Abstract
Human metapneumovirus (hMPV) has been reported to cause both upper and lower respiratory tract diseases in susceptible populations, particularly in children and the elderly. In this study, we describe a hospital‐based epidemiological study of hMPV in patients presenting to a children's hospital and show the demographic and clinical characteristics associated with hMPV infection in China, retrospectively. Specimens were collected over a 2‐year period from children hospitalized with acute lower respiratory tract infections (ALRTI) and analyzed for the presence of hMPV using real‐time RT‐PCR assays. The presence of hMPV was detected in 227 (25.9%) of the 878 children studied and may circulate year‐round in the area, peaking during the winter–spring season. Younger children (aged less than 6 months) had the highest positive rate. Infections by hMPV showed similar epidemiology and clinical manifestations as for respiratory syncytial virus (RSV) and were found in high co‐infections with RSV. Subgroup A2 hMPV was the most predominant genotype identified during the study period. This study indicates that hMPV is one of the major respiratory pathogens found in children in southwest China and vaccine development should be under consideration. Pediatr. Pulmonol. 2010; 45:824–831. © 2010 Wiley‐Liss, Inc.
Keywords: human metapneumovirus, respiratory syncytial virus, children, acute lower respiratory tract infections, epidemiology
Abbreviations:
hMPV, human metapneumovirus; ALRTI, acute lower respiratory tract infections; RSV, respiratory syncytial virus; NPAs, nasopharyngeal aspirates; cDNA, complementary DNA.
INTRODUCTION
Respiratory tract infections are a major cause of morbidity and mortality worldwide, particularly in developing countries. In children less than 5 years of age, respiratory tract infections rank second or third as the top causes of child death, regardless of geographic location.1 Previous researches have demonstrated that human respiratory syncytial virus (RSV), influenza virus types A and B, and parainfluenza virus types 1–3, and adenoviruses are important causes of acute lower respiratory tract infections (ALRTI) in China.2, 3 Human metapneumovirus (hMPV) was first isolated in 2001 from nasopharyngeal aspirates (NPAs) obtained from children with acute lower respiratory tract infections (ALRTI) in the Netherlands.4 Recently, it has been demonstrated that hMPV is a cause of respiratory tract infections with symptoms similar to those of respiratory syncytial virus (RSV). The clinical disease spectrum caused by hMPV varies from upper respiratory tract infection to bronchiolitis and pneumonia in infants and young children. Serological studies have shown that by 5 years of age, virtually all children in the Netherlands have been exposed to hMPV and that it has been circulating in humans for at least 50 years.4 Consistent with previous studies, we found that the vast majority (90.3%) of children were seropositive to hMPV by the age of six in Chongqing, a large city located in Southwest China.5 The high prevalence of hMPV indicates that it is an important respiratory pathogen in this temperate climate. A number of studies investigate the prevalence and clinical features of hMPV in China.6, 7 As these studies were not consecutive, and have geographic differences, and the enrolled population was quite limited, the epidemiological picture of hMPV in this most populous country in the world remains unclear. Thus, ongoing epidemiological surveillance in a consecutive manner will help assess the disease burden and provide information for the development of a vaccine strategy against hMPV.
Studies show that hMPV replicates slowly in host cells and, consequently, virus culture is rarely used as a diagnostic assay for epidemiological studies. Real‐time reverse transcription polymerase chain reaction (real‐time RT‐PCR) for hMPV, however, offers many advantages over other conventional methods.8 Therefore, this study first validated the reliability and reproducibility of real‐time RT‐PCR experiments targeting the hMPV F gene in serially diluted recombinant hMPV virus infected cell supernatants. The validated assay was then used to detect the presence of hMPV in NPAs from pediatric patients presenting at a large teaching children's hospital located in southwest China over a period of 2 years. Demographic, clinical and laboratory data were analyzed retrospectively to elucidate the epidemiology of hMPV in this area over time.
