Abstract
Clinical laboratories providing an etiological diagnosis of respiratory tract infections (RTI) have increasingly relied on nucleic acid amplification tests. Polymerase chain reaction‐based methods are becoming more standardized, and several have undergone the scrutiny of regulatory agencies mandated to assess the risks and benefits of implementing pathogen‐detection assays into diagnostic algorithms. Respiratory viruses lead to both upper and lower RTI and are implicated in exacerbations of chronic pulmonary conditions. Viruses from different taxonomic families present with overlapping clinical signs and symptoms, necessitating an accurate laboratory diagnosis. The clinical utility of diagnostic algorithms incorporating tests for respiratory viruses will depend on the breadth of pathogen coverage and the time to reliable and actionable results. This review covers strategies for detecting a panel of respiratory viruses employed over the last decade that have enabled an etiological diagnosis of RTI in a cost‐effective manner.
Keywords: respiratory virus, nucleic acid, laboratory, diagnosis, PCR, multiplex
Introduction
It has been widely reported that respiratory tract infections (RTI) are a significant cause of morbidity and mortality in developing and developed nations. 1 , 2 , 3 Timely laboratory diagnosis has proven to be an important part of the clinical algorithm in both an ambulatory and critical care setting.
The key objectives of an etiological diagnosis of RTI include (a) avoidance of empirical antibiotic treatment; (b) allowance of narrow spectrum–targeted antibiotic treatment; (c) appropriate use of antiviral drugs; (d) enablement of patient cohorting to limit nosocomial spread in the case of hospitalization; (e) provision of accurate epidemiological information to formulate preventive and therapeutic recommendations; and (f) general reductions in patient management costs. 4 Over the past two decades, technologies used in the laboratory diagnosis of RTI have evolved dramatically. Virus isolation via cell culture techniques has made substantial advances in terms of both time to result and pathogen coverage. 2 , 5 Antigen‐based assays have also been incorporated into clinical practice. 6 Notwithstanding advancements made in these testing methods, their limitations have led to unmet needs in the diagnostic laboratory being addressed by new tools, most notably nucleic acid–based assays developed for the detection of respiratory viruses. 2 , 4 , 7 , 8
As respiratory viruses are the causative agent in a significant number of patients presenting with RTI, clinicians are well served by a timely laboratory diagnosis that can either rule in or rule out viruses that have overlapping clinical presentations. 1 , 2 , 4 Additionally, the availability of sensitive and specific methods for respiratory virus detection aids in the management of (a) chronic pulmonary conditions and (b) medical complications associated with RTI. 9 , 10 , 11 Finally, the emergence of novel strains of respiratory viruses, most notably those responsible for significant outbreaks, epidemics, and pandemics, highlights the need for a comprehensive and flexible diagnostic algorithm. 12 , 13 , 14 , 15
This review aims to provide an overview of advances made in the diagnosis of RTI during the last two decades. It also discusses the specific role played by xTAG® RVP, a respiratory viral panel commercialized by Luminex Molecular Diagnostics (Toronto, Canada) and awarded the U.S. Prix‐Galien Medal in 2010.
Emergence of nucleic acid testing for the laboratory diagnosis of RTI
Since the mid 1980s, a variety of chemistries and detection technologies suitable for nucleic acid amplification tests (NAAT) have been evaluated and deployed by diagnostic laboratories. While technologies for direct detection of nucleic acids are suitable for some diagnostic applications, 16 most molecular diagnostic tests use amplification‐based technologies such as the polymerase chain reaction (PCR), nucleic acid sequence–based amplification (NASBA), and strand displacement amplification (SDA). 4 , 16 Considering the fact that microorganisms with RNA and DNA genomes are implicated in RTI, 2 , 4 , 8 PCR has become a widely deployed chemistry used to amplify genetic material from a variety of respiratory pathogens.
