Abstract
Ubiquitin‐specific proteases (USPs) emerge as key regulators of numerous cellular processes and account for the bulk of human deubiquitinating enzymes (DUBs). Their modular structure, mostly annotated by sequence homology, is believed to determine substrate recognition and subcellular localization. Currently, a large proportion of known human USP sequences are not annotated either structurally or functionally, including regions both within and flanking their catalytic cores. To extend the current understanding of human USPs, we applied consensus fold recognition to the unannotated content of the human USP family. The most interesting discovery was the marked presence of reliably predicted ubiquitin‐like (UBL) domains in this family of enzymes. The UBL domain thus appears to be the most frequently occurring domain in the human USP family, after the characteristic catalytic domain. The presence of multiple UBL domains per USP protein, as well as of UBL domains embedded in the USP catalytic core, add to the structural complexity currently recognized for many DUBs. Possible functional roles of the newly uncovered UBL domains of human USPs, including proteasome binding, and substrate and protein target specificities, are discussed. Proteins 2007. © 2007 Wiley‐Liss, Inc.
Keywords: consensus fold recognition, deubiquitination, proteasome, ubiquitin, UBL, USP
INTRODUCTION
Post‐translational ubiquitination of proteins in eukaryotes governs cellular activities ranging from selective protein degradation by proteasomes to membrane protein trafficking, signal transduction, transcription, nuclear transport, autophagy, and immune responses.1, 2, 3, 4 Protein ubiquitination is catalyzed by the sequential action of E1, E2, and E3 enzymes that activate and transfer ubiquitin or ubiquitin‐like modifiers to the ε‐amino group of an internal lysine residue of target proteins.5, 6 Ubiquitination is a reversible process. The isopeptide bond between ubiquitin and a substrate protein, or between ubiquitin molecules in a polyubiquitin chain, can be cleaved by deubiquitinating enzymes (DUBs), which are also responsible for the activation of ubiquitin and ubiquitin‐like modifiers by C‐terminal processing of their precursors.7 A large number of DUBs have been discovered and represent an emerging class of ubiquitin pathway regulators, predominantly from eukaryotes,8 but also of bacterial and viral origins.9, 10, 11
New insights into molecular structures, biochemical activities, substrate specificities and functions have been gained for the current inventory of DUBs over the past decade.7, 8 Most known cellular DUBs are cysteine proteases, including those from the ubiquitin‐specific protease (USP) structural class, which represents the bulk (over 50) of DUBs encoded in the human genome.8, 12 Little is known about the physiological function of most human USPs, and specific substrates remain elusive. The current view is that the modular, multidomain architecture of USPs contributes to their specificity with respect to the type of ubiquitin polymer and modifier, but perhaps more importantly, to the target protein part of the substrate.8 Human USPs have highly variable amino acid sequences upstream and/or downstream of the catalytic core. A number of domains have been annotated in these regions based on sequence homology,8, 12 some already confirmed experimentally, for example, the TRAF‐like domain of human USP7, the DUSP domain of human USP15, and the CS domain of human USP19. However, a large proportion of the N‐ and C‐terminal extensions of human USPs remain structurally and functionally unannotated. Also, the size of their catalytic core domains varies from ∼300 to 800 residues due to large sequences uncharacterized structurally, which may play functional roles.
Given the currently known and expected important cellular roles of USPs, a detailed structural annotation of individual family members of this class of DUBs is an important step toward elucidating their molecular functions in human health and disease. On this account, we have subjected the currently unannotated content of human USP family to advanced structural bioinformatics techniques. The most impressive finding of this prediction exercise is the abundance of ubiquitin‐like (UBL) domains in this family of enzymes, both within and outside USP catalytic core domains. The newly uncovered UBL domains are likely to play important functional roles toward the substrate and target protein specificities of human USPs.
