Abstract
BACKGROUND: Bemisia tabaci, the sweetpotato whitefly, is a globally invasive pest that causes serious agricultural damage by transmitting plant viruses. This pest forms a cryptic species complex that displays morphologically indistinguishable biotypes. Among them, the B and Q biotypes are the most important pests worldwide. Because they have different levels of insecticide resistance, these biotypes must be identified in order to achieve proper pest control. Therefore, a convenient, rapid and specific detection method for identifying the two biotypes is necessary.
RESULTS: Loop‐mediated isothermal amplification (LAMP) was employed for rapid identification of B. tabaci B and Q biotypes. By combining a quick DNA extraction method, identification of the two biotypes was achieved within 1 h of detection time. The LAMP assay was applied to study the dynamics of B. tabaci biotypes both in the field and in greenhouses. It was found that, while temperature may be important for population dynamics of the whitefly in the field, population dynamics in greenhouse conditions may be influenced by the types of insecticide.
CONCLUSION: The newly designed LAMP assay is a simple, rapid and accurate method for identifying the B and Q biotypes. It can be conducted by non‐specialists and can contribute to pest management. Copyright © 2012 Society of Chemical Industry
Keywords: LAMP, pest, molecular marker, whitefly, COI
INTRODUCTION
The whitefly, Bemisia tabaci (Gennadius) (Hemiptera: Aleyrodidae), is a globally invasive pest spread by poinsettia plants; it is known as a superbug in the agricultural sector and causes massive damage.1, 2 This polyphagous pest transmits more than 110 plant viruses, most of them geminiviruses. Whiteflies are the main vector for Begomovirus, which causes serious diseases in tomato, melon, squash, etc.3, 4 High genetic diversity and morphologically indistinguishable populations are observed in B. tabaci, suggesting that it forms a species complex.
Based on mitochondrial differences and geographic distributions, B. tabaci is divided into 24 biotypes and 12 genetic groups.1, 2 Different courtship behaviours have been observed among biotypes, suggesting the existence of reproductive barriers among them,5 and B. tabaci is a species complex with many cryptic species. Within this species complex, the B and Q biotypes are the most important pests worldwide. While the B biotype has high fecundity and a broad host range, the Q biotype has high‐temperature tolerance.6, 7 In addition, they are sometimes sympatrically distributed8, 9, 10 and have different levels of insecticide resistance, which may influence their population dynamics.7, 10, 11, 12 Therefore, the development of a rapid identification method to distinguish them is needed for pest control.
Many molecular tools have been developed to identify B. tabaci biotypes. These include random amplification polymorphism of DNA‐polymerase chain reaction (RAPD‐PCR), restriction fragment length polymorphism (RFLP), sequence‐characterised amplified region (SCAR) and real‐time PCR technology.3, 13, 14, 15, 16, 17 Nevertheless, these methods rely on specialised equipment and well‐trained technicians, neither of which is commonly available in the field. In addition, they are expensive and time consuming, which precludes their use in large‐scale surveys. More importantly, these methods might not be capable of providing real‐time information for pest control strategies. In the present paper, a loop‐mediated isothermal amplification (LAMP) assay was applied for the rapid identification of B. tabaci biotypes. The LAMP assay was developed for rapid, efficient and specific amplification of DNA, and it is performed under isothermal conditions that require a water bath or a heat block to catalyse the reaction. This method requires no specialised equipment or specially trained technicians and is more cost effective than real‐time PCR. Therefore, LAMP is suitable for large‐scale field surveys and can provide useful information for pest control strategies. LAMP is a popular and useful method for rapid pathogen/pest detection and identification. For instance, it was applied to detect the severe acute respiratory syndrome (SARS) coronavirus, a serious human disease pathogen associated with pneumonia.18 In previous studies, a whitefly‐transmitted geminivirus was diagnosed with LAMP in infected plants.19, 20 The LAMP assay has also been applied to identify malaria mosquito vectors and the Mediterranean fruit fly, an important insect pest.21, 22
Mitochondrial cytochrome oxidase subunit I (COI) gene was selected to design LAMP primers specific for B. tabaci B and Q biotypes. The COI gene is commonly used to study and identify different biotypes of the B. tabaci species complex.2, 13, 23 COI sequences of different biotypes were downloaded from GenBank, and two sets of primers were designed for the LAMP assay. It was shown that the present assay could specifically and accurately identify the B and Q biotypes. Applying LAMP assays to monitor the population dynamics of whitefly populations revealed that, while temperature may be important for population dynamics of whiteflies in the field, population dynamics in greenhouse conditions might be influenced by the types of insecticide. Therefore, LAMP assays would be useful in identifying B and Q biotypes to help with pest management.
MATERIALS AND METHODS
Whitefly samples and DNA extraction
Whiteflies were collected from Taiwan or obtained from other researchers (Table 1). Adult whiteflies were preserved in 95% ethanol and stored at − 20 °C for later use in this experiment. Genomic DNA was extracted using the BuccalAMP™ DNA extraction kit (Epicentre, Madison, WI). An adult whitefly was homogenised in a mortar and pestle, incubated with 50 µL of quick extraction solution at 65 °C for 6 min and heated to 98 °C for 2 min to terminate the reaction. The extract, which contained genomic DNA, was either immediately used for LAMP assays or stored at − 20 °C for further experiments.