MATERIALS AND METHODS
Patients and Clinical Samples
NPAs from pediatric patients presenting with ALRTI in the daytime were collected at the Respiratory Ward, Children's Hospital of Chongqing Medical University, on three fixed days each week over a 2‐year period from April 2006 to March 2008. NPAs (1–2 ml) were taken from the newly admitted patients, added to 2 ml phosphate‐buffered saline on ice immediately according to a standard protocol and transported within 4 hr to the virology laboratory.9 NPAs were clarified by centrifugation at 800g for 10 min at 4°C to separate the cells and cell debris and the cell‐free fluid stored in aliquots at −80°C until RNA extraction.
The study was approved by Ethical Committee, Children's Hospital of Chongqing Medical University. The informed consent was verbal from pediatric patient guardian.
RNA Extraction and cDNA Synthesis
Total RNA was extracted from NPAs using the QIAamp viral RNA mini‐kit (Qiagen, Hilden, Germany), and complementary DNA (cDNA) was synthesized in the PrimeScript RT Reagent Kit (TaKaRa, Dalian, China), according to the manufacturer's instructions.
PCR Assays and Sequencing
All NPAs were tested for RSV by RT‐PCR and hMPV by real‐time PCR. RSV forward primer P1 and reverse primer P2 were used to amplify the RSV G gene of subgroup A (GenBank accession: M11486). RSV forward primer P1 and reverse primer P3 were used to amplify the RSV G gene of subgroup B (GenBank accession: AY333364) (Table 1). Real‐time PCR primers and probe sequences to detect the hMPV F gene (Table 1) were used for the diagnosis of hMPV infection and were selected based on sequence homology with the prototype strain from China (GenBank accession: DQ336144). Viral cDNA was amplified using a real‐time PCR procedure with the Premix Ex Taq Kit (Perfect Real Time, TaKaRa) in a LightCycler instrument (Roche Diagnostics, Indianapolis, IN). The positive determination criterion was set at cycle threshold (Ct) ≤36. Each set of real‐time PCR experiments contained appropriate negative controls.
Table 1.
Sequences of Oligonucleotide Primers and Probes Used for the Detection of hMPV and RSV in Patient Samples
| Target | Primer/probe name | Primer/probe sequence (5′–3′) |
|---|---|---|
| hMPVa | FF | CGTTTCTTACATGCCGACATCTG |
| hMPVa | FR | GCTCCCGTAGACCCCTATCAG |
| hMPVa | FP | (FAM)CCCTTTCTTCGCACCATCGCACGG(Eclipse) |
| hMPVb | 450F | CTTTGGACTTAATGACAGATG |
| hMPVb | 450R | GTCTTCCTGTGCTAACTTTG |
| hMPVb | GF | GAGAACATTCGRRCRATAGAYATG |
| hMPVb | GR | AGATAGACATTRACAGTGGATTCA |
| RSV | P1 | TGGGACACTCTTAATCAT |
| RSVc | P2 | TGATTCCAAGCTGAGGAT |
| RSVd | P3 | GTTGTATGGTGTGTTTC |
Nucleotide sequences for hMPV according to GenBank accession number DQ336144.
Nucleotide sequences for hMPV according to GenBank accession number AF371337.
Nucleotide sequences for RSV prototype strain A2 according to GenBank accession number M11486.
Nucleotide sequences for RSV prototype strain B (strain 18537) according to GenBank accession number M17213.
Recombinant NL 00‐1 stain hMPV was recovered from cell cultures transfected with a cDNA clone containing the entire viral genome (a kind gift from Dr. Fushier, Erasmus Institute, The Netherlands) and used as positive control for verification of the real‐time PCR. The sensitivity of the real‐time PCR assay was demonstrated to be able to detect a minimum of 1 TCID50/ml recombinant virus and showed good reproducibility. To verify the presence and sequence of the virus, traditional RT‐PCR primers 450F and 450R (Table 1), previously described by Peret et al.,10 were used to amplify the hMPV F gene to ensure reproducibility (GenBank accession: AF371337). GF and GR primers (Table 1), previously described by Ludewick et al.,11 were used to amplify the hMPV G gene (GenBank accession: AF371337) and the sequences were used for phylogenetic analysis. All PCR assays were verified to be able to amplify F and G gene of the recombinant hMPV positive control.
PCR products were purified from agarose gels using the QIAquick gel extraction kit (Qiagen) and sequenced in both directions using the same PCR primers for amplification. Sequence analysis of the dye‐terminator products was performed on an ABI 3100 sequencer.