MATERIALS AND METHODS
Sequences of the currently known human USPs corresponding to the C19 family of the MEROPS peptidase database (http://merops.sanger.ac.uk/) were collected from the GenBank (http://www.ncbi.nlm.nih.gov/Genbank/) and SwissProt (http://www.expasy.org/sprot/) databases. Only one sequence was selected from those of multiple isoforms reported for some USPs (generally nearly identical mutation isoforms, otherwise the longest sequence was selected), thus leading to a nonredundant set of 54 distinct human USP sequences (see Supplementary Material). The boundaries of their catalytic core, as well as all their currently annotated domains outside this core domain were obtained from the Pfam (http://www.sanger.ac.uk/Software/Pfam/) and InterPro (http://www.ebi.ac.uk/interpro/) databases and confirmed, whenever available, with actual structures retrieved from the Protein Data Bank (PDB, http://www.rcsb.org/). The remaining unannotated sequence content was therefore defined by the sequences flanking or between the currently annotated domains, as well as inserted in the catalytic core. In the latter case, locating such insertions required (i) a multiple sequence alignment of all 54 catalytic core sequences, which was performed with the MAFFT5 algorithm,13 and (ii) comparisons with the minimal catalytic core domain delineated by its available crystal structures from several human USPs (with PDB IDs): 2 (2HD5), 7 (1NB8, 1NBF, 2F1Z), 8 (2GFO), and 14 (2AYN, 2AYO).
Structural domain detection of the currently unannotated content of the human USP family was carried out at the Structure Prediction Meta Server (http://bioinfo.pl/), which assembles state‐of‐the‐art fold recognition methods, and provides a consensus scoring of the three‐dimensional structure predictions generated for a given query sequence by independent algorithms, using the 3D‐Jury meta‐predictor.14 Short sequence stretches (<40 residues) were not considered. Overly long contiguous sequences (>800 residues) were split into shorter fragments prior to fold recognition calculations. This splitting was done in two ways: (i) generating three equal‐length sequences corresponding to the N‐ and C‐terminal halves plus the central region of the same length, and (ii) following the consensus predictions of domain boundaries generated by the Meta‐DP meta‐server (http://meta-dp.cse.buffalo.edu/).15 Considering the possibility of embedded domain folds, newly identified domains were excised out of the original query sequence (typically longer), and the resulting flanking regions were merged and subjected to a new round of fold detection. Finally, the excised sequences of all newly mapped domains were resubmitted to the Structure Prediction Meta Server to obtain the final template ranking, reliability indicators, query‐to‐template sequence alignments and secondary structure predictions.
The reliability of fold assignment was based primarily on the 3D‐Jury confidence score, which was calculated using the standard settings under which the score was found to correlate to the number of correctly predicted residues.16 Accordingly, a confidence threshold of 50 for the 3D‐Jury score translates into a prediction reliability of over 90%. For shorter sequences (<100 residues), the 3D‐Jury confidence cut‐off was lowered to 40. A qualitative evaluation of the query‐to‐template sequence and secondary structure alignments was also carried out to support the assessment of each top‐ranked structural assignment.
Secondary structure predictions were based on four methods: PROFsec,17 PSI‐PRED,18 and SAM‐T02 with DSSP and STRIDE alphabets.19 A consensus was then derived for each sequence by (i) majority voting over all four methods for α‐helices and β‐strands, and (ii) SAM‐T02 predictions of G‐helices, a secondary structure not available from the other prediction methods. The multiple sequence alignment of the identified UBL domains was assembled starting from individual query‐to‐template sequence alignments top‐ranked by 3D‐Jury consensus fold recognition. This preliminary alignment was further refined by: (i) considering the structure‐based sequence alignment between the top‐ranked UBL templates, which was generated with the Expresso (3D‐Coffee) program,20 and (ii) minor local improvements in the sequence and secondary structure alignments among predicted UBL domains. Sequence homology‐based clustering of predicted UBL domains of USPs was derived with the Clustal W program21 using the PAM350 scoring matrix, given the sequence divergence of the UBL fold.
RESULTS
One approach toward extending the current sequence‐homology‐based domain annotation of human USPs is to detect structural relationships that have only remote or no underlying sequence homology. This is the objective of fold recognition methods. Thus, we subjected the unannotated sequence content of the human USP family to the consensus protein structure prediction method 3D‐Jury.14, 16 This widely used meta‐predictor performs consensus scoring over the 3D models generated by state‐of‐the‐art fold recognition algorithms, and ranked as a top‐performer at the latest CASP, CAFASP, and LiveBench prediction contests.22, 23 We also have recently employed 3D‐Jury to predict the USP‐like structure and infer the deubiquitinating activity for the SARS coronavirus papain‐like protease,24, 25 predictions which were experimentally confirmed both functionally and structurally.26, 27, 28
One of the most interesting results stemming out of this analysis was the prediction of ubiquitin‐like (UBL) domains in an unexpectedly high number of human USPs (Fig. 1A). These UBL domains were predicted with high reliability as judged by the statistically significant 3D‐Jury scores obtained for the corresponding USP sequences against numerous UBL templates (see Supplementary Material). Consistent with the fold recognition data, the newly identified UBL domains follow the consensus secondary structure and the common fingerprint sequence characteristic to the ubiquitin superfold (Fig. 1B).29
Figure 1.