Table 1.
Samples of Bemisia tabaci biotypes subjected to LAMP
| Biotype | Location | Collector |
|---|---|---|
| B | Taiwan | CH Hsieh |
| China | CH Hsieh | |
| Japan | KI Honda and S Ueda | |
| Korea | SJ Suh | |
| Spain | S Pascual | |
| Q | Taiwan | CH Hsieh |
| China | D Chu | |
| Japan | KI Honda and S Ueda | |
| Korea | YH Chiang | |
| Spain | S Pascual | |
| AsiaI | Taiwan | CC Ko |
| AsiaII | Taiwan | CH Hsieh |
| SSAa | Spain | I Beford |
| NWb | United States | JK Brown |
SSA, Sub‐Saharan Africa non‐silverleafing.
NW, New World.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
LAMP primer design
The mitochondrial COI gene sequences of B. tabaci biotypes were downloaded from GenBank (Table 2) to design LAMP‐specific primers for the B and Q biotypes. All sequences were aligned using the Clustal X 2.0 program.24 LAMP primers were designed on the basis of biotype‐specific single‐nucleotide polymorphic (SNP) sites (Fig. 1), using PrimerExplorer v.4 (http://primerexplorer.jp/e/). LAMP primer sets specific for the B and Q biotypes are given in Table 3 and Fig. 1.
Table 2.
GenBank accession numbers of COI sequences from Bemisia tabaci biotypes
| Biotype | Acronym | Location | GenBank accession number |
|---|---|---|---|
| B | TaiwanB | Tainan, Taiwan | DQ174533 |
| ArizonaB | Arizona, United States | AY057123 | |
| ChinaBeijingB | Beijing, China | DQ989523 | |
| FranceB | Antibes, France | AJ550170 | |
| IsraelBF17 | Israel | DQ174536 | |
| JapanHonshuB | Japan | DQ989532 | |
| KoreaGoyangB | Korea | DQ989531 | |
| MoroccoB | Saidia, Morocco | AJ517768 | |
| PakistanB | Sindh, Pakistan | AJ510081 | |
| SouthAfricaB | South Africa | AY057140 | |
| SpainB | Spain | DQ989551 | |
| Q | TaiwanTNQ | Tainan, Taiwan | DQ989547 |
| ChinaZhejiangQ | Zhejiang, China | DQ473394 | |
| FranceQ | Var, Franca | AM691063 | |
| GreeceQ | Agrinio, Greece | DQ365865 | |
| JapanOKCQ | Kagoshima, Japan | AB204587 | |
| KoreaGeojeQ | Geoje, Korea | DQ462585 | |
| MoroccoQ | Morocco | AJ517769 | |
| NetherlandQ | Netherland | DQ174541 | |
| SpianQ | Spain | AF342775 | |
| USAGQ | Georgia, United States | EF080823 | |
| AsiaI | TaiwanAsiaI | Chiayi, Taiwan | DQ174525 |
| AsiaII | TaiwanAsiaII | Kaohsiung, Taiwan | DQ174521 |
| Australia | AustraliaAus | Australia | DQ174529 |
| Italy | ItalyT | Puglia, Italy | AY827595 |
| New World | USAA | United States | DQ174542 |
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
Figure 1.
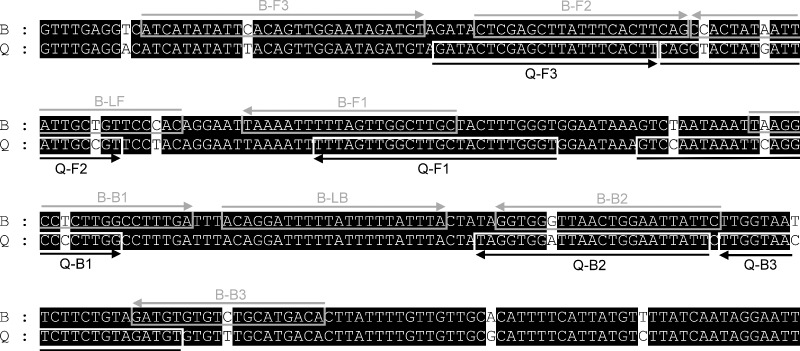
Mitochondrial cytochrome oxidase I (COI) gene sequences were used to design LAMP primers for Bemisia tabaci biotypes B and Q. LAMP primer sets comprise external primers (F3 and B3), internal primers (FIP and BIP) and loop primers (LF and LB).
Table 3.