Phylogenetic Analysis of hMPV Strains
Nucleotide sequences of the amplified F and G gene PCR products were aligned using CLUSTALW (version 1.74). Phylogenetic trees were constructed by the neighbor‐joining method in the MEGA software (version 4.0). The G gene sequence of the avian pneumovirus subgroup C (GenBank accession: AY590688) was chosen as the outgroup for rooting the tree. The phylogenetic analysis included hMPV archived sequences from GenBank belonging to four different subgroups: subgroup A1: NL00‐1 (AF371337) and JPS03‐180 (AY530092); subgroup A2: CAN 97‐83 (AY297749), JPS03‐240 (AY530095), and BJ1887 (DQ843659); subgroup B1: JPS03‐194 (AY530094) and NL/1/99 (AY525843); subgroup B2: CAN98‐75 (AY297748) and BJ1816 (DQ843658).
Analysis of Clinical Data
The clinical diagnosis of ALRTI was based on the presence of cough, tachypnea, chest indrawing, or wheeze of <7 days' duration, and followed the World Health Organization's standard protocol for research on ALRTI.12 Medical charts were reviewed by a senior pediatrician. Demographic data (sex, age, hospital day), clinical symptoms (such as body temperature, cough, wheezing, and cyanosis), laboratory findings (white blood count, levels of C‐reactive protein and chest X‐ray results), and treatments (administration of antibiotics and corticosteroids, and use of bronchodilators) were recorded and analyzed. The pediatrician and radiologists involved in this study were all blinded to the virological data.
Statistical Analysis
Univariate procedures were used to examine the distribution of illness characteristics and laboratory results. Statistical tests of comparison included chi square tests for dichotomous variables, t tests for normally distributed continuous variables. All statistical analyses were performed using SPSS software (version 11.5). P < 0.05 was considered statistically significant.
RESULTS
Detection of hMPV Infection
From April 2006 to March 2008, total 6,296 patients (4,054 males, 2,242 females) with ALRTI were hospitalized. NPAs were obtained from 878 children (592 males, 286 females; age range: 29 days to 15.9 years) who came to the hospital in daytime. The enrolled patients accounted for 13.9% (878/6296) of the total admissions with ALRTI during the same period. There was no statistical difference detected regarding the age, sex distribution and the main clinical diagnose between the total 6,296 patients and selected study population (P > 0.05).
NPAs from these children were tested for the presence of hMPV by real‐time PCR with a primer set specific for the F gene that can detect both genotypes of the virus and 25.9% (227 of 878) were positive. A total of 878 NPAs were tested for RSV by RT‐PCR and 35.6% (313 of 878) were positive. Among the 227 hMPV‐positive samples, 41.9% (95 of 227) showed evidence of co‐infection by hMPV and RSV.
Patients diagnosed with hMPV infection were aged from 29 days to 8.8 years, with the youngest patients having the highest positive rate. The overall age distribution of hMPV‐positive patients was found to be similar to that of RSV‐positive patients in this cohort (Table 2, P > 0.05), except that hMPV infection occurred more frequently than RSV in children from 2 to 6 years old (Table 2, P = 0.02). The prevalence of hMPV and RSV infection was similar between genders (Table 2, P > 0.05). The mean duration of hospitalization was 8.0 ± 4.6 days for hMPV‐positive children, similar to 7.8 ± 4.3 days for RSV‐positive children and 7.8 ± 3.2 days for hMPV and RSV‐positive children (Table 2, P = 0.571 and 0.569, respectively), but higher than 7.3 ± 3.7 days for hMPV and RSV‐negative children (Table 2, P = 0.016).
Table 2.
