 52 (perhaps without anomalous immune expression profiles), it is sometimes the case that infectious agents emerging from wildlife reservoirs may do so via an intermediate ‘amplifier’ host that does succumb to significant disease. Such hosts may shed more infective particles into the environment than the endemic reservoir and also show signs of acute immune responses. For example, putative amplifier hosts may have been involved in the emergence of the SARS coronavirus. Here, the ultimate natural reservoir appears to include horseshoe bats (Rhinolophus spp.), but the infection was likely transmitted to human hosts indirectly via other wild mammals, including the palm civet (Paguma larvata) 53, which is highly susceptible to the virus and sheds high titres of infective particles 54. In scenarios where the aim is to identify emergent disease risks before any infectious agent is specifically identified, immune expression studies may provide a way to filter potential diseased amplifier individuals from natural populations and focus attention on these for further study. It is also likely that the pattern of immune expression may be indicative of the type of pathogen involved [as is increasingly being exploited in medical diagnostics 55] and that this may help target subsequent efforts at identification.
Expanding Basic Understanding of the Immune System
Studies in wild nonmodel rodents may also yield unexpected general insights into the fundamental biology of the mammalian immune system. Thus, the laboratory model of mouse immunity cannot properly address many aspects of environmental variation seen in nature, and studies in humans are also limited in this way and by what tissues may be sampled or experimental approaches undertaken. On the other hand, immune responses in wild animals, once they can be interpreted using post‐genomic and systems biology approaches, may reveal functional pathways and interaction networks that were not previously understood, precisely because they relate to stressful environmental conditions that cannot ethically be replicated in laboratory or domestic animals, or in humans. An example where a focus on natural populations may feed‐back insights into general immunology is given in the section below dealing with immunodynamics in field vole populations 56.
Included in the fundamental biology of the immune system might be the costs of immunity embodied in classical ecological immunology 16, 57. This revolves around the concept of trade‐offs: that immune responses impose a penalty in the form of competition for energy allocation to, or functional interference with, other life‐history traits. Understanding these costs, and the interaction of immunity with other organismal traits, is an active and exciting area of research 16 that may generate insights relevant to human health and biomedical science. There is also relevance to agriculture, where production traits reflecting reproductive or growth parameters might interact with immunity in ways that are not yet fully appreciated. Whilst many advances have been made in the field relating to costs of immunity 16, further studies in natural populations using the type of holistic, genomewide approach possible with transcriptomic methods hold the promise of further advances.
What Study Systems and Measurements to Use?
Why not just well‐studied laboratory or farmed species?
Some studies may necessarily focus on particular species or local faunas because of specific concerns about infectious disease risks. Where more general questions are to be answered, though, an obvious starting point for studies of immune function in natural systems would be to use the wild counterparts of standard laboratory models or other well‐studied domesticated species. In particular, house mice (covered elsewhere in this special issue) would seem an obvious choice given their status as a central model in immunology. This would allow the use of robust measurements based on pre‐existing antibody reagents (see next section) and would also allow interpretation of immunological patterns in the context of a very detailed organism‐specific knowledge base.
Looking beyond the small number of laboratory and domesticated species, though, several considerations make it advisable to additionally consider other naturally occurring species. Firstly, as they occur in the ‘wild’ today, rats and mice may have patchy population structures that are narrowly focussed on unnatural anthropogenic environments and are the result of a long history of anthropogenically linked dispersal. In the case of the mouse, for example, dispersal is believed to have occurred from the fertile crescent in the near east 58 across most of the globe following the expansion of human civilizations 59. This history of co‐habitation and dispersal means that house mice may harbour symbiont assemblages restricted by lineage sorting (extinction) during dispersal/colonization events and, as invasive organisms, they may have acquired new infections in their relatively recent evolutionary history. In response to selective forces acting during dispersal and gain/loss of infectious threats, such invasive species may also have undergone rapid genetically based changes in immune function 43, 60. These considerations (lack of a natural symbiont flora/fauna, an unusual evolutionary history of dispersal and frequent occupation of anthropogenic habitats) make rats and mice weaker natural models to assess ‘hygiene hypothesis’ or ‘old friends’ ideas. Here, where the hypothesis is that immunopathological phenotypes in anthropogenic environments arise from a lack of stimuli from co‐evolved symbionts, models are needed where the host co‐exists in its natural setting with a complete assemblage of co‐evolved symbionts.
In contrast to the house mouse and Norway rat, common naturally occurring murine and microtine rodent species (e.g. Apodemus, Myodes and Microtus spp. in Europe), whose ecology has been well studied, often occur in relatively extensive, evenly distributed and persistent populations. These are likelier to have been stably linked in the long term with natural or quasi‐natural habitats and with a diverse, co‐adapted symbiont assemblage.
Transcriptomic studies are facilitated by a previously annotated genome, and this may affect the choice of study species. Increasing genomic information is becoming available for several well‐studied taxa, including the European species Apodemus sylvaticus, Microtus agrestis and M. glareolus, and annotated genomes have been assembled for Peromyscus maniculatus and Microtus ochrogaster in north America. However, transcriptomic studies can be carried out through de novo assembly without a pre‐existing species‐specific genome 61 and allow the exploitation of almost any species for ecological immunology studies. Naturally occurring rodents recently used for ecological immunological studies in other parts of the world include, for example, organisms as diverse as the capybara (Hydrochoerus hydrochaeris) 62, 63 or the pallid Atlantic forest rat (Delomys sublineatus) 64 in South America.
In general, naturally occurring rodents represent unparalleled models for work in ecological immunology and immunogenetics. Whilst their close relation to the laboratory mouse allows interpretation in terms of modern mechanistic immunology, their high population densities, amenability to longitudinal sampling and experimental manipulation in the field, short generation times, relatively spatially static populations and, ultimately, their adaptability to the laboratory environment, make these organisms uniquely tractable study systems. Given the diversity and abundance of rodents, it is likely that most researchers will have, at close hand, naturally occurring rodent systems that may serve as useful ecological models or that are practically relevant as reservoirs of transmissible infectious disease.
Finally, it should not be forgotten that there is very likely to be value, for its own sake, in carrying out studies across a wide diversity of host systems 16. This will be useful in revealing the generalities in immune function across species and will also provide insights from the specialized adaptations that individual species use to meet specific sets of circumstances.