LAMP primer sets specific for Bemisia tabaci biotypes B and Q
| Biotype | Primer | Sequence (5′–3′) |
|---|---|---|
| B | B‐F3 | ATATTCACAGTTGGAATAGATGT |
| B‐B3c | TGTCATGCAGACACACATC | |
| B‐FIP (B‐F1c + B‐F2) | CCAAAGTAGCAAGCCAACTAAAAATA‐CTCGAGCTTATTTCACTTCAG | |
| B‐BIP (B‐B1 + B‐B2c) | TAAGGCCTCTTGGCCTTTGA‐GAATAATTCCAGTTAACCCACC | |
| B‐LFc | GTGGGAACAGCAATAATTATAGTGG | |
| B‐LB | ACAGGATTTTTATTTTTTATTTA | |
| Q | Q‐F3 | GATACTCGAGCTTATTTCACTT |
| Q‐B3c | ACATCTACAGAAGAGTTACCAA | |
| Q‐FIP (Q‐F1c + Q‐F2) | ACCCAAAGTAGCAAGCCAACTAAA‐CAGCTACTATGATTATTGCCGT | |
| Q‐BIP (Q‐B1 + Q‐B2c) | GTCCAATAAATTCAGGCCCCTTGG‐AATAATTCCAGTTAATCCACCTA |
c: reverse‐complement sequence.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
LAMP assays
The LAMP reaction mixture (25 µL in total) contained 3 µL of template DNA (∼100 ng), 8 U of Bst DNA polymerase (New England Biolabs, Ipswich, MA), 1× reaction mix (20 mM of Tris‐HCl (pH 8.8), 10 mM of KCl, 8 mM of MgSO4, 10 mM of (NH4)2SO4, 0.1% Tween 20, 0.1% Triton X‐100, 0.8 M of betaine and 1.4 mM of each dNTP), 0.4 µM of each external primer (F3 and B3), 4 µM of each internal primer (FIP and BIP) and 2 µM of each loop primer (LF and LB). The reaction mixture was incubated at 63 °C for 30, 45, 60 or 90 min, and the reaction was terminated at 80 °C for 5 min. The amplification reaction was carried out in a GeneAmp® PCR System 2700 thermal cycler (Applied Biosystems, Foster City, CA).
The LAMP products were examined by electrophoresis using a 2% agarose gel with ethidium bromide (EtBr). Fluorescence visualisation was accomplished by adding 1 µL of 50× diluted SYBR Green I stain (Bio Basic, Ontario, Canada) and observing the sample under both visible and ultraviolet (UV) light.
Application of LAMP to monitor the dynamics of B. tabaci biotypes B and Q in Taiwan
Whiteflies were collected from fields (n = 555) and poinsettia greenhouses (n = 489) in Taiwan from 2005 to 2010. The collection sites were located in four counties (Nantou, Miaoli, Taoyuan and Yilan). The LAMP method was used to identify the two B. tabaci biotypes, and the proportion of each genotype was estimated in order to characterise the dynamics between biotypes in each county over time. The Pearson correlation coefficient (r) was used to measure the correlation of whiteflies between fields and greenhouses.
RESULTS
Three LAMP primer sets were designed for the B biotype, including two outer primers (F3 and B3), two inner primers (FIP and BIP) and two loop primers (LF and LR) (Table 3, Fig. 1). The loop primers were designed on the basis of a dumbbell‐like structure in order to increase the number of starting points for DNA synthesis. No suitable loop primer was designed for the Q biotype. LAMP primers for the Q biotype included only outer and inner primers (Table 3, Fig. 1).
LAMP assays were carried out at 63 °C for 30, 45, 60 or 90 min, and results were visualised using gel electrophoresis. The LAMP assay specific for the B biotype showed a positive result as early as at 30 min, and signals became stronger as the reaction time increased from 30 to 90 min. In contrast, other biotypes—including Q, AsiaI and AsiaII—were still negative after 90 min of reacting (Fig. 2A). The LAMP assay specific for the Q biotype revealed positive results at 45–90 min, but no amplification was observed for the other biotypes, even at 90 min (Fig. 2B).
Figure 2.
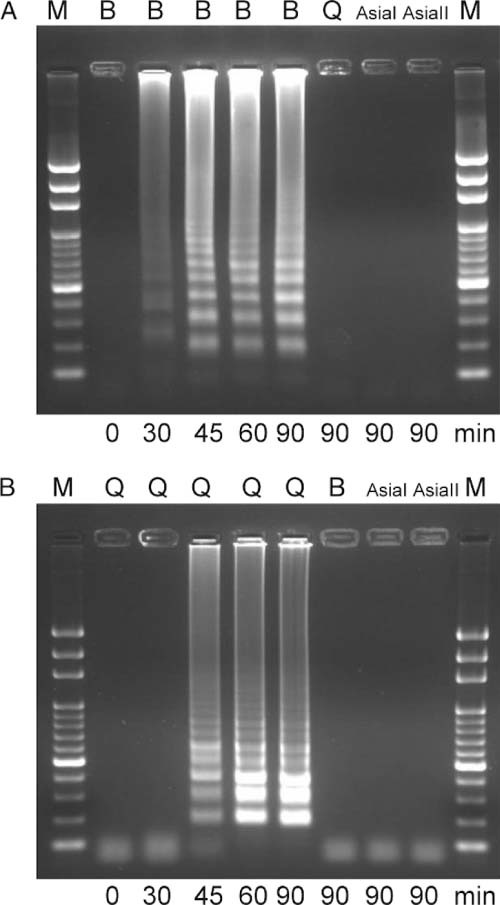
Detection of LAMP products by electrophoresis. A: LAMP assay specific for B biotypes. B: LAMP assay specific for Q biotypes. The test specimens included Bemisia tabaci B, Q, AsiaI and AsiaII biotypes. The LAMP reaction times were 0–90 min. M: Bio100 DNA Ladder™.