If a pathogen has an RNA genome, the PCR reaction must be preceded by a reverse transcription (RT) reaction that converts RNA to complementary DNA (cDNA). The RT‐PCR amplification/detection reaction can be modified in various ways depending on the assay development needs. 16 Diagnostics employing PCR fall into one of two broad categories: real‐time (q)PCR and end‐point PCR. 16 Due to its quantitative nature, qPCR is suitable for applications requiring an assessment of pathogen levels. This typically is not the case for assays that aid in the diagnosis of RTI. One feature of qPCR that is well suited to the detection of respiratory viruses is that most steps in the assay take place in a closed system. This feature limits the potential for carryover contamination and, thus, the potential for inaccurate results. 15 While detection of a single analyte can be achieved with relative ease, detection of multiple analytes (“multiplexing”) via qPCR requires a variety of workflow manipulations due to limitations inherent to detection platforms used with this chemistry. 2 , 16 Assays incorporating end‐point PCR generally have a greater multiplexing capacity than those incorporating qPCR.
Multiplexing approaches for PCR‐based tests for respiratory viruses
The need to test for a panel of respiratory viruses to aid in the diagnosis of RTI emerges from the fact that multiple etiological agents present with overlapping clinical signs and symptoms. 2 , 8 Multiplexing strategies that employ PCR chemistry are well suited for RTI diagnosis, particularly when considering the fact that many recently discovered respiratory viruses have proven difficult to detect by traditional methods. 2 , 4 , 5 Advances in real‐time amplification systems and liquid‐bead arrays have greatly improved the diagnostic yield of clinical algorithms over the last decade. 2 , 18
The migration toward multiplex detection strategies for respiratory viruses has led to significant, cost‐effective improvements in detection rates. 2 , 4 Nonetheless, each multiplexing strategy has inherent limitations. For instance, attempts to expand the diagnostic capacity of qPCR‐based tests through the introduction of multiple primer and probe sets targeting different viral genomes are met with a variety of technical and logistical challenges that can negatively affect both the cost and accuracy of testing. 2 , 7 , 19 At a certain point, the simplicity afforded through the closed‐tube feature of qPCR is outweighed by the adaptations required to increase multiplexing capacity. Several detection platforms that can accommodate multiplex applications have reached the diagnostic laboratory in recent years. 20 Each of these has advantages and disadvantages in terms of the ability to help laboratories achieve the goals of an etiological diagnosis of RTI. As laboratories continue to evaluate emerging technologies, considerations relating to cost and accuracy of diagnosis should remain at the forefront.
Multiplexing advantages afforded through xMAP® and xTAG® technology
Two significant advances in multiplexing that have enabled accurate, reproducible, flexible, and cost‐effective analyte detection involve technologies patented by Luminex Corporation (http://www.luminexcorp.com).
The first innovation is a solution‐phase array composed of spectrally distinct microspheres (“beads”) known as xMAP® detection. Early reports on this technology supported its potential for inclusion into systems designed for both protein and nucleic acid detection. 21 Since then, numerous research and diagnostic applications have incorporated xMAP detection and three platforms utilizing this technology are now commercially available. One has the ability to simultaneously detect 100 analytes in a single sample (i.e., reaction well on a microtiter plate). The second expands the multiplexing capability to 500 analytes per reaction well. The latest platform commercialized has demonstrated improvements in ease of use and cost‐efficiency for lower‐plex applications designed to simultaneously detect up to 50 analytes in a single reaction well.
The second significant advance in bead‐based arrays was initially reported in 2004. 22 This patented innovation involves universal, minimally cross‐hybridizing, complementary oligonucleotide sequences (“tags” and “antitags”). These tags and antitags were designed to maximize signal‐to‐noise ratios observed at the detection step of array‐based, nucleic acid, applications (Fig. 1). The advantage of this “Universal Array” is that assay‐specific hybridization optimization is not required because optimized tag/antitag hybridization conditions can be applied to any nucleic acid–based application. This is particularly noteworthy when considering the potential for nonspecific interactions in a multiplex reaction mixture. When Universal Array sorting is coupled to xMAP detection, enzymatic steps required for the detection and identification of analytes occur in solution phase. This is kinetically more advantageous when compared with solid‐phase arrays and is a key factor underlying the superior performance of applications incorporating these two innovations. The Universal Array is commercially available through Luminex Corporation; it is also the foundation of xTAG applications commercialized by Luminex Molecular Diagnostics. a
Figure 1.