Patient Demographic Data
| Parameter | No.(%) of children hMPV‐positive | No.(%) of children RSV‐positive | No.(%) of children hMPV‐ and RSV‐positive | No.(%) of children hMPV‐ and RSV‐negative | P |
|---|---|---|---|---|---|
| Median age(months) | 11.5 ± 15.6 | 9.0 ± 9.8 | 8.72 ± 10.3 | 17.1 ± 24.10 | <0.05 a, b |
| Age distribution | |||||
| <6months | 119/227 (52.4) | 175/313 (55.9) | 54/95 (56.9) | 188/433 (43.4) | <0.05 b |
| 6 months‐2 years | 72/227 (31.7) | 111/313 (35.5) | 31/95 (32.6) | 148/433 (11.0) | <0.05 b |
| 2 years‐6 years | 34/227 (15.0) | 27/313 (8.6) | 10/95 (10.5) | 79/433 (18.2) | <0.05 a |
| >6 years | 2/227 (0.9) | 0/313 (0) | 0/95 (0) | 18/433 (4.1) | <0.05 b |
| Sex | |||||
| male | 148/227 (65.2) | 213/313 (68.1) | 57/95 (60) | 285/433 (65.8) | NS |
| female | 79/227 (34.6) | 100/313 (31.9) | 38/95 (40) | 148/433 (34.2) | NS |
| Clinical sign | |||||
| Feverd | 82/227 (36.1) | 121/313 (38.7) | 31/95 (32.6) | 157/433 (36.3) | NS |
| cough | 225/227 (99.1) | 311/313 (99.4) | 94/95 (98.9) | 423/433 (97.7) | NS |
| wheezing | 167/227 (73.6) | 228/313 (72.8) | 78/95 (82.1) | 225/433 (52.0) | <0.05 b |
| cyanosis | 193/227 (85.0) | 279/313 (89.1) | 92/95 (96.8) | 340/433 (78.5) | <0.05 b, c |
| respiratory failure | 18/227 (7.9) | 31/313 (9.9) | 11/95 (11.6) | 14/433 (3.2) | <0.05b |
| mechanical ventilation | 1/227 (0.4) | 1/313 (0.3) | 0/95 (0) | 1/433 (0.2) | NS |
| Lab examination | |||||
| WBC >10 × 109/L(num) | 86/224 (38.4) | 113/310 (36.5) | 37/94 (39.4) | 133/421 (31.6) | NS |
| CRP > 8mg/L(num) | 15/170 (8.8) | 9/229 (3.9) | 3/72 (4.2) | 34/323 (10.5) | NS |
| Hospital stay (days) | 8.0 ± 4.6 | 7.8 ± 4.3 | 7.8 ± 3.2 | 7.3 ± 3.7 | <0.05b |
hMPV: human metapneumovirus; RSV: respiratory syncytial virus; NS: non‐significant; WBC: white blood count; num: numbers; CRP: C‐reative protein.
Comparison of hMPV‐positive to RSV‐positive patients.
Comparison of hMPV‐positive to hMPV and RSV‐negative patients.
Comparison of hMPV‐positive to hMPV and RSV co‐infection patients.
Body temperature of ≥38 °C.
Seasonal Distribution of hMPV Infection
The specimens were collected continuously during the 2 years from April 2006 to March 2008 on every Monday, Wednesday, and Friday. A total of 390 children diagnosed with ALRTI were enrolled in the first year (April 2006 to March 2007) and 488 in the second year (April 2007 to March 2008). Generally, 18.5% (72 of 390) of NPAs were positive for hMPV by real‐time RT‐PCR in the first year and 31.7% (155 of 488) were positive in the second year. Significant difference was shown in terms of the positive rate of hMPV infection during the 2 years (P < 0.0001). It was clear that hMPV circulated predominantly during the late winter and spring period (Fig. 1).
Figure 1.
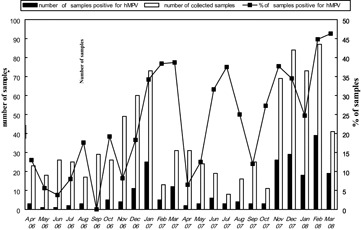
The number of specimens collected, hMPV‐positive samples and the percent positive per month for the 2‐year period from April 2006 to March 2008.
Clinical Manifestations of hMPV Infection
The clinical symptoms observed for hMPV‐infected children included fever (body temperature ≥38 °C; 36.1%, 82 of 227), cough (99.1%, 225 of 227), wheezing (73.6%, 167 of 227), and cyanosis (85.0%, 193 of 227). These symptoms were observed with similar frequency in the group of RSV‐infected children (Table 2, P > 0.05). As compared to the hMPV and RSV negative patients, hMPV‐infected children showed a higher frequency of wheezing and cyanosis (Table 2, P < 0.05).