In order to accelerate the speed of identification, SYBR Green I was added to the reaction, with which a colour transformation from orange to green under visible light indicated positive amplification. In the LAMP assay for the B biotype, positive results for the B biotype, as indicated by a green colour, were observed at 30–90 min (Fig. 3A). This was in contrast to the results for AsiaI, AsiaII and Q biotypes, which remained orange at this time interval. However, positive LAMP results for the Q biotype, as indicated by a green colour, were observed at 45–90 min, while the other biotypes still had negative results, as indicated by their orange colour (Fig. 3B). The colour change under UV light conditions was also visualised (Fig. 4), and the results were identical.
Figure 3.
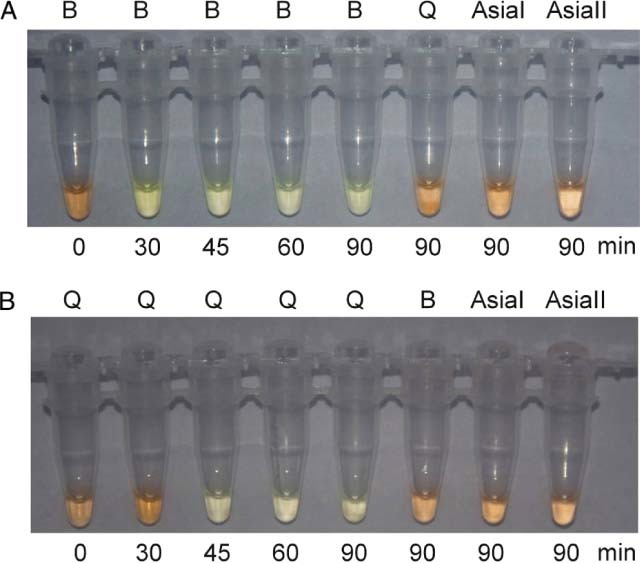
Detection of LAMP products with SYBR Green I staining. A: LAMP assay specific for B biotypes. B: LAMP assay specific for Q biotypes. Green colour indicates a positive result, and orange a negative result. The test specimens included Bemisia tabaci B, Q, AsiaI and AsiaII biotypes. The LAMP reaction times were 0–90 min.
Figure 4.
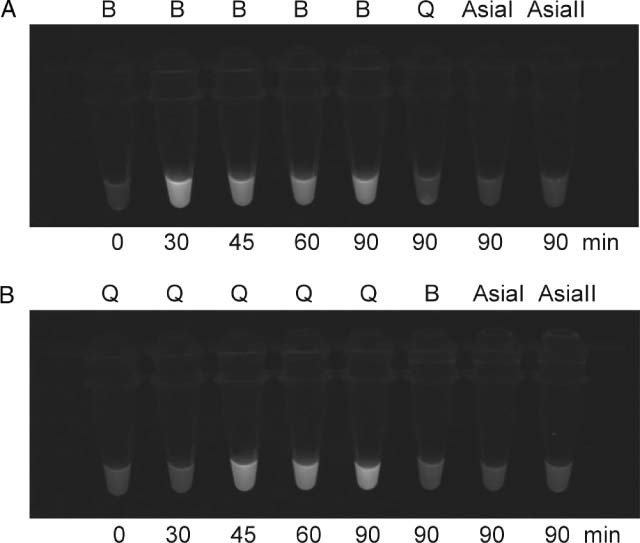
Detection of LAMP products with SYBR Green I staining under UV light. A: LAMP assay specific for B biotypes. B: LAMP assay specific for Q biotypes. Fluorescence emission indicates a positive result, and no emission indicates a negative result. The test specimens included Bemisia tabaci B, Q, AsiaI and AsiaII biotypes. The LAMP reaction times were 0–90 min.
The sensitivity of the assays was tested by serial tenfold dilutions of whitefly DNA. LAMP reactions were carried out at 63 °C for 60 min, and positive results were revealed for reactions with input DNA from 100 ng to 1 pg for both B (Fig. 5A) and Q (Fig. 5B) biotypes. The stability and specificity of the present LAMP assays were further tested for B. tabaci B and Q biotypes collected worldwide (Table 1). LAMP reactions were carried out at 63 °C for 45 min. As shown in Fig. 6, the present assays were able specifically to amplify the B (Fig. 6A) and Q (Fig. 6B) biotypes collected from different countries around the world.
Figure 5.