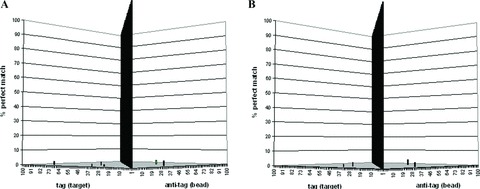
Universal array validation results. Of the possible 9,900 mismatch hybridization events that could have occurred when each of the 100 complementary tag (target) sequences was hybridized individually to the pool of 100 bead‐immobilized universal antitags, only six events were observed in run 1 (A). Similarly, in run 2 (B), only five events were observed. Of these 11 events, 4 were common to both runs, the highest mismatch hybridization event generating a signal equivalent to 3.7% of the signal observed for the perfectly matched pair (i.e., specific hybridization event). For each validation run, the randomly dispersed bars represent the mismatch hybridization events expressed as the percentage of perfect matches. The center wall represents the 100 perfectly matched pairs. Reproduced from Mahony et al. 22
The advantages of nucleic acid–based tests incorporating xMAP and xTAG technologies are numerous. A priori, a test has significant multiplexing capacity not achievable through qPCR chemistries currently used in diagnostic applications. 4 Furthermore, negligible cross‐reactivity among tags and antitags that are not specifically paired elevates the baseline for diagnostic sensitivity and specificity and, consequently, simplifies optimization efforts during the assay development process. Luminex technologies have been extensively adopted in both research and diagnostic applications and have been comprehensively assessed by authorities such as the U.S. Centers for Disease Control and Prevention (CDC) and the U.S. Food and Drug Administration (FDA).
Introduction of xTAG RVP into diagnostic algorithms
By the end of 2006, xTAG RVP was commercialized in the European Union as a diagnostic kit (CE‐IVD) that used xMAP detection. Early in 2008, U.S. and Canadian versions of this in vitro diagnostic (IVD) device received regulatory clearances from the FDA and Health Canada, respectively. The CE‐IVD kit provides the most comprehensive viral coverage in the xTAG RVP IVD product line through its ability to simultaneously detect the following viruses:
-
1
Human influenza (Flu) A
-
2
Seasonal H1 subtype of Flu A (Flu A‐H1)
-
3
Seasonal H3 subtype of Flu A (Flu A‐H3)
-
4
Highly pathogenic avian influenza A H5N1 (Flu A‐H5)
-
5
Flu B
-
6
Respiratory syncytial virus (RSV) A
-
7
RSV B
-
8
Parainfluenza (Para) 1
-
9
Para 2
-
10
Para 3
-
11
Para 4
-
12
Coronavirus (CoV) OC43
-
13
CoV 229E
-
14
CoV NL63
-
15
CoV HKU1
-
16
CoV SARS
-
17
Human metapneumovirus (hMPV)
-
18
Adenovirus (Adeno)
-
19
Enterovirus (Entero)
-
20
Rhinovirus (Rhino)
The test provides a unique result for 18 of the 20 viruses listed above. A single result is provided for the closely related members of the picornaviridae family, enterovirus and rhinovirus, due to the high degree of nucleic acid sequence homology across different strains of these viruses. In addition to detecting the aforementioned viruses in a single reaction well, there are two unique results generated for controls detected in the same multiplex reaction. The first (Escherichia coli phage MS2) is an internal control that is added to each patient sample. This internal control monitors all steps in the assay: (1) nucleic acid extraction; (2) RT; (3) multiplex PCR; (4) multiplex target specific primer extension (TSPE); (5) Universal Array sorting; and (6) xMAP detection. The second (bacteriophage lambda) is assayed in a separate well and controls for a subset of these steps (PCR, TSPE, sorting, and detection). By assessing results for these two controls together with results for targeted analytes, the end‐user is able to ascertain whether assay failures are due to a failure in the nucleic acid extraction/RT step or in the subsequent assay steps (PCR/TSPE/sorting/detection).
The diagnostic sensitivity and specificity of the CE‐IVD kit was established through an evaluation of 1,464 prospectively and retrospectively collected clinical samples. The demographic features of this study cohort are summarized in Table 1. It is worthwhile to note that several of these patients presented with a variety of coinfections that were detected by xTAG RVP but that would otherwise be missed by traditional methods. Of these 1,464 patient samples, 544 were prospectively collected nasopharyngeal (NP) swabs used to establish clinical performance characteristics for the U.S. version of the kit (Table 2). A second‐generation IVD (xTAG RVP FAST) was introduced in Europe and Canada late in 2009. This line extension has a streamlined workflow, shorter time to result, and modified analyte coverage when compared to xTAG RVP.