Two hundred four of 227(89.9%) hMPV‐positive patients received chest radiographs during their illnesses. Radiologic evidence of pneumonia or pulmonary infiltration was found in 199 of 204 (97.5%) total hMPV‐positive patients, while 126 (61.8%) had evidence of hyperinflation, three had evidence of atelectasis and one had pleural effusion. The remaining patients included five children with a normal radiogram and 23 were not examined. The ratio of abnormal white blood counts and levels of C‐reactive protein patients were not significantly different among these four groups (P < 0.05).
Diagnoses upon admission were obtained for all patients enrolled in the study. The main clinical diagnoses of ALRTI in hMPV‐positive patients were acute laryngotracheobronchitis (1 of 227, 0.4%), asthmatic bronchitis (1 of 227, 0.4%), bronchitis (3 of 227, 1.3%), bronchiolitis (65 of 227, 28.6%) and pneumonia (148 of 227, 65.2%). The presence of hMPV in 11% (25 of 227) of asthmatic children presented with an acute exacerbation. Underlying medical conditions were common in hMPV‐positive children (23.8%, 54 of 227), including severe asthma (10.6%, 24 of 227), congenital heart disease (10.1%, 23 of 227), congenital laryngeal stridor (1.8%, 4 of 227) and bronchomalacia (1.8%, 4 of 227) (one patient with severe asthma and congenital heart disease). Among 227 hMPV‐positive patients, two patients had recurrent respiratory tract infection and two patients had midrange malnutrition. It is noteworthy that 18 patients had serious respiratory failure and required supplemental oxygen and eleven (61.1%) of these had evidence of co‐infection with hMPV and RSV. Two patients with serious respiratory failure were admitted to intensive care units and one patient required mechanical ventilation. All children with hMPV were given antibiotics during their hospitalization, however, not all patients had specimens collected for bacterial culture. Corticosteroids were given to 189 of 227 (83.3%) hMPV‐positive patients and bronchodilators were given to 203 of 227 (89.4%) patients during hospitalization.
Phylogenetic Analysis of the hMPV F and G Gene
We selected 10 hMPV samples by using random digits table to both confirm the circulation of hMPV and analyze F gene nucleotide sequences. The nucleotide identity of sequenced F gene fragments for nine strains with the subgroup A2 reference sequence, CAN97‐83, was 95–98%; whereas, one strain showed an F gene nucleotide identity of 96% with the subgroup B2 reference sequence, CAN98‐75.
Phylogenetic trees were constructed based on the G gene sequences from 14 hMPV Chongqing strains. The phylogenetic tree of the hMPV strains recovered during 2006–2008 showed two major groups or clusters, each of which can be divided into two subgroups. The majority of strains grouped with subgroup A2, except one strain that grouped with subgroup B2. The nucleotide identity of sequenced G gene fragments of the 13 subgroup A2 strains and the prototypical subgroup A2 sequence, CAN97‐83, was 90–95%; whereas, one subgroup B2 strain showed a G gene nucleotide identity of 92% with the subgroup B2 prototype strain, CAN98‐75 (Fig. 2).
Figure 2.
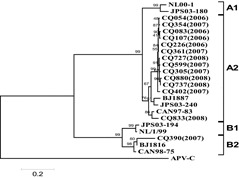
Phylogenetic analysis of hMPV G gene sequences. The tree was constructed by the neighbor‐joining method. The Avian pneumovirus subgroup C (APV‐C) outgroup and hMPV subgroups A1, A2, B1, and B2 are indicated.