Measurement: protein vs. nucleic acids
There is a strong emphasis in modern immunology on the use of antibody reagents against immunological biomolecules for robust molecular phenotypic measurements. Gene expression measurements at the mRNA level, in comparison, are generally considered a more problematical approach that is ‘resorted to’ if necessary, particularly in the case of analyses of individual genes by real‐time PCR (QPCR). This is based primarily on the fact that the expression of bioactive protein may not always track upstream mRNA concentrations 65. Complex kinetics in the pathway between mRNA and protein, and the stability of the proteins themselves, can prevent a simple relationship. Often there is poor concordance of matched transcriptomic and proteomic data sets across a range of organisms 66. Such a lack of gene‐by‐gene correlation, though, is much less relevant than the information content that transcriptomic profiles carry in relation to the biological status of the individual. This information content is attested to by the increasing use of transcriptomic approaches in modern biology. More specifically, early indications in wild rodents 1, 47, 56, 64 suggest that gene expression measurements, especially if interpreted with the complexity of post‐transcriptional dynamics in mind, do contain much useful information.
Practical issues restrict the usefulness of antibody‐based methods in nonmodel rodents. Due to structural variability in many immune molecules (especially canonical cytokines and cell surface markers), the transferability of commercially available off‐the‐shelf reagents amongst rodents, even within the subfamily Murinae, is limited. Whilst some commercial assays and reagents may indeed be found to cross‐react amongst rodent species (for example, many commercial antibodies targeting immunoglobulins), this approach may involve trialling dozens of others unsuccessfully. This has been the experience of researchers targeting cytokines in the cricetid, Peromyscus maniculatus, in North America 67, or even the murine, Apodemus sylvaticus, in Western Europe 49, 68. Thus, developing a broad panel of assays may require the de novo production of monoclonal and/or polyclonal antibodies, which is technically feasible but is costly and time‐consuming 4, 5, 6, especially given that two separate antibodies may be required per target (if setting up a microplate ELISA, for example).
A promising and more economical strategy will be to explore further the potential of established and emerging nucleic acids measurement 4, 6, 69. This approach can focus on panels of candidate genes, selected on the basis of the existing knowledge base for mouse immunological pathways. Even more powerfully, RNAseq 70, 71, because it encompasses the whole transcriptome (10s of thousands of genes, depending on sequencing coverage), largely removes concerns associated with measuring single genes as it allows a focus on whole pathways whose concerted variation is much more likely to reflect the phenotype. This is a technique that, although very expensive per sample, can be used in an unbiased way to identify informative and reliable marker genes that can then be measured by cheaper technologies in larger sample sizes. As an approach directed at the entire transcriptome, it is likely to supersede oligonucleotide microarray methods, due to general technical superiority 72 and, particularly in the case of studies in nonmodel species, the requirement of microarrays for prior sequence information. Targeting of single genes (or small panels of genes) may be carried out by QPCR (quantitative real‐time PCR, often abbreviated as RT‐PCR) or other emerging technologies that measure nucleic acids more directly, for example digital PCR 73 or new developments of microarray‐like systems 74. The latter technologies are likely to be technically more robust than QPCR, which although still invaluable, suffers from sensitivity to variable reaction kinetics amongst samples; however, they are currently more expensive, and in the case of digital PCR less applicable to high‐throughput applications.
Finally, the point should be made that, whatever molecular measurement approach taken (protein or RNA‐based), this should ultimately be cross‐referenced to functional measures of immunity (such as susceptibility to pathogens). In natural populations, this cross‐referencing can be achieved either by observational epidemiological studies, or, more powerfully (and with greater difficulty), by manipulative experimental infectious challenges.
Forward vs. reverse engineering complex biological systems
The advent of genomewide transcriptomic (and other high throughput) measurements arguably allows rapid progress in the study of immunity in natural systems through exploratory, data‐led approaches that might loosely be covered by the term ‘reverse engineering’ 75. Biologists often attempt to explain natural systems using a forward‐engineering‐like philosophy, where a high level model or concept is used to direct the interpretation of data. Although this approach (in some form or other) is likely to remain indispensible and also corresponds to many biologists idea of the basic scientific method, it is vulnerable to arbitrary choice of starting model and is not necessarily the exclusive, or most direct, route to understand complex systems (especially when starting from a low knowledge base). In contrast, in reverse engineering, large sets of responses (as generated by, for example, transcriptomic or multiplexed protein measurements) can be correlated with environmental and organismal variables of interest across perturbations (either natural or experimental), allowing interaction networks to be inferred 3. This can help establish how molecular pathways interact with each other and the environment to generate the observed phenotype. A reverse‐engineering‐like philosophy has, in part, featured in the examples dealt with below, and although these work with limited panels of measurements, they illustrate the potential for future studies using broader transcriptomic approaches.
Case Studies: Two Recent Focuses on Naturally Occurring Nonmodel Rodents
Macroparasites and innate antimicrobial responses
Background
The immune system has co‐evolved with commensal microbes to the extent that it requires cues from these organisms to programme its normal development 76, 77. Moreover, as a result of this, immunopathological phenotypes are to be expected where microbial exposures occur that are outside the parameters within which natural selection has operated during evolutionary history 77, 78. This developmental interaction between microbiota and immune system is, in part, channelled by toll‐like receptors (TLRs) of the innate immune system 76, 79, 80. The specificities of these receptors 81 and the inflammatory programmes they recruit are essentially directed against microbes. However, recent studies in wild rodents, which will briefly be reviewed here, suggest that natural exposures to macroparasites modify systemic TLR‐mediated responses to bacterial molecular patterns. Macroparasites, given their widespread occurrence in natural vertebrate populations, may thus be part of an extended co‐evolved interaction network, involving commensal microbes and innate antimicrobial responses, which can drive the development and homeostasis of the immune system.