Table 1.
General demographic data for a study cohort used to establish performance characteristics of xTAG RVP (N = 1,464)
| Sex | Number of subjects |
|---|---|
| Male | 750 (51.2%) |
| Female | 667 (45.6%) |
| Not determined | 47 (3.2%) |
| Age (years) | Number of subjects |
| 0–1 | 463 (31.6%) |
| >1–5 | 229 (15.6%) |
| >5–21 | 177 (12.1%) |
| >21–65 | 333 (22.7%) |
| >65 | 214 (14.6%) |
| Not determined | 48 (3.3%) |
| Subject status | |
| Outpatients | 559 (38.2%) |
| Hospitalized | 495 (33.8%) |
| Emergency department | 216 (14.8%) |
| Extended care facility | 42 (2.9%) |
| Not determined | 152 (10.4%) |
Table 2.
Sensitivity and specificity of xTAG RVP in 544 NP swabs prospectively collected during the 2005/2006 flu season
| Virus | Diagnostic sensitivity | Diagnostic specificity | ||||
|---|---|---|---|---|---|---|
| TP/(TP+FN)a | Percent | 95% Confidence interval for sensitivity | TN/(TN+FP)a | Percent | 95% Confidence interval for specificity | |
| Flu A | 81/84 | 96.4% | 89.9–99.3% | 441/460 | 95.9% | 93.6–97.5% |
| Flu A‐H1b | 6/6 | 100% | 54.1–100% | 532/532 | 100% | 99.3–100% |
| Flu A‐H3b | 66/72 | 91.7% | 82.7–96.9% | 463/469 | 98.7% | 97.2–99.5% |
| Flu B | 54/59 | 91.5% | 81.3–97.2% | 469/485 | 96.7% | 94.7–98.1% |
| RSV A | 23/23 | 100% | 85.2–100% | 501/509 | 98.4% | 96.9–99.3% |
| RSV B | 33/33 | 100% | 89.4–100% | 492/505 | 97.4% | 95.6–98.6% |
| Para 1 | 3/3 | 100% | 29.2–100% | 540/541 | 99.8% | 99.0–100% |
| Para 2 | 6/6 | 100% | 54.1–100% | 537/538 | 99.8% | 99.0–100% |
| Para 3 | 16/19 | 84.2% | 60.4–96.6% | 523/525 | 99.6% | 98.6–100% |
| Rhinoc | 43/43 | 100% | 91.8–100% | 168/183 | 91.8% | 86.8–95.3% |
| Adenod | 18/23 | 78.3% | 56.3–92.5% | 520/520 | 100% | 99.3–100% |
| hMPVe | 24/25 | 96% | 79.7–99.9% | 320/324 | 98.8% | 96.9–99.7% |
aTP, true positive (i.e., xTAG RVP positive result is concordant with comparator method result); FP, false positive (i.e., xTAG RVP positive result is discordant with comparator method result); TN, true negative (i.e., xTAG RVP negative result is concordant with comparator method result); FN, false negative (i.e., xTAG RVP negative result is discordant with comparator method result). Depending on the virus, the comparator method was one, or a combination of culture, DFA, qRT PCR, and/or sequencing.
bInfluenza A prevalence data reported on http://www.cdc.gov for the 2005/06 flu season indicate that the dominant strains circulating at that time were classified as the seasonal Flu A‐H3 subtype, with significantly fewer cases classified as seasonal Flu A‐H1 being reported.
cDue to a high degree of sequence homology, significant cross‐reactivity between enterovirus and rhinovirus is expected.
dData from reference strains together with sequence analysis of clinical samples included in this dataset suggest that the overall sensitivity values for adenovirus were negatively impacted by poor detection of serotypes falling within the adenovirus C species.
ehMPV sensitivity and specificity was established against a composite comparator method (culture and PCR followed by bidirectional sequencing or qRT PCR).