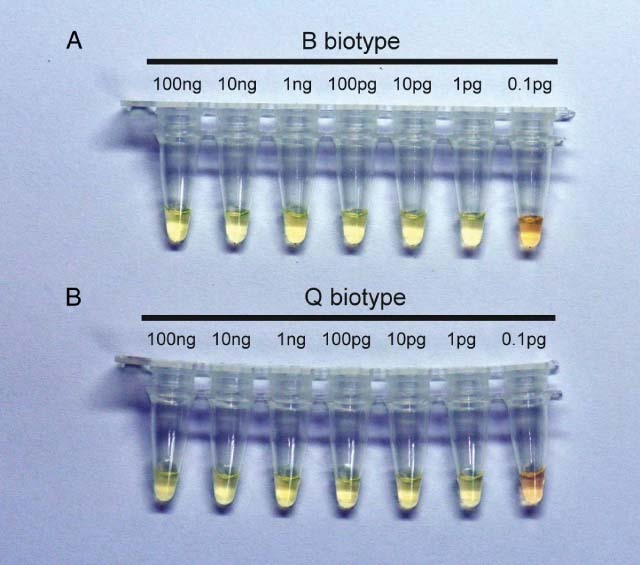
The sensitivity of LAMP assays for (A) B and (B) Q biotypes. A serial tenfold dilution of whitefly DNA was from 100 ng to 0.1 pg.
Figure 6.
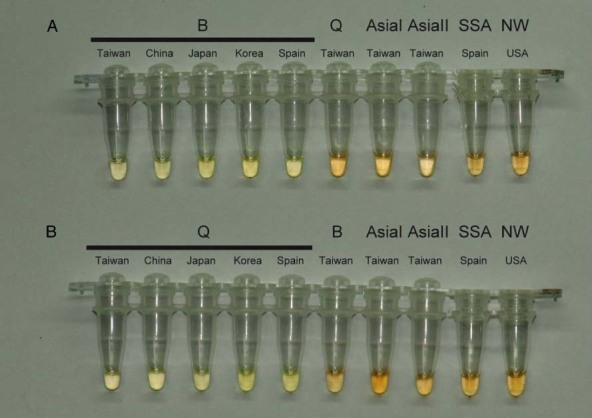
Detection of LAMP specificity for target biotypes with SYBR Green I staining. A: LAMP assay specific for B biotypes. B: LAMP assay specific for Q biotypes. Green colour indicates a positive result, and orange a negative result. The test specimens included Bemisia tabaci B, Q, AsiaI and AsiaII biotypes from Taiwan, China, Japan, Korea and Spain.
The specificity of the newly designed LAMP primers for different biotypes was demonstrated. In addition, by adding SYBR Green I to the reaction mixture, the results could be visualised under visible light 45 min after the reaction began. These LAMP assays were then applied to survey the distribution of the B and Q biotypes in the field and in poinsettia greenhouses. The results showed that biotype B was the most common one found in the fields. Its proportion gradually increased during the sampling period in Miaoli, Taoyuan and Yilan counties (Fig. 7) and comprised more than 50% of B. tabaci individuals collected in these areas. Interestingly, the proportion of biotype B collected in the field decreased from 75% in 2005 to 15% in 2009 in Nantou County. The Q biotype was only found in poinsettia greenhouses during the present survey. Although the B and Q biotypes are the two major biotypes, comprising more than 75% of B. tabaci collected in most greenhouses, their relative proportions varied greatly by year. For example, in Miaoli County, the proportion of Q to B was 25%/60% in 2006, 90%/10% in 2007, 100%/0% in 2008, 33%/33% in 2009 and 60%/35% in 2010. Great variations in proportions of the B and Q biotypes were also observed in Nantou, Yilan and Taoyuan counties. The frequencies of biotype B collected from fields and greenhouses revealed no correlation (r = 0.26, P = 0.60).
Figure 7.
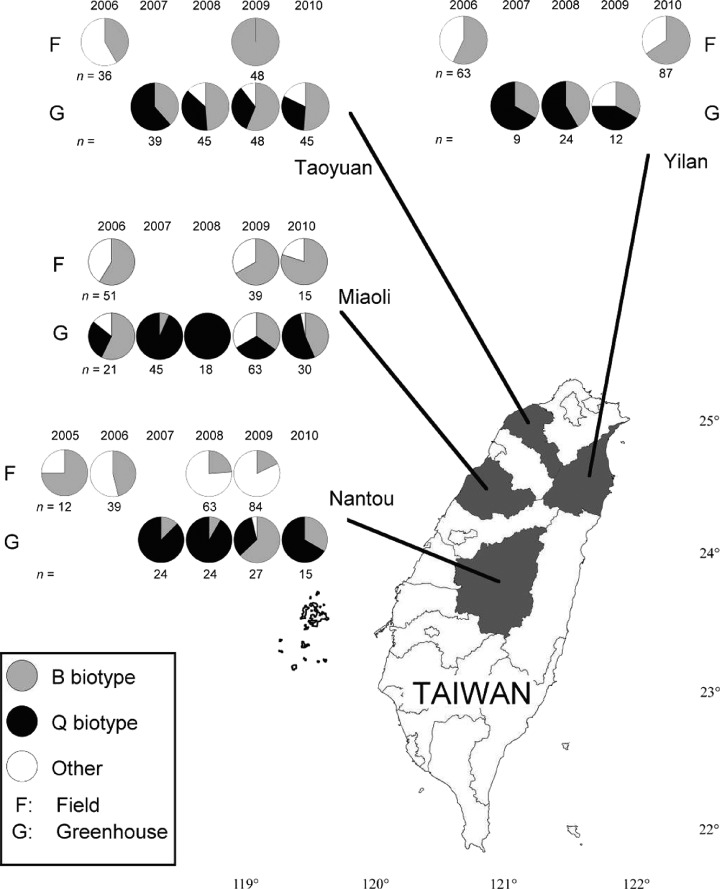
Application of LAMP to estimate proportions of Bemisia tabaci biotypes from fields and greenhouses in Taiwan from 2005 to 2010. In total, 1044 individuals were collected in fields (n = 555) and greenhouses (n = 489) from four counties (Nantou, Miaoli, Taoyuan and Yilan).