TLR‐mediated responses in Apodemus sylvaticus at Cotgrave Forest, Nottinghamshire
A wood mouse population at Cotgrave Forest, Nottinghamshire, was monitored 49, 68, 82 over time, with a series of cross‐sectional samples between 2006 and 2008. A range of ex vivo TLR‐mediated responses (to defined TLR agonists) were measured in cultured splenocytes from subsets of animals. Due to a pattern of positive covariation amongst TLR‐mediated tumour necrosis factor alpha (TNF‐α) protein responses (TLRs 2, 3–5, 7, 9) measured in the early part of the sample series, and the especially strong associations of TLR2‐mediated TNF‐α production with measures of macroparasite infection, this last response in particular was chosen to be measured in all samples.
Focussing on TLR2‐mediated response across the whole study period, this was found to be associated with certain macroparasites, especially the gastrointestinal heligmosomatid nematode, Heligmosomoides polygyrus, and the blood‐sucking ectoparasitic louse, Polyplax serrata. The magnitude and direction of the associations, though, changed across the study period. The changing associations corresponded to different environmental conditions following a perturbation (population crash) in the mouse population during the middle part of the study. Early in the study, the abundances of mice and macroparasites were high, but following the mouse population crash, the later part of the study was characterized by lower macroparasite abundance. Corresponding changes occurred in the association between macroparasites (P. serrata and H. polygyrus) and TLR2‐mediated TNF‐α responses, with a strong negative coefficient before the perturbation (at high infection levels) and a positive one subsequently (at lower infection levels).
Complementary laboratory experiments using the mouse‐Heligmosomoides bakeri model supported a causal effect of nematode infection upon TLR‐mediated responses. This model is relevant because H. bakeri is a very close relative 59, 83 of the H. polygyrus occurring in wood mice. Previously naïve inbred mice exposed to single (CBA, BALB/c, C57BL/6, SWR) and trickle H. bakeri infections (BALB/c, C57BL/6), typically up‐regulated TLR responses signalling through myeloid differentiation primary response gene 88 (MyD88) at some point during the infection course, usually coinciding with peak standing worm burdens. This is consistent with a permissive effect of MyD88 signalling on gastrointestinal nematode infection demonstrated through experiments with Myd88 −/− and interleukin one receptor one (Il1r1) −/− mice 84, 85. It is interesting, though, that in the trickle infection experiments, resistance developed in both BALB/c and C57BL/6. This contrasts with more permissive infection phenotypes previously achieved under similar experimental regimens, where the susceptible strain C57BL/6 supports chronic infection and can continue accumulating worms to the point of lethality 86. Furthermore, in wild wood mice there is a linear accumulation of H. polygyrus with age and no indication of acquired immunity in the form of abundance as a decelerating function of age indicators 82. It may also be significant that the laboratory experiments, in initially naïve animals, produced results consistent with the positive abundance – TLR response association seen in the wood mouse population during times of low parasite abundance, but were not consistent with the negative association seen at times of high parasite abundance. The latter negative association (and perhaps the permissive infection phenotypes noted above) could be accounted for by the well‐known immunosuppressive effects of chronic heligmosomatid infections. These effects would likely involve adaptive regulatory T‐cell responses that could feedback negatively onto innate immune responses 87.
Taken together, all of this information indicates that macroparasite infection exposures may have significant and context‐dependent effects on the innate responses that mediate interactions of the immune system with microbes. The mechanism for this remains to be determined, but as discussed by Friberg et al. 5, the possibilities include effects on TLR signalling by worms that are direct, via secreted immunomodulators, or indirect, via feedback from adaptive immune responses. Other indirect effects could be mediated by altered exposure to microbes or innate damage signals resulting from the activities of macroparasites at their site of infection. There is some reason to believe that one or both of these last mechanisms might be important. Thus, heligmosomatid excretory–secretory products 88, and the regulatory 87 and Th2 89 immune responses that these parasites typically trigger, generally reduce TLR‐mediated signalling. In the laboratory experiments, though, heligmosomatid infections actually increased TLR responses (perhaps consistent with activation by TLR ligands, which are primarily microbial‐ or damage‐associated patterns). This re‐emphasizes the possibility that co‐infections with macroparasites contribute to the network of interactions between commensal microbes and the immune system. Moreover, due to the epidemiological association of ectoparasitic lice with systemic responses in the Cotgrave Forest study, it seems that antiparasite responses at peripheral sites beyond the gut 78 may also be involved. The existence of such an extended interaction network is highly relevant to our understanding of how microbiotal exposures programme immunity. Because macroparasites are often absent in anthropogenic environments 5, this may contribute to the disruption of co‐evolved interactions (cf. the hygiene hypothesis). Thus, studies in a wild system have pointed towards the need for further work to establish the role of natural macroparasite communities in the formation of microbiotal assemblages and the contribution of this to health.
Gene expression signatures of antipathogen strategies and their life‐history correlates
Background
Evolutionary fitness in the natural environment is not measureable in the laboratory, and so studies of host–pathogen community dynamics in the wild are essential to fully understand immune responses in their wider context – as components of antipathogen strategies that maximize fitness. Studies in wild field voles, briefly reviewed below, have aimed to identify distributional infection patterns associated with different antipathogen strategies in natural populations and to link these to expression signatures in immune‐relevant genes. Such gene expression markers can then be used to track the life‐history correlates of different putative strategies and may also give insights into the immunological mechanisms involved.
When considering adaptations to infection exposures, two strategies are available to a host and these may often be deployed together, although there may be some emphasis on one relative to the other. These strategies are resistance, where the host prevents infection (denies access to the pathogen), and tolerance, where the host allows access to the pathogen whilst actively mitigating the negative effects of infection. Identifying patterns of tolerance and resistance in natural populations is problematical but can be approached using a general framework like that summarized in Jackson et al. 56. In cross‐sectional samples (‘snapshot’ destructive samples of individuals), tolerance in identifiable groups of animals may be measured as the regression slopes (reaction norms) of fitness measures (for example, body condition) against infection load. Here there is the problem that, in individuals from natural populations, the infection dose and the time course of infection are not standardized. Reaction norms, though, can also be supplemented by consideration of the phase curves (temporal trajectories of fitness in relation to infection load) of infection courses in longitudinally sampled individual animals. These allow a known infection load to be related to fitness measurements at a later time point. In the study briefly reviewed below, both approaches were used, with initial identification of a tolerance‐like pattern in cross‐sectional samples, which was then corroborated and extended by focussed analyses in longitudinal samples.