In addition to performance validations required for regulatory applications, there have been numerous third‐party validations of the xTAG RVP product line. Several of these have involved comparisons to real‐time applications and support the conclusion of improved diagnostic coverage and cost‐efficiency being afforded through xTAG/xMAP detection. 19 , 23 , 24
The introduction of respiratory viral panels built on xTAG/xMAP technologies has served as a catalyst for NAAT adoption in diagnostic algorithms. In the United States, the January 3, 2008 clearance of xTAG RVP by the FDA represented the first clearance of a respiratory viral panel utilizing nucleic acid–based detection. 25 This device was also the first to receive FDA clearance for a recently discovered respiratory virus (hMPV) and for Flu A subtyping. 25 The ability to specifically identify seasonal subtypes of influenza A proved to be an invaluable tool during the novel influenza A/H1N1 pandemic that ensued the following year.
Role of xTAG RVP in the 2009 influenza A/H1N1 pandemic
Of the three known types of influenza (influenza A, B, and C), only influenza A viruses are further classified by subtypes based on two main surface glycoproteins: hemagglutinin and neuraminidase (http://www.cdc.gov/flu/avian/gen-info/flu-viruses.htm). Observations to date suggest that influenza A and B continuously evolve through antigenic drift, while only influenza A evolves through antigenic shift (http://www.cdc.gov/flu/avian/gen-info/flu-viruses.htm). Antigenic drift is a gradual process of change via point mutations that lead to novel strains of viruses. It explains why people and animals can be infected by the same virus type (e.g., influenza B) more than once. Namely, the host response to an infecting strain may not grant immunity to new strains that have emerged through antigenic drift (http://www.cdc.gov/flu/avian/gen-info/flu-viruses.htm). Antigenic shift, on the other hand, is a more dramatic process resulting from genetic reassortment, which occurs when influenza viruses exchange genetic material. This abrupt process can occur when two or more influenza A viruses infect a single human or animal host (http://www.cdc.gov/h1n1flu/information_h1n1_virus_qa.htm). Antigenic shift can lead to a global influenza pandemic if three conditions are met: (1) a new influenza A subtype is introduced into the human population, (2) the virus causes serious illness in humans, and (3) the virus can easily spread from person to person in a sustainable manner (http://www.cdc.gov/flu/avian/gen-info/flu-viruses.htm). This is exactly what happened with the novel influenza A/H1N1 subtype that emerged from Mexico in the spring of 2009.
A key innovation in the design of xTAG RVP that proved to be invaluable during the 2009 pandemic is the manner in which it detects influenza A. Specifically, this multiplex test simultaneously probes a region in the influenza A matrix gene conserved across influenza A subtypes together with subtype specific regions in the variable hemagglutinin gene. Accordingly, an influenza A virus that cannot be recognized by the subtype‐specific probes in xTAG RVP will generate a “flu A‐unsubtypeable” result. This feature of the test was addressed in the labeling cleared by regulatory agencies prior to the emergence of 2009 influenza A/H1N1.
In the early days of the 2009 pandemic, a cluster of cases presented themselves in the New York City metropolitan area. 14 The availability of xTAG RVP at the affected hospital was a key element of the surge response. 14 Specifically, several diagnostic testing options were evaluated during this initial outbreak and xTAG RVP proved to be the best tool available for the emerging pandemic. 26 At the time, the only FDA sanctioned test for the specific identification of novel influenza A/H1N1 was one developed by the CDC and made available through a federal Emergency Use Authorization. Samples collected in the initial New York City outbreak and confirmed positive for novel influenza A/H1N1 by the New York State Department of Health all generated a “flu A‐unsubtypeable” result with xTAG RVP. 27 In other words, xTAG RVP enabled clinicians to rapidly discriminate infections caused by this novel influenza A strain from those caused by seasonal strains of influenza and by other viruses included in the panel.