DISCUSSION
The newly designed LAMP assays were successfully applied to identify and distinguish B. tabaci B and Q biotypes, which are two globally invasive pests that cannot be morphologically distinguished. These assays are highly sensitive, as the detection limit can be as low as 1 pg DNA in a total of 25 µL of reaction mixture. The specificities of the LAMP assays were demonstrated using samples collected worldwide. In a comparison of the reactions between the B and Q biotypes, the LAMP reaction for the B biotype in the presence of loop primers required only 30 min, which was 15 min less than the time taken for LAMP reaction of the Q biotype with no loop primers. This result confirms that the use of loop primers efficiently reduces reaction times. Furthermore, adding SYBR Green I to the LAMP assay increased the sensitivity of detection. While there was only weak amplification detected by gel electrophoresis after 30 min of reaction, a strong positive signal was revealed by SYBR Green I staining at the same time point.
Although there are several methods available for the identification of different B. tabaci biotypes, the present assays have several advantages over others. Firstly, unlike PCR‐based methods, which require special equipment and reagents, the present method requires only a thermostat (such as a water or dry bath) with limited kinds of chemical. Secondly, when combined with quick DNA extraction methods, which can be done in 10 min, the total reaction takes only 1 h. Thirdly, the LAMP assay is cost effective, as one reaction costs less than $US 1 (DNA extraction, $US 0.25; primers, $US 0.07; Bst polymerase, $US 0.5; dNTP, $US 0.04; SYBR, $US 0.04). Fourthly, the result is easily visualised by a colour change, and no sophisticated analytical tool is necessary for interpretation. Finally, the procedures described here can be performed by non‐specialists who have limited knowledge of whiteflies or molecular biology. All of the above make it possible to conduct large‐scale surveys in the field. In addition, the current design provides real‐time monitoring for biotypes B and Q. Because these two biotypes are known to have different levels of insecticide resistance, the information could be useful in whitefly control.
The present methods were applied to monitor the population dynamics of B. tabaci biotypes in Taiwan. The B biotype was first introduced into Taiwan in the early 1990s,25 but its proportions among whitefly populations over time remain unclear. In the present data, the proportion of biotype B gradually increased over the course of the season in whitefly populations collected from the field in Miaoli, Taoyuan and Yilan counties. The increasing trend of B biotypes in these locations is interesting, because it seems to be consistent with the invasive theory of asymmetric mating interactions,26 in which the invasive whitefly of B. tabaci biotype B is widely displacing indigenous biotypes in the field. However, in Nantou County, the opposite trend of a decreasing B biotype population was observed, suggesting the existence of unknown factors restricting population growth in that area. Because there was only limited correlation of B biotype abundance between samples collected from greenhouses and fields, population dynamics of biotype B in the field cannot be explained by the influence of whiteflies from greenhouses. According to the life‐table study, environmental temperature strongly influences the development and reproduction of biotype B. The average adult longevity at 15, 20 and 28 °C was 5.5, 15.8 and 15.4 days respectively, and the fecundity levels at the above temperatures were 2, 54 and 109 eggs respectively. It seems that 20 °C is a critical temperature for the development and reproduction of biotype B. The average elevation of Nantou County is higher than those of the other three counties, and the average annual temperature in Nantou is 18.9 °C (the average monthly temperatures range from 15.4 to 22.5 °C), which is lower than the 22.7 °C in Yilan (19.8–26.4 °C), the 21.7 °C in Taoyuan (19.0–24.9 °C) and the 20.2 °C in Miaoli (17.5–24.2 °C). It was hypothesised that the lower average annual temperature may have limited the population growth and dispersal of biotype B in Nantou County.
The Q biotype of B. tabaci is an emerging, globally invasive pest and was reported from poinsettia greenhouses in Taiwan only in recent years.27 In the present study, the Q biotype was only restricted to greenhouses. In addition, the relative proportions of B and Q biotypes varied greatly by year. The environmental conditions in greenhouses are more stable than in the field, and thus the temperature might not be the most important factor influencing population dynamics in this setting. In addition, it was shown that biotype Q has higher mortality and lower fecundity than biotype B, and the latter has a competitive advantage over the former in greenhouse conditions.6, 28 It is expected that biotype Q will be replaced by biotype B in greenhouses if there is no other intervention. Nevertheless, complex population dynamics between biotypes B and Q was observed in different years. After interviewing growers, it was found that different types of insecticide, including organophosphates, imidacloprid of the neonicotinoids, pyrethroids and pyriproxyfen, or combinations of them, were applied for whitefly control in different greenhouses. In addition, a grower would use different insecticides at different times of the year. It was proposed that different levels of insecticide resistance for B and Q biotypes might influence the dynamics and distribution of the two biotypes.10, 28 Further, neonicotinoid resistance of the Q biotype was observed in Taiwan and elsewhere.7, 10, 27 It was hypothesised that the kind of insecticide used might influence the population dynamics of the two biotypes in greenhouses in Taiwan.