Immune gene expression profiles in field voles at Kielder Forest, Northumberland
Gene expression measurements are increasingly being used in studies of infectious disease in nonmodel rodent systems 1, 6, 49, 64, 90, 91, 92. In work carried out in the well‐studied Kielder voles (M. agrestis) system 39, measurements of a panel of candidate genes (representing different immunological pathways) were taken in sets of cross‐sectional and longitudinal samples from individual voles at two sites in each of two seasons (2008–2010). This design aimed to capitalize on the respective strengths of the two sampling modes: the greater range and precision of measurement in destructively sampled ‘snapshot’ cross‐sectional samples (where more tissues can be interrogated and manipulated in the laboratory), also the stronger inference of causality in time series data from repeat‐sampled individually marked animals (longitudinal samples). In cross‐sectional samples, expression profiles were measured in ex vivo stimulatory assays of cultured splenocytes (with stimulants including TLR2 and TLR7 agonists and mitogen), whilst in longitudinal samples, constitutive expression was recorded in peripheral blood. In addition, a range of infection and condition measures were recorded in the sample animals.
Early analyses in a partial cross‐sectional data set for immune gene expression (2008–2009), and without considering pathogen data, suggested the value of the measurement approach through the existence of significant variation of expression in relation to season, life‐history stage and individual condition 1. Further analyses, on the full data set, searched for patterns of resistance or tolerance to pathogens. Initially focussing on the more detailed cross‐sectional data, and using body and organ condition (weight adjusted for standard length) as a fitness measure, it was possible to recognize a predominant pattern indicative of tolerance to macroparasite infection in mature males (where macroparasites accumulated with age indicators and were associated with increasing body condition) and a pattern indicative of resistance in immature males (where macroparasite abundances were decelerating functions of age indicators and not associated with body condition) 56. Although unexpected, the positive association of body condition with macroparasite infection in mature males was in the context of negative changes in other life‐history components and not inconsistent with a negative overall impact of infection exposure on fitness.
High expression of the transcription factor GATA‐binding factor 3 (Gata3) in mitogen‐stimulated splenocytes was found to mark both tolerant animals (amongst mature males) and resistant animals (amongst immature males) in the cross‐sectional set. Furthermore, analyses of time‐lagged associations in mature males in the longitudinal data suggested that macroparasite infections triggered Gata3 responses [as might be expected in laboratory models 93, 94, 95], which in turn gave rise to increases in body condition. This corroborated the cross‐sectional analyses and supported a hypothesis that Gata3 activity stimulated by macroparasites is part of a complex of (tolerance) responses leading to the readjustment of body condition in mature males. Constitutive Gata3 expression in peripheral blood also correlated with survival in longitudinally monitored animals, with high relative expression of Gata3 predicting poorer survival in younger animals but having a progressively more positive effect on survivorship with increasing age.
In this observational field study, the existence of confounding processes that might produce the cross‐sectional patterns attributed to tolerance should be considered. Indeed, the multifaceted nature of the Kielder study, with cross‐sectional and longitudinal components providing a range of measurements in different population strata, increases the opportunities for comparing predictions to data. Two main confounding processes might be relevant in terms of their potential to generate tolerance‐like patterns. One of these is differential mortality (DM): where, amongst animals heavily infected with macroparasites, those in poor condition die more quickly, biasing the average condition of the survivors upwards. Another possibility is correlated risk (CR): where parasite load may be linked to good condition because hosts in good condition forage more or range more and encounter more parasite infective stages as a result.
The DM and CR scenarios were poorer explanations for the observed patterns from a number of perspectives. Under CR, increased condition would precede increased acquisition of parasites and the Gata3 responses they trigger; but in temporal series for individual mature males, these two sets of events, in reality, occurred in the reversed sequence. This observed sequence and the direction of association also contradict the prediction of DM of a negative effect of macroparasite infection (and the Gata3 responses triggered) on subsequent condition (which follows if infection is a major cause of mortality) – in reality there was a positive association. Perhaps most importantly, the DM and CR scenarios do not explain the age‐specific changes in the relationship between Gata3 expression and survival in longitudinal data, or in the relationship between Gata3, condition and macroparasite infection in the cross‐sectional data. Thus, if DM were true, Gata3 expression in peripheral blood would be expected to show a consistent negative association with individual survival, especially in classes of animal with an apparent Gata3‐associated tolerance pattern. However, in reality Gata3 expression decreases survival in smaller males (where a tolerance pattern is not seen) but tends to increase survival in larger animals (which do show the Gata3‐associated tolerance pattern). Furthermore, if CR were true, better conditioned animals would generally have higher parasite exposure and higher Gata3 expression; but in reality, this is only seen in mature males and not in immature males. It might also be argued that a special case of CR, which could explain the latter age‐specific pattern, is that better conditioned mature males range more, or have more social contacts, due to increased breeding activity. Even in this case, though, the wide‐ranging males would be expected to have better testis condition (if undergoing increased behaviours associated with breeding); but in our study, Gata3 expression was negatively correlated with testis condition. Thus, the details of the study were consistent with a hypothesis of elevated Gata3 expression mediating resistance in immature males and tolerance in mature males; there were major inconsistencies with alternative DM and CR interpretations.
Gata3 is a master transcription factor involved in the differentiation and development of Th2 (T‐helper type 2 cell) cells 96 and would be expected to mark Th2 activity in the splenocyte cultures studied. The seemingly dual aspect of Gata3 (involved in tolerance and resistance) is not biologically implausible, given that Th2 responses have long been associated with resistance to macroparasite infections in laboratory models but are also linked to wound‐healing mechanisms that might be involved in tolerance 97. It would seem possible, however, that Gata3‐expressing Th2 cells might drive different downstream effector responses in resistance and tolerance, and this is one aspect that is worthy of further study. Whilst regulatory components of the immune system have previously been considered as possible mediators of tolerance 98, through their ability to dampen effector responses, this was not supported in the Kielder study. Thus, the anti‐inflammatory cytokines interleukin (IL) 10 and transforming growth factor beta one (TGF‐β1) and the transcription factor forkhead box P3 (FoxP3), whose expression characterizes many regulatory T‐cell subsets, were measured but were not associated with tolerance patterns.