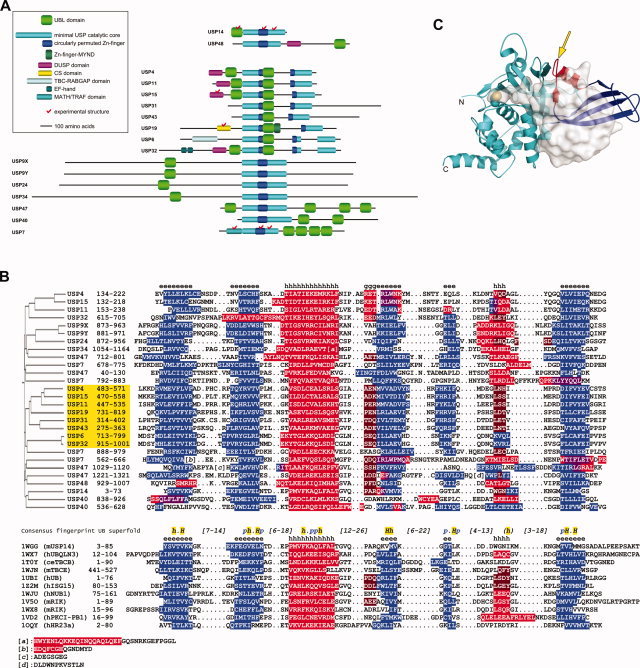
Novel UBL domains of human USPs predicted by structural bioinformatics. (A) Schematic domain organization of human USPs with predicted UBL domains (approximate scale). Note the split in the circularly permuted Zn‐finger‐like domain (blue) and the catalytic core (cyan) due to sizable insertions in some family members. Other domains of these USPs, according to their currently available public annotations, are also shown (see insert). Human USPs 14 and 48, with UBL domains previously annotated based on sequence homology, are shown at the top. (B) Sequence and secondary structure alignment between UBL domains predicted for human USPs (upper part) and selected members from the ubiquitin superfold (lower part, identified primarily by their PDB codes). See Materials and Methods for details, and Supplementary Material for other UBL structures identified by consensus fold recognition as statistically significant templates for the newly detected UBL domains, including the full query‐to‐template assignment. Secondary structure elements, predicted for USP sequences, and observed for the other UBL domains, are highlighted in blue – β‐strand, red – α‐helix, brown – G‐helix, and violet – ambivalent α/β predictions. Consensus secondary structures over the query and template alignments are indicated by e – β‐strand, h – α‐helix, and g – G‐helix. The common fingerprint sequence of the ubiquitin superfold is taken from Ref.29, where conserved hydrophobic residues are denoted by h (90% conserved) and H (100% conserved) and conserved polar residues are denoted by p (90% conserved). A sequence homology‐based clustering of the UBL domains from human USPs is shown on the left. The UBL domains inserted in USP catalytic core domains are highlighted in yellow. (C) Location of UBL domain insertion (arrow) in the minimal USP catalytic core domain, exemplified here from human USP7 (PDB code 1NBF). The papain‐like protease fold is colored in cyan. The nested circularly permuted Zn‐finger‐like domain is in blue, except for its β3‐α1 region (in red) grafted onto the β‐ribbon and attached to the papain‐like domain. The catalytic Cys side chain is rendered as space‐filled model. Bound ubiquitin aldehyde is displayed with translucent molecular surface.
As shown schematically in Figure 1A, the previously unannotated UBL domains detected for various human USPs by our structural bioinformatics analysis are present both inside and outside their catalytic core domains. Ubiquitin‐like domains nested inside catalytic core domains are found in the human USPs 4, 6, 11, 15, 19, 31, 32, and 43. In all these enzymes, the UBL domain insertion occurs at highly homologous positions, specifically, in the middle of the circularly permuted Zn‐finger‐like domain, itself nested within the catalytic core between the two sub‐domains of the papain‐like fold.30 The nested UBL domain would be inserted in these USPs between the β‐strand and the α‐helix that are grafted onto the four‐stranded β‐ribbon of the circularly permuted Zn‐finger and are utilized for its attachment to the C‐terminal sub‐domain of the papain‐like fold (Fig. 1C), as observed in the crystal structures of several USPs.31, 32, 33 In each case, the inserted UBL domain is directly followed by a region of about 170‐240 residues (depending on the enzyme) before the remainder of the circularly permuted Zn‐finger‐like fold (Fig. 1A). No fold similarity could be detected for any of these regions, which, for most parts, lack predicted secondary structure elements. An interesting variation is observed for USP19, where an annotated MYND Zn‐finger domain of about 45 residues is intercalated immediately after the nested UBL domain and before the large, mostly unstructured, region.