DISCUSSION
Respiratory tract infections are the major cause of admission to hospital of young children during the winter months, worldwide. Viruses most frequently associated with respiratory infections include rhinoviruses, coronaviruses, influenza viruses, parainfluenza viruses, RSV and adenoviruses. In 2001, a new member of the Paramyxoviridae associated with respiratory illness, hMPV, was described in the Netherlands.4 Data from that report suggested that the illnesses caused by hMPV were similar to RSV, with both typically causing diseases ranging from mild cold to bronchiolitis and pneumonia during the winter months in children.13 Some previous studies have suggested that hMPV is an important causative agent for ALRTI in children in China. Yet, despite this knowledge, the epidemiological picture of hMPV infection in the most populous country in the world remains largely unknown due to the limits of these studies.6, 7
The present study enrolled 878 pediatric patients with ALRTI who were admitted to Chongqing Children's Hospital of Chongqing Medical University over a 2‐year period from April 2006 to March 2008. The enrolled patients accounted for a significant proportion of the total hospitalizations due to ALRTI during that period. Although we designed to enroll all newly admitted patients in three fixed days every week, the study results were not necessarily free from selection bias because we only collected NPAs in the daytime and some of the admitted patients refused to provide NPAs. The age, sex distribution and main clinical diagnoses between enrolled 878 patients in this study and the whole 6,296 admissions during the same period, though, was not statistically different, reflecting the selected subjects may thus be relatively representative. In previous studies of different ethnic and regional populations, the hMPV positive rate in NPAs from children with unknown etiology varied from 1.5% to 43%.4, 9, 14, 15, 16, 17 The present study used real‐time PCR targeting the hMPV F gene and found up to 25.9% of ALRTI could be attributed to hMPV infection from non‐selected NPAs. The general prevalence of hMPV infection was relatively high as compared with some other regions, probably due to a higher population density in this city. Thus, hMPV appeared to cause a significant disease burden in Chongqing and was a common prevalent viral agent for ALRTI. Previous reports showed multivirus infections have a high occurrence in patients with ALRTI in china.2, 3 In our study, hMPV co‐infection with RSV was relatively common. The reason for high co‐infection rate remains unclear by far, probably also partially due to a higher population density in this city.
The age distribution of hMPV infection was similar to RSV infection, and hMPV infection has been found to cause ALRTI in infants and younger children, with a more severe disease manifestation in young children less than 2 years of age, immunocompromized subjects, and frail, elderly individuals.4, 18 Primary hMPV infection occurred at a young age (<1 year) and several seroprevalence studies have previously shown that 90–100% of children are infected by the ages of 5–10 years.4, 19, 20 Similar result was got in our study that the majority (84.1%, 191 of 227) of patients were under 2 years of age. The frequency of hMPV particularly in little children might be caused by their immune status.
HMPV cause seasonal outbreaks of respiratory infections in children and the epidemic peak for hMPV infection have been demonstrated to be during the winter‐spring season in most areas of the world.21, 22 In the present study, hMPV was identified every month from April in 2006 to March in 2008 (except September in 2006) suggesting that hMPV may circulate year‐round, and hMPV outbreak activity in Chongqing was high in the winter‐spring season, similar to other studies of the temperate zone.21, 22 Although a relatively high prevalence (25–37.5%) was seen throughout June to August 2007, this may not indicate that summer is the prevalent season for hMPV in Chongqing as there were a limited number of specimens collected during that period. The Chinese spring festival may have an impact on the prevalence for February 2007 when many patients are not likely to be hospitalized. Limited sample collection is likely to reduce the reliability of prevalence rates. Previous studies found hMPV infection rate may vary greatly during the course of successive respiratory infection seasons.9, 11, 17, 23 We also found a significant difference in the prevalence of hMPV infection in continuous 2 years, and the monthly prevalence of hMPV infection during the second year was higher than that during the first year, particularly in winter–spring seasons. The reason is not clear yet, while that may indicate hMPV has a periodic circulating pattern probably due to the immunity of the local population. Apparently, more data for a longer study period needed to elucidate the real pattern of hMPV circulation in this region.