Gata3 expression in tolerant males was associated with a complex of life‐history readjustments likely to impact fitness (represented by schematic phase curves in Figure 1). Apart from the increase in body condition, there was a decrease in a fecundity indicator (testis condition) and increasing survival as animals aged. Possibly, then, the tolerance strategy increases fitness via improvements in residual reproductive value (potential for future reproduction) during macroparasite infection. This is epidemiologically significant, as the transmission of infectious agents may become focussed through tolerant subsets of the population with consequences for the population dynamics, life histories and co‐evolutionary dynamics of the interacting organisms. But there are also insights that can be fed back into laboratory immunology. The results raise the possibility that Th2 cell activity may show ontogenetic changes, sometimes mediating resistance and at other times mediating tolerance. This dichotomy could be relevant to the laboratory as, typically, laboratory experiments are carried out in restricted life‐history stages under restricted environmental conditions, and this may have skewed our view of what responses are likely to occur in systems outside of the laboratory. There is also a relevance to vaccination strategies in real‐world situations, which may need to encompass the possibility that, under some conditions and in some subsets of a population, similar immunogenic stimuli may result in resistance or tolerance responses (that might not be protective in terms of preventing infection, but could have some benefit in terms of ameliorating disease). This may be especially significant, given that most vaccine adjuvants used in practical contexts are Th2‐inducing aluminium salts (alums).
Figure 1.
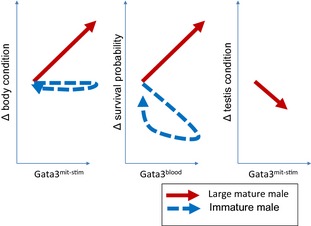
Life‐history responses in male field voles (Microtus agrestis) associated with Gata3 expression triggered by macroparasite exposures, represented schematically as phase curves (temporal tracks through the plotted parameter space). Gata3mit‐stim, Gata3 expression in cultured mitogen‐stimulated splenocytes; Gata3blood, constitutive Gata3 expression in peripheral blood.
Conclusions
Although much interesting work has been done in nonmodel rodents using narrow focusses on individual immune responses, a renewed effort addressing the immune system (and its interaction with wider organismal traits and the environment) in a more holistic way seems likely to pay dividends. The pivotal technological means to do this are now accessible, in the form of next‐generation sequencing analyses of the transcriptome. Early indications from studies using QPCR panels of candidate genes suggest that gene expression measurements in natural populations do convey interpretable information about individual status, especially where combined with defined stimulatory treatments of cultured cell populations. Associations occur with season, life‐history stage and body condition. Furthermore, there are strong indications that infection pressures are key drivers of many aspects of expression in the immune system in nature and that, as a result of these pressures, wild mammals can be confidently predicted to adopt phenotypes very different to those seen in laboratory rodents. All of this supports the utility of immune gene expression measurements for ecologically and epidemiologically motivated studies; it also confirms the interest for our basic understanding of the immune system: where the diverse combinations of environmental conditions seen in the wild may reveal interaction networks that remain hidden under controlled laboratory conditions. Finally, studying the diversity of immune function in natural and anthropogenic environments (and, ultimately, further dissection of this variation under experimental conditions) will help us resolve the environmental stimuli (and genotype × environment interactions) that affect the formation of immunopathological phenotypes such as those responsible for so much variation in human health and animal welfare.
Acknowledgements
I gratefully acknowledge the colleagues with whom I have worked on nonmodel rodent systems and whose ideas have influenced my own thinking, also acknowledged is funding from the Leverhulme Trust (RPG‐301) and NERC (NE/L013517/1).
References
- 1. Jackson JA, Begon M, Birtles R, et al The analysis of immunological profiles in wild animals: a case study on immunodynamics in the field vole, Microtus agrestis . Mol Ecol 2011; 20: 893–909. [DOI] [PubMed] [Google Scholar]
- 2. Demas GE, Zysling DA, Beechler BR, Muehlenbein MP & French SS. Beyond phytohaemagglutinin: assessing vertebrate immune function across ecological contexts. J Anim Ecol 2011; 80: 710–730. [DOI] [PubMed] [Google Scholar]
- 3. Williams TD, Turan N, Diab AM, et al Towards a system level understanding of non‐model organisms sampled from the environment: a network biology approach. PLoS Comput Biol 2011; 7: e1002126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pedersen AB & Babayan SA. Wild immunology. Mol Ecol 2011; 20: 872–880. [DOI] [PubMed] [Google Scholar]
- 5. Friberg IM, Bradley JE & Jackson JA. Macroparasites, innate immunity and immunoregulation: developing natural models. Trends Parasitol 2010; 26: 540–549. [DOI] [PubMed] [Google Scholar]
- 6. Bradley JE & Jackson JA. Measuring immune system variation to help understand host‐pathogen community dynamics. Parasitology 2008; 135: 807–823. [DOI] [PubMed] [Google Scholar]
- 7. Rook GA. Regulation of the immune system by biodiversity from the natural environment: an ecosystem service essential to health. Proc Natl Acad Sci USA 2013; 110: 18360–18367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Coussens LM & Werb Z. Inflammation and cancer. Nature 2002; 420: 860–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Canivell S & Gomis R. Diagnosis and classification of autoimmune diabetes mellitus. Autoimmun Rev 2014; 13: 403–407. [DOI] [PubMed] [Google Scholar]
- 10. Legatzki A, Rosler B & von Mutius E. Microbiome diversity and asthma and allergy risk. Curr Allergy Asthma Rep 2014; 14: 466. [DOI] [PubMed] [Google Scholar]