The remainder of the newly identified UBL domains are located outside the boundaries of the catalytic core domain (Fig. 1A). Ubiquitin‐like domains N‐terminal to the catalytic core are detected in human USPs 4, 9X, 9Y, 11, 15, 24, 32, 34, and 47. Thus, USPs 4, 11, 15, and 32 feature two UBL domains, one inside and the other one immediately upstream to the catalytic core domain. Interestingly, the predicted N‐terminal UBL domain in all these four USPs is preceded by a DUSP domain.34 The single N‐terminal UBL domains of USPs 9X, 9Y, 24, and 34 are predicted to be flanked on both sides by all‐α‐helical domains (not shown). Multiple UBL domains were detected in the C‐terminal extensions relative to the catalytic core of human USPs 7 (four domains), 40 (two domains), and 47 (three domains; a fourth UBL domain is predicted upstream to the catalytic core).
DISCUSSION
Based on primary sequence homology, UBL domains have been previously detected only in the N‐terminal part of human USP14,35 and in the C‐terminal end of human USP48.12 In the former case, the solution NMR structure of the UBL domain of mouse USP14 (PDB ID: 1WGG; 97% sequence identity to the human domain) confirms this structural assignment. The exquisite promiscuity of the ubiquitin superfold to variations in primary sequence,29 may have precluded the detection of most UBL domains by simple applications of standard homology tools such as PSI‐BLAST,36 possibly leading to their under‐representation in the currently available public annotations of USPs. Supporting this idea, the only other previously reported UBL domain of a human USP, that of USP9Y, resulted from a fold recognition‐based annotation study targeted to the male‐specific region of the human Y chromosome.37
The present structural bioinformatics analysis of the currently unannotated content of the entire human USP family significantly augments the existing annotation with the addition of 26 UBL domains from 15 distinct human USPs. Thus, the UBL domain can be regarded as the most frequently occurring domain in the human USP family, after the characteristic protease core domain.8, 12 We cannot exclude the possibility that a few other UBL domains, perhaps more remotely related to the currently known members of the ubiquitin superfold,29 have escaped our fold detection employing the existing best‐performing algorithms and the current PDB content. For example, several other putative UBL domains from USPs 7, 40, and 47 were detected with scores below the significance threshold and therefore not reported here. The occurrence of multiple UBL domains per USP protein, as well as of UBL domains embedded in the USP catalytic core (hence comprising three distinct folds in a nested rather than successive type of assembly) add to the structural complexity that is currently being recognized for many DUBs.
The marked presence of UBL domains in human USPs is likely to have profound functional consequences in this class of enzymes, primarily via (i) modulation of enzymatic activity and specificity, and (ii) recruitment of nonsubstrate partners. In principle, due to structural similarities to ubiquitin, modulation of USP enzymatic activity by UBL domains could arise from the trivial action of competing directly with ubiquitinated substrates for the ubiquitin binding site on the Zn‐finger‐like domain, thus acting as auto‐inhibitory domains. The newly identified UBL domains may also affect the enzymatic specificities of their USPs toward the substrate protein part or toward the ubiquitin part of the substrate conjugate. Owing to their proximity to the Zn‐finger‐like domain implicated directly in ubiquitin docking,31, 33 the UBL domains nested within the catalytic core (Fig. 1A and 1C) may affect the specificities of these USPs with respect to the degree (mono/di/tetra/poly) and type (e.g., K48/K63, branched/linear) of polyubiquitination, or to the type of modifier (e.g., ubiquitin, ISG15, NEDD8). For instance, an intact metal‐chelating Zn‐finger‐like domain of human USP15, which features a nested UBL domain, is essential for degradation of K48‐branched polyubiquitin chains but not for hydrolysis of the ubiquitin‐GFP fusion.38 This, in light of the fact that USP7 can degrade polyubiquitin substrates,39 in the absence of the metal‐chelating ability of its Zn‐finger‐like domain and of a nested UBL domain, suggests that the metal center might stabilize or position the nested UBL domain in order to allow polyubiquitin recognition by some USPs.