Clinical symptoms associated with hMPV infection in young children are considered indistinguishable from those caused by RSV.18, 22, 24, 25, 26 Consistent with previous studies, we found the most common clinical signs of hMPV‐associated ALRTI were fever, cough, wheezing, and cyanosis. Comparison of the medical files of hMPV‐infected children with those of RSV‐infected children revealed that the clinical symptoms associated with these viruses were similar. This study found that a bulk of hMPV‐infected children who received chest radiographs showed abnormalities such as pneumonia or pulmonary infiltration, hyperinflation, atelectasis, and pleural effusion. The main clinical diagnoses ranged from acute laryngotracheobronchitis, asthmatic bronchitis, bronchitis, bronchiolitis, and pneumonia. We found hMPV infection was related to wheezing, asthma exacerbations or bronchiolitis, as previous study described.13, 23, 26, 27 Of 18 hMPV‐positive cases with a diagnosis of respiratory failure, 11 (61.1%) were identified as a co‐infection with RSV, indicating that hMPV and RSV co‐infection may lead to a more severe disease than single infections. Nevertheless, in a larger number of patients with both hMPV and RSV, co‐infection did not present as a severe condition for the duration of hospitalization. All hMPV‐positive patients were unfortunately given antibiotics mainly because no etiological agent had been identified at the time of hospitalization. The majority of the hMPV‐positive patients had corticosteroid and bronchodilators treatment for wheezing. These issues clearly reflect misuse of these medications. Application of reliable rapid diagnostics for not only hMPV but also for the common respiratory viruses may greatly help and be in priority.
HMPV is classified into two major groups, designated group A and B.23, 28 In this study, from 14 hMPV G gene sequences, the majority (13/14) grouped in A (subgroup A2), except one strain grouped in B (subgroup B2). HMPV group A showed a predominant circulation, in consistent with other studies,11, 13, 23, 24, 26, 27, 28, 29, 30 only one study showed a shift to a predominant B genotype.31 We used the primers set located on G gene, originally described by Ludewick et al.11 Due to greatest diversity of the G proteins between hMPV subgroups (37% identity) as reported by Biacchesi et al.,32 we cannot rule out that we may have missed some A or B strains during 2 years. There is no report whether these genotypes represent distinct serotypes of hMPV or genetic variants of a prototype strain until now. Similar to RSV epidemics,33 these findings indicate that hMPV from the two major groups could co‐circulate in the population in the same period. Although a report described that A strains may be more pathogenic than B strains,34 we found no significant differences in the severity of illness between any of the groups or between subgroups because we detected only one hMPV B strain.
In conclusion, hMPV appears to be an important viral agent for ALRTI and is likely to be found in high co‐infections with RSV in Chongqing, China. Given the high prevalence and disease burden associated with hMPV infection, vaccine development against this virus should be under consideration. As the present study was a retrospective one, a perspective study in larger scale for a longer period is needed to further illustrate epidemiology of hMPV in China.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (No. 30730098), the Program for the New Century Excellent Talents in University of China (No. NCET‐05‐0774). We thank Dr. Fushier (Department of Virology, Erasmus Medical Center, The Netherlands) for the kind gift of the cDNA clones to produce recombinant hMPV virus. We also thank Jin Ying Ge, Zhi Gao Bu, and Wang Yong (Harbin Veterinary research institute, Chinese Agricultural Academy of Science) for excellent technical assistance for the production of recombinant hMPV virus. We would also like to appreciate Mr. Qianglin Duan for critical reading of the manuscript.
REFERENCES
- 1. Murray CJL, Lopez AD, Mathers CD, Stein C. The global burden of disease 2000 project: aims, methods and data sources. In: Global programme on evidence for health policy. Geneva: World Health Organization; 2001. discussion Paper No. 36, 1–57. [Google Scholar]
- 2. Zhang HY, Li ZM, Zhang GL, Diao TT, Cao CX, Sun HQ. Respiratory viruses in hospitalized children with acute lower respiratory tract infections in harbin, China. Jpn J Infect Dis 2009; 62: 458–460. [PubMed] [Google Scholar]