General applications for multiplex respiratory viral panels
The clinical utility of xTAG RVP during the 2009 pandemic was not limited to its ability to detect influenza A. Of note is the fact that other viruses with overlapping clinical features were in circulation at that time. 19 , 26 By simultaneously probing for multiple viruses in a single patient sample, xTAG RVP has proven to be a cost‐effective solution for the laboratory diagnosis of RTI. 23 Multiplex detection of a panel of respiratory viruses also affords advantages to hospital surveillance programs that aim to limit nosocomial infections. 28 Clinically, conditions such as asthma and bronchiolitis benefit from the availability of multiplex respiratory viral panels, particularly in light of emerging evidence implicating more than one virus in these conditions. 29 , 30 , 31 Considering that respiratory syncytial virus is well established as a key risk factor for bronchiolitis in infants, having the ability to discriminate it from other viruses that may be the causative agent (e.g., rhinovirus) is a key element of the clinical algorithm. 1 , 3 Furthermore, dual infections have been noted in bronchiolitis, with one study demonstrating a higher clinical severity score and prolonged hospital stays in patients infected with more than one virus. 30 In light of different treatment courses available to physicians for the aforementioned clinical conditions, an accurate laboratory diagnosis can prove to be vital.
While conditions such as asthma and bronchiolitis are known to involve respiratory viruses, mechanisms by which these agents affect patient status are still under investigation. 32 , 33 , 34 The impact of respiratory viruses in immunocompromised patients and in patients with pneumonia is also an area of investigation. 11 , 18 , 35 , 36 , 37 , 38 Further studies are needed to fully elucidate the role of respiratory viruses in all of these conditions. These and other evaluations will surely benefit from the availability of panels such as xTAG RVP and xTAG RVP FAST. 24 , 39 Similarly, studies on the efficacy of novel vaccines, 40 such as ones being developed for rhinovirus, 41 will benefit from the availability of respiratory viral panels. In addition to the diagnostic and clinical research applications highlighted above, multiplex respiratory viral panels can have tremendous utility in surveillance programs. 28 , 42 The preventive and therapeutic strategies that emerge from epidemiological information gathered through such programs will be augmented by the extensive data afforded through multiplex panels.
Conclusions
Over the past two decades, the etiological diagnosis of RTI has proven to be an important element of the clinical algorithm. Timely and accurate data on the multitude of pathogens that have been implicated in RTI and associated clinical conditions are paramount. Traditional methods such as cell culture still have a role to play in diagnostic algorithms; however, advances in nucleic acid amplification technologies have led to an increase in their adoption by clinical laboratories. Innovations linked to both real‐time and end‐point PCR have been particularly noteworthy. The innovations underlying xTAG RVP are an example of how advances in technology can be translated into clinical utility.
Conflicts of interest
The corresponding author and coauthors are employed by Luminex Molecular Diagnostics Inc., a wholly owned subsidiary of Luminex Corporation.
Acknowledgments
We would like to thank Dr. C.C. Ginocchio, Dr. J.B. Mahony, Dr. T. Mazzulli, Dr. K.E. Templeton and their respective institutions and staff for their collaboration in performance evaluations required for regulatory filings.
Footnotes
xTAG® applications were formerly referred to as “Tag‐It™” or “ID‐Tag™” applications commercialized by Tm Bioscience Corporation (now Luminex Molecular Diagnostics Inc.).
References
- 1. Beck, E.T. & Henrickson K.J.. 2010. Molecular diagnosis of respiratory viruses. Future Microbiol. 5: 901–916. Review. [DOI] [PubMed] [Google Scholar]