More broadly, UBL domains can engage in specific interactions with domains of both substrate and nonsubstrate protein targets. In the latter case, such specific interactions will determine noncatalytic properties of USPs such as localization, trafficking, and participation in intracellular and signaling pathways, although they can also affect the USP enzymatic activity and specificity. For example, human USP14 and its yeast homolog Ubp6 bind through their N‐terminal UBL domain to the 26S proteasome, which also results in a dramatic increase of their catalytic activity.32, 40, 41, 42 Proteasome‐associated DUBs can act catalytically to remove (poly)ubiquitin before proteasomal degradation, thus serving in the editing of poorly targeted substrates and ubiquitin recycling, or noncatalytically to delay proteasomal degradation and regulate both the nature and magnitude of proteasomal activity.43 It is tempting to speculate that other human USPs would also have the ability to bind directly to proteasome subunits via some of their UBL domains identified here. The UBL domain of mouse USP14, as well as other UBL domains known to associate with proteasome, for example, from human ubiquilins 2 and 3, or DNA damage repair protein Rad23A,44, 45, 46 were retrieved among the high scoring structural templates for newly mapped UBL domains of several USPs (see Supplementary Material). In this context, it is also worth to note the association of human USP15 with the COP9‐signalosome (CSN), which has subunits similar to components of the 26S proteasome lid complex.38 Detection of the UBL fold in USP15 suggests that the mechanism of CSN‐association of USP15 might resemble proteasome‐association of USP14, that is, direct interactions via UBL domains.
Functions other than proteasome and CSN binding can also be anticipated, at least in some cases, given the notorious functional variability within the ubiquitin domain superfold, comprising ubiquitin, ubiquitin‐like modifiers and internal UBL structures.29 Generally, the interaction surface seems not to be conserved within the ubiquitin superfold and almost every element of the fold is used in protein recognition, although interactions with one protein family tend to use the same surface.29, 47, 48, 49 A vast repertoire of ubiquitin‐binding domains (UBDs) is known to interact with members of the ubiquitin superfold. Ubiquitin‐dependent signaling pathways include UIM, UBA, UBL, CUE, GAT, GLUE, and various types of Zn‐fingers such as ZnF‐A20, NZF, and ZnF‐UBP, among other UBDs.48 Outside the ubiquitin pathways, small GTPases represent the prevalent fold interacting with UBL domains. The presence of UBDs can be diagnostic of putative substrate or nonsubstrate protein targets, largely unknown for most UBL domain‐containing human USPs.
While the ubiquitin‐like fold was abundantly detected in the relatively large family (54 members) of human USPs, its presence in USPs from older eukaryotes may indicate that UBL domains represent important functional features that were conserved during evolution in this class of enzymes. Indeed, a fold recognition analysis of the unannotated sequence content of the USP family from the Saccharomyces cerevisiae yeast (16 enzymes, Ubp1 to Ubp16), detects UBL domains in Ubp12 and Ubp15, the yeast homologs of human USP15 and USP7, respectively, in addition to the known UBL domain in Ubp6 (see Supplementary Material). Another compelling example suggesting the importance of the UBL domain as a key structural and putatively regulatory module in the USP class of DUBs is provided by the SARS coronavirus papain‐like protease. This viral enzyme, whose primary function is in viral replication via polyprotein processing, not only acquired the USP molecular architecture and deubiquitinating activity common to the corresponding host cell enzymes,11, 26, 27 but it also features a UBL domain (albeit about 20 residues smaller than ubiquitin).28
The UBL domains detected in human USPs do not show, in the sequence following their C‐terminal β‐strand, similarity to the ubiquitin precursor processing site, which occurs after the ubiquitin C‐terminal sequence 73LRGG76. We therefore conclude that they are internal noncleavable UBL domains. However, a structural relationship to ubiquitin has been recently proposed for human USP1, which is inactivated by auto‐cleavage after an internal ubiquitin‐like di‐glycine motif.50 The cleavage occurs in the middle of a 140‐residue insertion in the minimal USP catalytic core domain, based on the available crystal structures of USPs 2, 7, 8, and 14. However, we could not detect the UBL fold in the 70‐residue inserted sequence preceding the processing site. It is possible that in the case of USP1, key substrate interactions between its catalytic groove S 6‐S 1 subsites and its residues 666IGLLGG671 preceding the cleavage site, which are homologous to the ubiquitin C‐terminal sequence 71LRLRGG76, are sufficient for cleavage without the need of additional interactions from the rest of the ubiquitin fold. Like the acquisition of noncleavable UBL domains, the utilization of a cleavable internal ubiquitin‐like C‐terminal motif in USP1 provides another fascinating example for the reuse of structural elements specific to the ubiquitin signaling pathways towards increasing their own regulatory capabilities and functional diversity. The structural bioinformatics analysis reported here provides valuable information that can spur further structure‐function characterization studies in this class of deubiqutinating enzymes.