- 11. Kannarkat GT, Boss JM & Tansey MG. The role of innate and adaptive immunity in Parkinson's disease. J Parkinsons Dis 2013; 3: 493–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ellwardt E & Zipp F. Molecular mechanisms linking neuroinflammation and neurodegeneration in MS. Exp Neurol 2014; 262PA: 8–17. [DOI] [PubMed] [Google Scholar]
- 13. Rook GA, Raison CL & Lowry CA. Microbiota, immunoregulatory old friends and psychiatric disorders. Adv Exp Med Biol 2014; 817: 319–356. [DOI] [PubMed] [Google Scholar]
- 14. Rook GA. Hygiene hypothesis and autoimmune diseases. Clin Rev Allergy Immunol 2012; 42: 5–15. [DOI] [PubMed] [Google Scholar]
- 15. Turner AK & Paterson S. Wild rodents as a model to discover genes and pathways underlying natural variation in infectious disease susceptibility. Parasite Immunol 2013; 35: 386–395. [DOI] [PubMed] [Google Scholar]
- 16. Brock PM, Murdock CC & Martin LB. The history of ecoimmunology and its integration with disease ecology. Integr Comp Biol 2014; 54: 353–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jackson JA, Friberg IM, Little S & Bradley JE. Review series on helminths, immune modulation and the hygiene hypothesis: immunity against helminths and immunological phenomena in modern human populations: coevolutionary legacies? Immunology 2009; 126: 18–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chia R, Achilli F, Festing MFW & Fisher EMC. The origins and uses of mouse outbred stocks. Nat Genet 2005; 37: 1181–1186. [DOI] [PubMed] [Google Scholar]
- 19. Babay BEC, Louzir H, Kebaïer C, Boubaker S, Dellagi K & Cazenave P‐A. Inbred strains derived from feral mice reveal new pathogenic mechanisms of experimental Leishmaniasis due to Leishmania major. Infect Immun 2004; 72: 4603–4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Harper J. Wild‐derived mouse stocks: an underappreciated tool for aging research. Age (Dordr) 2008; 30: 135–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gabriel SE, Brigman KN, Koller BH, Boucher RC & Stutts MJ. Cystic fibrosis heterozygote resistance to cholera toxin in the cystic fibrosis mouse model. Science 1994; 266: 107–109. [DOI] [PubMed] [Google Scholar]
- 22. Pasvol G, Weatherall DJ & Wilson RJ. Cellular mechanism for the protective effect of haemoglobin S against P. falciparum malaria. Nature 1978; 274: 701–703. [DOI] [PubMed] [Google Scholar]
- 23. Willcocks LC, Carr EJ, Niederer HA, et al A defunctioning polymorphism in FCGR2B is associated with protection against malaria but susceptibility to systemic lupus erythematosus. Proc Natl Acad Sci USA 2010; 107: 7881–7885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Woolf LI. The heterozygote advantage in phenylketonuria. Am J Hum Genet 1986; 38: 773–775. [PMC free article] [PubMed] [Google Scholar]
- 25. Lee JC, Espeli M, Anderson CA, et al Human SNP links differential outcomes in inflammatory and infectious disease to a FOXO3‐regulated pathway. Cell 2013; 155: 57–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fumagalli M, Sironi M, Pozzoli U, Ferrer‐Admetlla A, Pattini L & Nielsen R. Signatures of environmental genetic adaptation pinpoint pathogens as the main selective pressure through human evolution. PLoS Genet 2011; 7: e1002355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Forni D, Cagliani R, Tresoldi C, et al An evolutionary analysis of antigen processing and presentation across different timescales reveals pervasive selection. PLoS Genet 2014; 10: e1004189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Leffler EM, Gao Z, Pfeifer S, et al Multiple instances of ancient balancing selection shared between humans and chimpanzees. Science 2013; 339: 1578–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Morger J, Bajnok J, Boyce K, et al Naturally occurring Toll‐like receptor 11 (TLR11) and Toll‐like receptor 12 (TLR12) polymorphisms are not associated with Toxoplasma gondii infection in wild wood mice. Infect Genet Evol 2014; 26: 180–184. [DOI] [PubMed] [Google Scholar]
- 30. Tschirren B, Andersson M, Scherman K, Westerdahl H, Mittl PR & Raberg L. Polymorphisms at the innate immune receptor TLR2 are associated with Borrelia infection in a wild rodent population. Proc Biol Sci 2013; 280: 20130364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tschirren B, Raberg L & Westerdahl H. Signatures of selection acting on the innate immunity gene Toll‐like receptor 2 (TLR2) during the evolutionary history of rodents. J Evol Biol 2011; 24: 1232–1240. [DOI] [PubMed] [Google Scholar]
- 32. Turner AK, Begon M, Jackson JA, Bradley JE & Paterson S. Genetic diversity in cytokines associated with immune variation and resistance to multiple pathogens in a natural rodent population. PLoS Genet 2011; 7: e1002343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Turner AK, Begon M, Jackson JA & Paterson S. Evidence for selection at cytokine loci in a natural population of field voles (Microtus agrestis). Mol Ecol 2012; 21: 1632–1646. [DOI] [PubMed] [Google Scholar]
- 34. Graham AL, Shuker DM, Pollitt LC, Auld S, Wilson AJ & Little TJ. Fitness consequences of immune responses: strengthening the empirical framework for ecoimmunology. Funct Ecol 2011; 25: 5–17. [Google Scholar]
- 35. Meerburg BG, Singleton GR & Kijlstra A. Rodent‐borne diseases and their risks for public health. Crit Rev Microbiol 2009; 35: 221–270. [DOI] [PubMed] [Google Scholar]
- 36. Davis S, Calvet E & Leirs H. Fluctuating rodent populations and risk to humans from rodent‐borne zoonoses. Vector Borne Zoonotic Dis 2005; 5: 305–314. [DOI] [PubMed] [Google Scholar]
- 37. Lloyd‐Smith JO, Schreiber SJ, Kopp PE & Getz WM. Superspreading and the effect of individual variation on disease emergence. Nature 2005; 438: 355–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Graham AL, Cattadori IM, Lloyd‐Smith JO, Ferrari MJ & Bjornstad ON. Transmission consequences of coinfection: cytokines writ large? Trends Parasitol 2007; 23: 284–291. [DOI] [PubMed] [Google Scholar]
- 39. Turner AK, Beldomenico PM, Bown K, et al Host‐parasite biology in the real world: the field voles of Kielder. Parasitology 2014; 141: 997–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Demas GE & Nelson RJ. Photoperiod, ambient temperature, and food availability interact to affect reproductive and immune function in adult male deer mice (Peromyscus maniculatus). J Biol Rhythms 1998; 13: 253–262. [DOI] [PubMed] [Google Scholar]