- 2. Mahony, J.B. 2008. Detection of respiratory viruses by molecular methods. Clin. Microbiol. Rev. 21: 716–747. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tregoning, J.S. & Schwarze J.. 2010. Respiratory viral infections in infants: causes, clinical symptoms, virology, and immunology. Clin. Microbiol. Rev. 23: 74–98. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ieven, M. 2007. Currently used nucleic acid amplification tests for the detection of viruses and atypicals in acute respiratory infections. J. Clin. Virol. 40: 259–276. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. McAdam, A.J. & Riley A.M.. 2009. Developments in tissue culture detection of respiratory viruses. Clin. Lab. Med. 29: 623–634. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Principi, N. & Esposito S.. 2009. Antigen‐based assays for the identification of influenza virus and respiratory syncytial virus; why and how to use them in pediatric practice. Clin. Lab. Med. 29: 649–660. Review. [DOI] [PubMed] [Google Scholar]
- 7. Fox, J.D. 2007. Nucleic acid amplification tests for detection of respiratory viruses. J. Clin. Virol. 40 (Suppl 1): S15–S23. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kehl, S.C. & Kumar S.. 2009. Utilization of nucleic acid amplification assays for the detection of respiratory viruses. Clin. Lab. Med. 29: 661–671. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mohan, A. , Chandra S., Agarwal D., et al 2010. Prevalence of viral infection detected by PCR and RT‐PCR in patients with acute exacerbation of COPD: a systematic review. Respirology 15: 536–542. Review. Erratum in: Respirology July 2010 15(5):871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Martinez, F.D. 2009. The origins of asthma and chronic obstructive pulmonary disease in early life. Proc. Am. Thorac. Soc. 6: 272–277. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wark, P. 2010. Viral and bacterial interactions in pneumonia. Expert Rev. Respir. Med. 4: 221–228. Review. [DOI] [PubMed] [Google Scholar]
- 12. Cowling, B.J. , Ho L.M. & Leung G.M.. 2008. Effectiveness of control measures during the SARS epidemic in Beijing: a comparison of the Rt curve and the epidemic curve. Epidemiol. Infect. 136: 562–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Baric, R.S. 2008. SARS‐CoV: lessons for global health. Virus Res. 133: 1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Crawford, J.M. , Stallone R., Zhang F., et al 2010. Laboratory surge response to pandemic (H1N1) 2009 outbreak, New York City metropolitan area, USA. Emerg. Infect. Dis. 16: 8–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dinh, P.N. , Long H.T., Tien N.T., et al ; World Health Organization/Global Outbreak Alert and Response Network Avian Influenza Investigation Team in Vietnam . 2006. Risk factors for human infection with avian influenza A H5N1, Vietnam, 2004. Emerg. Infect. Dis. 12: 1841–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. O’Connor, L. & Glynn B.. 2010. Recent advances in the development of nucleic acid diagnostics. Expert Rev. Med. Devices 7: 529–539. [DOI] [PubMed] [Google Scholar]
- 17. Jothikumar, P. , Hill V. & Narayanan J.. 2009. Design of FRET‐TaqMan probes for multiplex real‐time PCR using an internal positive control. Biotechniques 46: 519–524. [DOI] [PubMed] [Google Scholar]
- 18. Nolte, F.S. 2008. Molecular diagnostics for detection of bacterial and viral pathogens in community‐acquired pneumonia. Clin. Infect. Dis. 47(Suppl 3): S123–S126. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pabbaraju, K. , Tokaryk K.L., Wong S. & Fox J.D.. 2008. Comparison of the Luminex xTAG respiratory viral panel with in‐house nucleic acid amplification tests for diagnosis of respiratory virus infections. J. Clin. Microbiol. 46: 3056–3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wu, W. & Tang Y.W.. 2009. Emerging molecular assays for detection and characterization of respiratory viruses. Clin. Lab. Med. 29: 673–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Earley, M.C. , R.F. Vogt, Jr ., Shapiro H.M., et al Report from a workshop on multianalyte microsphere assays. Division of Laboratory Sciences, National Center for Environmental Health, Centers for Disease Control and Prevention, Atlanta, Georgia 30341–3724, USA. mee9@cdc.gov.