Supporting information
The Supplementary Material referred to in this article can be found online at http://www.interscience.wiley.com/jpages/0887-3585/suppmat/
Supporting Information file jws‐prot.21546.pdf
Acknowledgements
We thank Dr. Holger A. Lindner for useful discussions and critical reading of the manuscript. X.Z. thanks Dr. Enrico O. Purisima for securing financial support to carry out this work.
REFERENCES
- 1. Glickman MH,Ciechanover A. The ubiquitin‐proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev 2002; 82: 373–428. [DOI] [PubMed] [Google Scholar]
- 2. d'Azzo A,Bongiovanni A,Nastasi T. E3 ubiquitin ligases as regulators of membrane protein trafficking and degradation. Traffic 2005; 6: 429–441. [DOI] [PubMed] [Google Scholar]
- 3. Haglund K,Dikic I. Ubiquitylation and cell signaling. EMBO J 2006; 24: 3353–3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Welchman RL,Gordon C,Mayer RJ. Ubiquitin and ubiquitin‐like proteins as multifunctional signals. Nat Rev Mol Cell Biol 2005; 6: 599–609. [DOI] [PubMed] [Google Scholar]
- 5. Hershko A,Ciechanover A. The ubiquitin system. Annu Rev Biochem 1998; 67: 425–479. [DOI] [PubMed] [Google Scholar]
- 6. Pickart CM,Eddins MJ. Ubiquitin: structures, functions, mechanisms. Biochim Biophys Acta 2004; 1695: 55–72. [DOI] [PubMed] [Google Scholar]
- 7. Amerik AY,Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochim Biophys Acta 2004; 1695: 189–207. [DOI] [PubMed] [Google Scholar]
- 8. Nijman SMB,Luna‐Vargas MPA,Velds A,Brummelkamp TR,Dirac AMG,Sixma TK,Bernards R. A genomic and functional inventory of deubiquitinating enzymes. Cell 2005; 123: 773–786. [DOI] [PubMed] [Google Scholar]
- 9. Zhou H,Monack DM,Kayagaki N,Wertz I,Yin J,Wolf B,Dixit VM. Yersinia virulence factor YopJ acts as a deubiquitinase to inhibit NF‐kappaB activation. J Exp Med 2005; 202: 1327–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Misaghi S,Balsara ZR,Catic A,Spooner E,Ploegh HL,Starnbach MN. Chlamydia trachomatis‐derived deubiquitinating enzymes in mammalian cells during infection. Mol Microbiol 2006; 61: 142–150. [DOI] [PubMed] [Google Scholar]
- 11. Sulea T,Lindner HA,Menard R. Structural aspects of recently discovered viral deubiquitinating activities. Biol Chem 2006; 387: 853–862. [DOI] [PubMed] [Google Scholar]
- 12. Quesada V,Diaz‐Perales A,Gutierrez‐Fernandez A,Garabaya C,Cal S,Lopez‐Otin C. Cloning and enzymatic analysis of 22 novel human ubiquitin‐specific proteases. Biochem Biophys Res Commun 2004; 314: 54–62. [DOI] [PubMed] [Google Scholar]
- 13. Katoh K,Kuma Ki,Toh H,Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res 2005; 33: 511–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ginalski K,Elofsson A,Fischer D,Rychlewski L. 3D‐Jury: a simple approach to improve protein structure predictions. Bioinformatics 2003; 19: 1015–1018. [DOI] [PubMed] [Google Scholar]
- 15. Saini HK,Fischer D. Meta‐DP: domain prediction meta‐server. Bioinformatics 2005; 21: 2917–2920. [DOI] [PubMed] [Google Scholar]
- 16. Ginalski K,Rychlewski L. Detection of reliable and unexpected protein fold predictions using 3D‐Jury. Nucleic Acids Res 2003; 31: 3291–3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rost B. Protein secondary structure prediction continues to rise. J Struct Biol 2001; 134: 204–218. [DOI] [PubMed] [Google Scholar]
- 18. Jones DT. Protein secondary structure prediction based on position‐specific scoring matrices. J Mol Biol 1999; 292: 195–202. [DOI] [PubMed] [Google Scholar]