Therefore, although the B biotype may have a competitive advantage over the Q biotype in greenhouse conditions, this may be due to a higher level of insecticide resistance.7, 10 The combination of these factors may explain the observed complex population dynamics of biotypes B and Q in greenhouses in Taiwan. Most poinsettia greenhouse owners applied insecticides for whitefly control without prior knowledge of the biotypes. By applying the present LAMP method, greenhouse owners may be able to monitor the population dynamics of different biotypes in real time, which would be valuable for whitefly management.
In conclusion, LAMP assays have been designed to identify biotypes B and Q of B. tabaci. The specificity and sensitivity of the methods has been demonstrated. A proof‐of‐concept study has been performed in which the population dynamics of whiteflies in both the field and greenhouses was monitored. One of the most important implications of the present results is that the LAMP assays can be conducted by non‐specialists, using limited equipment. In the most extreme case, the assays can be done in a refitted automobile equipped with a thermostat and a fridge. Thus, farmers could test whitefly biotypes in their greenhouses before determining their pest control strategies.
Acknowledgements
The authors thank HT Fang, YH Chiang and CG Huang (Department of Entomology, National Taiwan University, Taipei, Taiwan) and MC Cheng (Bureau of Animal and Plant Health Inspection and Quarantine, Taipei, Taiwan) for their assistance, and CC Yang (Master Program for Plant Medicine, National Taiwan University, Taipei, Taiwan) for comments on this manuscript. They also thank SJ Suh (National Plant Quarantine Service, Goyang, South Korea), KI Honda and S Ueda (National Institute of Vegetable and Tea Science, Japan), D Chu (High‐Tech Research Centre, Shandong Academy of Agricultural Sciences, and the Key Laboratory for Genetic Improvement of Crop, Animal and Poultry of Shandong Province, Shandong, China), JK Brown (University of Arizona, Tucson, AZ), I Bedford (John Innes Centre, Norwich, UK) and S. Pascual (INIA, Departamento de Proteccion Vegetal, Madrid, Spain) for their assistance with whitefly materials. This paper was supported in part by grants from the National Science Council (NSC97‐2621‐B‐002‐008‐MY3, NSC100‐2313‐B‐002‐014) to CC Ko and (NSC 97‐2321‐B‐002‐027, NSC 98‐2321‐B‐002‐005‐, NSC 99‐2321‐B‐002‐004‐) to HY Wang, and also the grant from Bureau of Animal and Plant Health Inspection and Quarantine, Taiwan (99AS‐9.3.1‐BQ‐B2).
Contributor Information
Hurng‐Yi Wang, Email: hurngyi@ntu.edu.tw.
Chiun‐Cheng Ko, Email: kocc2501@ntu.edu.tw.
REFERENCES
- 1. Perring TM, The Bemisia tabaci species complex. Crop Prot 20: 725–737 (2001). [Google Scholar]
- 2. Boykin LM, Shatters RG, Rosell RC, McKenzie CL, Bagnall RA, De Barro P, et al, Global relationships of Bemisia tabaci (Hemiptera: Aleyrodidae) revealed using Bayesian analysis of mitochondrial COI DNA sequences. Mol Phylogenet Evol 44: 1306–1319 (2007). [DOI] [PubMed] [Google Scholar]
- 3. Jones DR, Plant viruses transmitted by whiteflies. Eur J Plant Pathol 109: 195–219 (2003). [Google Scholar]
- 4. Fauquet CM and Stanley J, Geminivirus classification and nomenclature: progress and problems. Ann Appl Biol 142: 165–189 (2003). [Google Scholar]
- 5. Xu J, De Barro PJ and Liu SS, Reproductive incompatibility among genetic groups of Bemisia tabaci supports the proposition that the whitefly is a cryptic species complex. Bull Entomol Res 100: 359–366 (2010). [DOI] [PubMed] [Google Scholar]
- 6. Pascual S and Callejas C, Intra‐ and interspecific competition between biotypes B and Q of Bemisia tabaci (Hemiptera: Aleyrodidae) from Spain. Bull Entomol Res 94: 369–375 (2004). [DOI] [PubMed] [Google Scholar]
- 7. Horowitz AR, Kontsedalov S, Khasdan V and Ishaaya I, Biotypes B and Q of Bemisia tabaci and their relevance to neonicotinoid and pyriproxyfen resistance. Arch Insect Biochem Physiol 58: 216–225 (2005). [DOI] [PubMed] [Google Scholar]
- 8. Dalton R, Whitefly infestations—the Christmas invasion. Nature 443: 898–900 (2006). [DOI] [PubMed] [Google Scholar]
- 9. Hsieh CH, Wang CH and Ko CC, Evidence from molecular markers and population genetic analyses suggests recent invasions of the Western North Pacific region by biotypes B and Q of Bemisia tabaci (Gennadius). Environ Entomol 36: 952–961 (2007). [DOI] [PubMed] [Google Scholar]