- 22. Bortolin, S. , Black M., Modi H., et al 2004. Analytical validation of the Tag‐It high throughput microsphere‐based universal array genotyping platform: application to the multiplex detection of a panel of thrombophilia associated single‐nucleotide polymorphisms. Clin. Chem. 50: 2028–2036. [DOI] [PubMed] [Google Scholar]
- 23. Mahony, J.B. , Blackhouse G., Babwah J., et al 2009. Cost analysis of multiplex PCR testing for diagnosing respiratory virus infections. J. Clin. Microbiol. 47: 2812–2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gadsby, N.J. , Hardie A., Claas E.C. & Templeton K.E.. 2010. Comparison of the Luminex respiratory virus panel fast assay with in‐house real‐time PCR for respiratory viral infection diagnosis. J. Clin. Microbiol. 48: 2213–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Food and Drug Administration, HHS . 2009. Medical devices; immunology and microbiology devices; classification of respiratory viral panel multiplex nucleic acid assay. Final rule. Fed. Regist. 74: 52136–52138. [PubMed] [Google Scholar]
- 26. Ginocchio, C.C. , Zhang F., Manji R., et al 2009. Evaluation of multiple test methods for the detection of the novel 2009 influenza A (H1N1) during the New York City outbreak. J. Clin. Virol. 45: 191–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ginocchio, C.C. & St George K.. 2009. Likelihood that an unsubtypeable influenza A virus result obtained with the Luminex xTAG respiratory virus panel is indicative of infection with novel A/H1N1 (swine‐like) influenza virus. J. Clin. Microbiol. 47: 2347–2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Maltezou, H.C. 2008. Nosocomial influenza: new concepts and practice. Curr. Opin. Infect. Dis. 21: 337–343. [DOI] [PubMed] [Google Scholar]
- 29. Leung, T.F. , To M.Y., Yeung A.C., et al 2010. Multiplex molecular detection of respiratory pathogens in children with asthma exacerbation. Chest 137: 348–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Midulla, F. , Scagnolari C., Bonci E., et al 2010. Respiratory syncytial virus, human bocavirus and rhinovirus bronchiolitis in infants. Arch. Dis. Child. 95: 35–41. [DOI] [PubMed] [Google Scholar]
- 31. Kaur, C. , Chohan S., Khare S. & Puliyel J.M.. 2010. Respiratory viruses in acute bronchiolitis in Delhi. Indian Pediatr. 47: 342–343. [PubMed] [Google Scholar]
- 32. Bochkov, Y.A. , Hanson K.M., Keles S., et al 2010. Rhinovirus‐induced modulation of gene expression in bronchial epithelial cells from subjects with asthma. Mucosal. Immunol. 3: 69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jackson, D.J. 2010. The role of rhinovirus infections in the development of early childhood asthma. Curr. Opin. Allergy Clin. Immunol. 10: 133–138. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Scagnolari, C. , Midulla F., Pierangeli A., et al 2009. Gene expression of nucleic acid‐sensing pattern recognition receptors in children hospitalized for respiratory syncytial virus‐associated acute bronchiolitis. Clin. Vaccine Immunol. 16: 816–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Woo, P.C. , Lau S.K. & Yuen K.Y.. 2009. Clinical features and molecular epidemiology of coronavirus‐HKU1‐associated community‐acquired pneumonia. Hong Kong Med. J. 15(Suppl 9): 46–47. [PubMed] [Google Scholar]
- 36. Chisti, M.J. , Tebruegge M., La Vincente S., et al 2009. Pneumonia in severely malnourished children in developing countries—mortality risk, aetiology and validity of WHO clinical signs: a systematic review. Trop. Med. Int. Health 14: 1173–1189. Review. [DOI] [PubMed] [Google Scholar]
- 37. Gaunt, E.R. , Hardie A., Claas E.C., et al 2010. Epidemiology and clinical presentations of the four human coronaviruses 229E, HKU1, NL63 and OC43 detected over 3 years using a novel multiplex real‐time PCR method. J. Clin. Microbiol. 48: 2940–2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pinsky, B.A. , Mix S., Rowe J., et al 2010. Long‐term shedding of influenza A virus in stool of immunocompromised child. Emerg. Infect. Dis. 16: 1165–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kumar, D. , Husain S., Chen M.H., et al 2010. A prospective molecular surveillance study evaluating the clinical impact of community‐acquired respiratory viruses in lung transplant recipients. Transplantation 89: 1028–1033. [DOI] [PubMed] [Google Scholar]
- 40. Gillim‐Ross, L. & Subbarao K.. 2006. Emerging respiratory viruses: challenges and vaccine strategies. Clin. Microbiol. Rev. 19: 614–636. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Edlmayr, J. , Niespodziana K., Linhart B., et al 2009. A combination vaccine for allergy and rhinovirus infections based on rhinovirus‐derived surface protein VP1 and a nonallergenic peptide of the major timothy grass pollen allergen Phl p 1. J. Immunol. 182: 6298–6306. [DOI] [PubMed] [Google Scholar]
- 42. Washington, C. , Metzgar D., Hazbón M.H., et al 2010. Multiplexed Luminex xMAP assay for detection and identification of five adenovirus serotypes associated with epidemics of respiratory disease in adults. J. Clin. Microbiol. 48: 2217–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]