- 19. Karplus K,Karchin R,Draper J,Casper J,Mandel‐Gutfreund Y,Diekhans M,Hughey R. Combining local‐structure, fold‐recognition, and new fold methods for protein structure prediction. Proteins 2006; 53: 491–496. [DOI] [PubMed] [Google Scholar]
- 20. Armougom F,Moretti S,Poirot O,Audic S,Dumas P,Schaeli B,Keduas V,Notredame C. Expresso: automatic incorporation of structural information in multiple sequence alignments using 3D‐Coffee. Nucleic Acids Res 2006; 34: W604–W608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chenna R,Sugawara H,Koike T,Lopez R,Gibson TJ,Higgins DG,Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res 2003; 31: 3497–3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ginalski K. Comparative modeling for protein structure prediction. Curr Opin Struct Biol 2006; 16: 172–177. [DOI] [PubMed] [Google Scholar]
- 23. Fischer D. Servers for protein structure prediction. Curr Opin Struct Biol 2006; 16: 178–182. [DOI] [PubMed] [Google Scholar]
- 24. Sulea T,Lindner HA,Purisima EO,Menard R. Deubiquitination, a new function of the severe acute respiratory syndrome coronavirus papain‐like protease? J Virol 2005; 79: 4550–4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sulea T,Lindner HA,Purisima EO,Menard R. Binding site based classification of coronaviral papain‐like proteases. Proteins 2006; 62: 760–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Barretto N,Jukneliene D,Ratia K,Chen Z,Mesecar AD,Baker SC. The papain‐like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J Virol 2005; 79: 15189–15198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lindner HA,Fotouhi‐Ardakani N,Lytvyn V,Lachance P,Sulea T,Menard R. The papain‐like protease from the severe acute respiratory syndrome coronavirus is a deubiquitinating enzyme. J Virol 2005; 79: 15199–15208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ratia K,Saikatendu KS,Santarsiero BD,Barretto N,Baker SC,Stevens RC,Mesecar AD. Severe acute respiratory syndrome coronavirus papain‐like protease: structure of a viral deubiquitinating enzyme. Proc Natl Acad Sci USA 2006; 103: 5717–5722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kiel C,Serrano L. The ubiquitin domain superfold: structure‐based sequence alignments and characterization of binding epitopes. J Mol Biol 2006; 355: 821–844. [DOI] [PubMed] [Google Scholar]
- 30. Krishna SS,Grishin NV. The finger domain of the human deubiquitinating enzyme HAUSP is a zinc ribbon. Cell Cycle 2004; 3: 1046–1049. [PubMed] [Google Scholar]
- 31. Hu M,Li P,Li M,Li W,Yao T,Wu JW,Gu W,Cohen RE,Shi Y. Crystal structure of a UBP‐family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde. Cell 2002; 111: 1041–1054. [DOI] [PubMed] [Google Scholar]
- 32. Hu M,Li P,Sing L,Jeffrey PD,Chenova TA,Wilkinson KD,Cohen RE,Shi Y. Structure and mechanisms of the proteasome‐associated deubiquitinating enzyme USP14. EMBO J 2005; 24: 3747–3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Renatus M,Parrado SG,D'Arcy A,Eidhoff U,Gerhartz B,Hassiepen U,Pierrat B,Riedl R,Vinzenz D,Worpenberg S,Kroemer M. Structural basis of ubiquitin recognition by the deubiquitinating protease USP2. Structure 2006; 14: 1293–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. de Jong RN,AB E,Diercks T,Truffault V,Daniels M,Kaptein R,Folkers GE. Solution structure of the human ubiquitin‐specific protease 15 DUSP domain. J Biol Chem 2006; 281: 5026–5031. [DOI] [PubMed] [Google Scholar]