- 10. Khasdan V, Levin I, Rosner A, Morin S, Kontsedalov S, Maslenin L, et al, DNA markers for identifying biotypes B and Q of Bemisia tabaci (Hemiptera: Aleyrodidae) and studying population dynamics. Bull Entomol Res 95: 605–613 (2005). [DOI] [PubMed] [Google Scholar]
- 11. Gorman K, Slater R, Blande JD, Clarke A, Wren J, McCaffery A, et al, Cross‐resistance relationships between neonicotinoids and pymetrozine in Bemisia tabaci (Hemiptera: Aleyrodidae). Pest Manag Sci 66: 1186–1190 (2010). [DOI] [PubMed] [Google Scholar]
- 12. Dennehy TJ, Degain BA, Harpold VS, Zaborac M, Morin S, Fabrick JA, et al, Extraordinary resistance to insecticides reveals exotic Q biotype of Bemisia tabaci in the New World. J Econ Entomol 103: 2174–2186 (2010). [DOI] [PubMed] [Google Scholar]
- 13. Hsieh CH, Wang CH and Ko CC, Analysis of Bemisia tabaci (Hemiptera: Aleyrodidae) species complex and distribution in eastern Asia based on mitochondrial DNA markers. Ann Entomol Soc Am 99: 768–775 (2006). [Google Scholar]
- 14. De Barro PJ and Driver F, Use of RAPD PCR to distinguish the B biotype from other biotypes of Bemisia tabaci (Gennadius) (Hemiptera: Aleyrodidae). Aust J Entomol 36: 149–152 (1997). [Google Scholar]
- 15. Abdullahi I, Winter S, Atiri GI and Thottappilly G, Molecular characterization of whitefly, Bemisia tabaci (Hemiptera: Aleyrodidae) populations infesting cassava. Bull Entomol Res 93: 97–106 (2003). [DOI] [PubMed] [Google Scholar]
- 16. Ko CC, Hung YC and Wang CH, Sequence characterized amplified region marker for identifying biotype of Bemisia tabaci (Hem., Aleyrodidae). J Appl Entomol 131: 542–547 (2007). [Google Scholar]
- 17. Jones CM, Gorman K, Denholm I and Williamson MS, High‐throughput allelic discrimination of B and Q biotypes of the whitefly, Bemisia tabaci, using TaqMan allele‐selective PCR. Pest Manag Sci 64: 12–15 (2008). [DOI] [PubMed] [Google Scholar]
- 18. Hong TC, Mai QL, Cuong DV, Parida M, Minekawa H, Notomi T, et al, Development and evaluation of a novel loop‐mediated isothermal amplification method for rapid detection of severe acute respiratory syndrome coronavirus. J Clin Microbiol 42: 1956–1961 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fukuta S, Kato S, Yoshida K, Mizukami Y, Ishida A, Ueda J, et al, Detection of tomato yellow leaf curl virus by loop‐mediated isothermal amplification reaction. J Virol Methods 112: 35–40 (2003). [DOI] [PubMed] [Google Scholar]
- 20. Kuan CP, Wu MT, Lu YL and Huang HC, Rapid detection of squash leaf curl virus by loop‐mediated isothermal amplification. J Virol Methods 169: 61–65 (2010). [DOI] [PubMed] [Google Scholar]
- 21. Bonizzoni M, Afrane Y and Yan GY, Loop‐mediated isothermal amplification (LAMP) for rapid identification of Anopheles gambiae and Anopheles arabiensis mosquitoes. Am J Trop Med Hyg 81: 1030–1034 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Huang CG, Hsu JC, Haymer DS, Lin GC and Wu WJ, Rapid identification of the mediterranean fruit fly (Diptera: Tephritidae) by loop‐mediated isothermal amplification. J Econ Entomol 102: 1239–1246 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dinsdale A, Cook L, Riginos C, Buckley YM and De Barro P, Refined global analysis of Bemisia tabaci (Hemiptera: Sternorrhyncha: Aleyrodoidea: Aleyrodidae) mitochondrial cytochrome oxidase 1 to identify species level genetic boundaries. Ann Entomol Soc Am 103: 196–208 (2010). [Google Scholar]
- 24. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al, Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 (2007). [DOI] [PubMed] [Google Scholar]
- 25. Lin FC, Occurrence of whiteflies on ornamental plants and their control. Plant Prot Bull Spec Pub New 2: 177–184 (1994). [Google Scholar]
- 26. Liu SS, De Barro PJ, Xu J, Luan JB, Zang LS, Ruan YM, et al, Asymmetric mating interactions drive widespread invasion and displacement in a whitefly. Science 318: 1769–1772 (2007). [DOI] [PubMed] [Google Scholar]
- 27. Hsieh CH, Chiang YH and Ko CC, Population genetic structure of the newly invasive Q biotype of Bemisia tabaci in Taiwan. Entomol Exp Appl 138: 263–271 (2011). [Google Scholar]
- 28. Dalmon A, Halkett F, Granier M, Delatte H and Peterschmitt M, Genetic structure of the invasive pest Bemisia tabaci: evidence of limited but persistent genetic differentiation in glasshouse populations. Heredity 100: 316–325 (2008). [DOI] [PubMed] [Google Scholar]