- 41. Martin LB, Weil ZM & Nelson RJ. Seasonal changes in vertebrate immune activity: mediation by physiological trade‐offs. Philos Trans R Soc Lond B Biol Sci 2008; 363: 321–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nelson RJ & Demas GE. Seasonal changes in immune function. Q Rev Biol 1996; 71: 511–548. [DOI] [PubMed] [Google Scholar]
- 43. Martin LB, Coon CA, Liebl AL & Schrey AW. Surveillance for microbes and range expansion in house sparrows. Proc Biol Sci 2013; 281: 20132690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. White TA, Perkins SE, Heckel G & Searle JB. Adaptive evolution during an ongoing range expansion: the invasive bank vole (Myodes glareolus) in Ireland. Mol Ecol 2013; 22: 2971–2985. [DOI] [PubMed] [Google Scholar]
- 45. Wang LF, Walker PJ & Poon LL. Mass extinctions, biodiversity and mitochondrial function: are bats ‘special’ as reservoirs for emerging viruses? Curr Opin Virol 2011; 1: 649–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Martin LB 2nd, Weil ZM & Nelson RJ. Immune defense and reproductive pace of life in Peromyscus mice. Ecology 2007; 88: 2516–2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Guivier E, Galan M, Henttonen H, Cosson JF & Charbonnel N. Landscape features and helminth co‐infection shape bank vole immunoheterogeneity, with consequences for Puumala virus epidemiology. Heredity (Edinb) 2014; 112: 274–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bidaisee S & Macpherson CN. Zoonoses and one health: a review of the literature. J Parasitol Res 2014; 2014: 874345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Friberg IM, Lowe A, Ralli C, Bradley JE & Jackson JA. Temporal anomalies in immunological gene expression in a time series of wild mice: signature of an epidemic? PLoS One 2011; 6: e20070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ueno K, Ishii A & Ito K. ELM: enhanced lowest common ancestor based method for detecting a pathogenic virus from a large sequence dataset. BMC Bioinformatics 2014; 15: 254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Liu S, Vijayendran D & Bonning BC. Next generation sequencing technologies for insect virus discovery. Viruses 2011; 3: 1849–1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhou P, Cowled C, Todd S, et al Type III IFNs in pteropid bats: differential expression patterns provide evidence for distinct roles in antiviral immunity. J Immunol 2011; 186: 3138–3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Shi Z & Hu Z. A review of studies on animal reservoirs of the SARS coronavirus. Virus Res 2008; 133: 74–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kan B, Wang M, Jing H, et al Molecular evolution analysis and geographic investigation of severe acute respiratory syndrome coronavirus‐like virus in palm civets at an animal market and on farms. J Virol 2005; 79: 11892–11900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lin CY, Roberts GW, Kift‐Morgan A, Donovan KL, Topley N & Eberl M. Pathogen‐specific local immune fingerprints diagnose bacterial infection in peritoneal dialysis patients. J Am Soc Nephrol 2013; 24: 2002–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Jackson JA, Hall AJ, Friberg IM, et al An immunological marker of tolerance to infection in wild rodents. PLoS Biol 2014; 12: e1001901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sheldon BC & Verhulst S. Ecological immunology: costly parasite defences and trade‐offs in evolutionary ecology. Trends Ecol Evol 1996; 11: 317–321. [DOI] [PubMed] [Google Scholar]
- 58. Rajabi‐Maham H, Orth A & Bonhomme F. Phylogeography and postglacial expansion of Mus musculus domesticus inferred from mitochondrial DNA coalescent, from Iran to Europe. Mol Ecol 2008; 17: 627–641. [DOI] [PubMed] [Google Scholar]
- 59. Behnke J & Harris PD. Heligmosomoides bakeri: a new name for an old worm? Trends Parasitol 2010; 26: 524–529. [DOI] [PubMed] [Google Scholar]
- 60. White TA & Perkins SE. The ecoimmunology of invasive species. Funct Ecol 2012; 26: 1313–1323. [Google Scholar]
- 61. Grabherr MG, Haas BJ, Yassour M, et al Full‐length transcriptome assembly from RNA‐Seq data without a reference genome. Nat Biotechnol 2011; 29: 644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Eberhardt AT, Costa SA, Marini MR, et al Parasitism and physiological trade‐offs in stressed capybaras. PLoS One 2013; 8: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Racca AL, Eberhardt AT, Moreno PG, Baldi C & Beldomenico PM. Differences in natural antibody titres comparing free‐ranging guanacos (Lama guanicoe) and capybaras (Hydrochoerus hydrochaeris). Vet J 2014; 199: 308–309. [DOI] [PubMed] [Google Scholar]
- 64. Schwensow N, Axtner J & Sommer S. Are associations of immune gene expression, body condition and parasite burden detectable in nature? A case study in an endemic rodent from the Brazilian Atlantic Forest. Infect Genet Evol 2011; 11: 23–30. [DOI] [PubMed] [Google Scholar]
- 65. Pradet‐Balade B, Boulmé F, Beug H, Müllner EW & Garcia‐Sanz JA. Translation control: bridging the gap between genomics and proteomics? Trends Biochem Sci 2001; 26: 225–229. [DOI] [PubMed] [Google Scholar]
- 66. Payne SH. The utility of protein and mRNA correlation. Trends Biochem Sci 2015; 40: 1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Oko L, Aduddell‐Swope B, Willis D, et al Profiling helper T cell subset gene expression in deer mice. BMC Immunol 2006; 7: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Jackson JA, Friberg IM, Bolch L, et al Immunomodulatory parasites and toll‐like receptor‐mediated tumour necrosis factor alpha responsiveness in wild mammals. BMC Biol 2009; 7: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Schountz T & Prescott J. Hantavirus immunology of rodent reservoirs: current status and future directions. Viruses 2014; 6: 1317–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Johnston PR & Rolff J. Immune‐ and wound‐dependent differential gene expression in an ancient insect. Dev Comp Immunol 2013; 40: 320–324. [DOI] [PubMed] [Google Scholar]