- 35. Wyndham AM,Baker RT,Chelvanayagam G. The Ubp6 family of deubiquitinating enzymes contains a ubiquitin‐like domain: SUb. Protein Sci 1999; 8: 1268–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Altschul SF,Madden TL,Schaffer AA,Zhang J,Zhang Z,Miller W,Lipman DJ. Gapped BLAST and PSI‐BLAST: a new generation of protein database search programs. Nucleic Acids Res 1997; 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ginalski K,Rychlewski L,Baker D,Grishin NV. Protein structure prediction for the male‐specific region of the human Y chromosome. Proc Natl Acad Sci USA 2004; 101: 2305–2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hetfeld BKJ,Helfrich A,Kapelari B,Scheel H,Hofmann K,Guterman A,Glickman M,Schade R,Kloetzel PM,Dubiel W. The zinc finger of the CSN‐associated deubiquitinating enzyme USP15 is essential to rescue the E3 ligase Rbx1. Curr Biol 2005; 15: 1217–1221. [DOI] [PubMed] [Google Scholar]
- 39. Canning M,Boutell C,Parkinson J,Everett RD. A RING finger ubiquitin ligase is protected from autocatalyzed ubiquitination and degradation by binding to ubiquitin‐specific protease USP7. J Biol Chem 2004; 279: 38160–38168. [DOI] [PubMed] [Google Scholar]
- 40. Borodovsky A,Kessler BM,Casagrande R,Overkleeft HS,Wilkinson KD,Ploegh HL. A novel active site‐directed probe specific for deubiquitylating enzymes reveals proteasome association of USP14. EMBO J 2001; 20: 5187–5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Leggett DS,Hanna J,Borodovsky A,Crosas B,Schmidt M,Baker RT,Walz T,Ploegh H,Finley D. Multiple associated proteins regulate proteasome structure and function. Mol Cell 2002; 10: 495–507. [DOI] [PubMed] [Google Scholar]
- 42. Borodovsky A,Ovaa H,Kolli N,Gan‐Erdene T,Wilkinson KD,Ploegh HL,Kessler BM. Chemistry‐based functional proteomics reveals novel members of the deubiquitinating enzyme family. Chem Biol 2002; 9: 1149–1159. [DOI] [PubMed] [Google Scholar]
- 43. Hanna J,Hathaway NA,Tone Y,Crosas B,Elsasser S,Kirkpatrick DDS,Leggett DS,Gygi SP,King RW,Finley D. Deubiquitinating enzyme Ubp6 functions noncatalytically to delay proteasomal degradation. Cell 2006; 127: 99–111. [DOI] [PubMed] [Google Scholar]
- 44. Walters KJ,Kleijnen MF,Goh AM,Wagner G,Howley PM. Structural studies of the interaction between ubiquitin family proteins and proteasome subunit S5a. Biochemistry 2002; 41: 1767–1777. [DOI] [PubMed] [Google Scholar]
- 45. Ko HS,Uehara T,Tsuruma K,Nomura Y. Ubiquilin interacts with ubiquitylated proteins and proteasome through its ubiquitin‐associated and ubiquitin‐like domains. FEBS Lett 2004; 566: 110–114. [DOI] [PubMed] [Google Scholar]
- 46. Walters KJ,Lech PJ,Goh AM,Wang Q,Howley PM. DNA‐repair protein hHR23a alters its protein structure upon binding proteasomal subunit S5a. Proc Natl Acad Sci USA 2003; 100: 12694–12699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Harper JW,Schulman BA. Structural complexity in ubiquitin recognition. Cell 2006; 124: 1133–1136. [DOI] [PubMed] [Google Scholar]
- 48. Hurley JH,Lee S,Prag G. Ubiquitin‐binding domains. Biochem J 2006; 399: 361–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Reverter D,Wu K,Erdene TG,Pan ZQ,Wilkinson KD,Lima CD. Structure of a complex between Nedd8 and the Ulp/Senp protease family member Den1. J Mol Biol 2005; 345: 141–151. [DOI] [PubMed] [Google Scholar]
- 50. Huang TT,Nijman SMB,Mirchandani KD,Galardy PJ,Cohn MA,Haas W,Gygi SP,Ploegh HL,Bernards R,D'Andrea AD. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat Cell Biol 2006; 8: 341–347. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The Supplementary Material referred to in this article can be found online at http://www.interscience.wiley.com/jpages/0887-3585/suppmat/
Supporting Information file jws‐prot.21546.pdf