- 71. Lenz TL, Eizaguirre C, Rotter B, Kalbe M & Milinski M. Exploring local immunological adaptation of two stickleback ecotypes by experimental infection and transcriptome‐wide digital gene expression analysis. Mol Ecol 2013; 22: 774–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. McLoughlin KE, Nalpas NC, Rue‐Albrecht K, et al RNA‐seq transcriptional profiling of peripheral blood leukocytes from cattle infected with Mycobacterium bovis . Front Immunol 2014; 5: 396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Day E, Dear PH & McCaughan F. Digital PCR strategies in the development and analysis of molecular biomarkers for personalized medicine. Methods 2013; 59: 101–107. [DOI] [PubMed] [Google Scholar]
- 74. Forreryd A, Johansson H, Albrekt AS & Lindstedt M. Evaluation of high throughput gene expression platforms using a genomic biomarker signature for prediction of skin sensitization. BMC Genom 2014; 15: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Villaverde AF & Banga JR. Reverse engineering and identification in systems biology: strategies, perspectives and challenges. J R Soc Interface 2014; 11: 20130505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Molloy MJ, Bouladoux N & Belkaid Y. Intestinal microbiota: shaping local and systemic immune responses. Semin Immunol 2012; 24: 58–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Hooper LV, Littman DR & Macpherson AJ. Interactions between the microbiota and the immune System. Science 2012; 336: 1268–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Hanski I, von Hertzen L, Fyhrquist N, et al Environmental biodiversity, human microbiota, and allergy are interrelated. Proc Natl Acad Sci USA 2012; 109: 8334–8339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Sayi A, Kohler E, Toller IM, et al TLR‐2‐activated B cells suppress Helicobacter‐induced preneoplastic gastric immunopathology by inducing T regulatory‐1 cells. J Immunol 2011; 186: 878–890. [DOI] [PubMed] [Google Scholar]
- 80. Vijay‐Kumar M, Aitken JD, Carvalho FA, et al Metabolic syndrome and altered gut microbiota in mice lacking toll‐like receptor 5. Science 2010; 328: 228–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Medzhitov R. TLR‐mediated innate immune recognition. Semin Immunol 2007; 19: 1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Friberg IM, Little S, Ralli C, et al Macroparasites at peripheral sites of infection are major and dynamic modifiers of systemic antimicrobial pattern recognition responses. Mol Ecol 2013; 22: 2810–2826. [DOI] [PubMed] [Google Scholar]
- 83. Maizels RM, Hewitson JP & Gause WC. Heligmosomoides polygyrus: one species still. Trends Parasitol 2011; 27: 100–101. [DOI] [PubMed] [Google Scholar]
- 84. Reynolds LA, Harcus Y, Smith KA, et al MyD88 signaling inhibits protective immunity to the gastrointestinal helminth parasite Heligmosomoides polygyrus . J Immunol 2014; 193: 2984–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Helmby H & Grencis RK. Essential role for TLR4 and MyD88 in the development of chronic intestinal nematode infection. Eur J Immunol 2003; 33: 2974–2979. [DOI] [PubMed] [Google Scholar]
- 86. Behnke JM, Mugambi JM, Clifford S, et al Genetic variation in resistance to repeated infections with Heligmosomoides polygyrus bakeri, in inbred mouse strains selected for the mouse genome project. Parasite Immunol 2006; 28: 85–94. [DOI] [PubMed] [Google Scholar]
- 87. Murphy TJ, Choileain NN, Zang Y, Mannick JA & Lederer JA. CD4+CD25+ regulatory T Cells control innate immune reactivity after injury. J Immunol 2005; 174: 2957–2963. [DOI] [PubMed] [Google Scholar]
- 88. Segura M, Su Z, Piccirillo C & Stevenson MM. Impairment of dendritic cell function by excretory‐secretory products: a potential mechanism for nematode‐induced immunosuppression. Eur J Immunol 2007; 37: 1887–1904. [DOI] [PubMed] [Google Scholar]
- 89. Mueller T, Terada T, Rosenberg IM, Shibolet O & Podolsky DK. Th2 cytokines down‐regulate TLR expression and function in human intestinal epithelial cells. J Immunol 2006; 176: 5805–5814. [DOI] [PubMed] [Google Scholar]
- 90. Guivier E, Galan M, Salvador AR, et al Tnf‐alpha expression and promoter sequences reflect the balance of tolerance/resistance to Puumala hantavirus infection in European bank vole populations. Infect Genet Evol 2010; 10: 1208–1217. [DOI] [PubMed] [Google Scholar]
- 91. Axtner J & Sommer S. Heligmosomoides polygyrus infection is associated with lower MHC class II gene expression in Apodemus flavicollis: indication for immune suppression? Infect Genet Evol 2011; 11: 2063–2071. [DOI] [PubMed] [Google Scholar]
- 92. Schountz T, Shaw TI, Glenn TC, Feldmann H & Prescott J. Expression profiling of lymph node cells from deer mice infected with Andes virus. BMC Immunol 2013; 14: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Boppana VD, Thangamani S, Adler AJ & Wikel SK. SAAG‐4 is a novel mosquito salivary protein that programmes host CD4(+) T cells to express IL‐4. Parasite Immunol 2009; 31: 287–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Boppana VD, Thangamani S, Alarcon‐Chaidez FJ, Adler AJ & Wikel SK. Blood feeding by the Rocky Mountain spotted fever vector, Dermacentor andersoni, induces interleukin‐4 expression by cognate antigen responding CD4(+) T cells. Parasit Vectors 2009; 2: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Gause WC, Urban JF & Stadecker MJ. The immune response to parasitic helminths: insights from murine models. Trends Immunol 2003; 24: 269–277. [DOI] [PubMed] [Google Scholar]
- 96. Ho IC, Tai TS & Pai SY. GATA3 and the T‐cell lineage: essential functions before and after T‐helper‐2‐cell differentiation. Nat Rev Immunol 2009; 9: 125–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Allen JE & Wynn TA. Evolution of Th2 immunity: a rapid repair response to tissue destructive pathogens. PLoS Pathog 2011; 7: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Raberg L, Graham AL & Read AF. Decomposing health: tolerance and resistance to parasites in animals. Philos Trans R Soc Lond B Biol Sci 2009; 364: 37–49. [DOI] [PMC free article] [PubMed] [Google Scholar]