- 3. Peng D, Zhao D, Liu J, Wang X, Yang K, Xicheng H, Li Y, Wang F. Multipathogen infections in hospitalized children with acute respiratory infections. Virol J 2009; 6: 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Van den Hoogen BG, de Jong JC, Groen J, Kuiken T, de Groot R, Fouchier RA, Osterhaus AD. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat Med 2001; 7: 719–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhang Q, Yang XQ, Zhao Y, Zhao XD. High seroprevalence of human metapneumovirus infection in children in Chongqing, China. Chin Med J (Engl) 2008; 121: 2162–2166. [PubMed] [Google Scholar]
- 6. Li XY, Chen JY, Kong M, Su X, Yi YP, Zou M, Zhang H. Prevalence of human metapneumovirus in hospitalized children with respiratory tract infections in Tianjin, China. Arch Virol 2009; 154: 1831–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ji W, Wang Y, Chen Z, Shao X, Ji Z, Xu J. Human metapneumovirus in children with acute respiratory tract infections in Suzhou, China 2005–2006. Scand J Infect Dis 2009; 41: 735–744. [DOI] [PubMed] [Google Scholar]
- 8. Côté S, Abed Y, Boivin G. Comparative evaluation of real‐time PCR assays for detection of the human metapneumovirus. J Clin Microbiol 2003; 41: 3631–3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Maggi F, Pifferi M, Vatteroni M, Fornai C, Tempestini E, Anzilotti S, Lanini L, Andreoli E, Ragazzo V, Pistello M, Specter S, Bendinelli M. Human metapneumovirus associated with respiratory tract infections in a 3‐year study of nasal swabs from infants in Italy. J Clin Microbiol 2003; 41: 2987–12991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Peret TC, Boivin G, Li Y, Couillard M, Humphrey C, Osterhaus AD, Erdman DD, Anderson LJ. Characterization of human metapneumoviruses isolated from patients in North America. J Infect Dis 2002; 185: 1660–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ludewick HP, Abed Y, van Niekerk N, Boivin G, Klugman KP, Madhi SA. Human metapneumovirus genetic variability, South Africa. Emerg Infect Dis 2005; 11: 1074–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cuevas LE, Nasser AM, Dove W, Gurgel RQ, Greensill J, Hart CA. Human metapneumovirus and respiratory syncytial virus, Brazil. Emerg Infect Dis 2003; 9: 1626–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Eihara T, Endo R, Kikuta H, Ishiguro N, Ishiko H, Hara M, Takahashi Y, Kobayashi K. Human metapneumovirus infection in Japanese children. J Clin Microbiol 2004; 42: 126–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nissen MD, Siebert DJ, Mackay IM, Sloots TP, Withers SJ. Evidence of human metapneumovirus in Australian children. Med J Aust 2002; 176: 188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Stockton J, Stephenson I, Fleming D, Zambon M. Human metapneumovirus as a cause of community‐acquired respiratory illness. Emerg Infect Dis 2002; 8: 897–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Viazov S, Ratjen F, Scheidhauer R, Fiedler M, Roggendorf M. High prevalence of human metapneumovirus infection in young children and genetic heterogeneity of the viral isolates. J Clin Microbiol 2003; 41: 3043–3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kim YK, Lee HJ. Human metapneumovirus‐associated lower respiratory tract infections in Korean infants and young children. Pediatr Infect Dis J 2005; 24: 1111–1112. [DOI] [PubMed] [Google Scholar]
- 18. Boivin G, Abed Y, Pelletier G, Ruel L, Moisan D, Côté S, Peret TC, Erdman DD, Anderson LJ. Virological features and clinical manifestations associated with human metapneumovirus: a new paramyxovirus responsible for acute respiratory‐tract infections in all age groups. J Infect Dis 2002; 186: 1330–1334. [DOI] [PubMed] [Google Scholar]
- 19. Ebihara T, Endo R, Kikuta H, Ishiguro N, Ishiko H, Hara M, Takahashi Y, Kobayashi K. Seroprevalence of human metapneumovirus in Japan. J Med Virol 2003; 70: 281–283. [DOI] [PubMed] [Google Scholar]
- 20. Wolf DG, Zakay‐Rones Z, Fadeela A, Greenberg D, Dagan R. High seroprevalence of human metapneumovirus among young children in Israel. J Infect Dis 2003; 188: 1865–1867. [DOI] [PubMed] [Google Scholar]
- 21. Freymouth F, Vabret A, Legrand L, Eterradossi N, Lafay‐Delaire F, Brouard J, Guillois B. Presence of the new human metapneumovirus in French children with bronchiolitis. Pediatr Infect Dis J 2003; 22: 92–94. [DOI] [PubMed] [Google Scholar]
- 22. Van den Hoogen BG, van Doornum GJ, Fockens JC, Cornelissen JJ, Beyer WE, de Groot R, Osterhaus AD, Fouchier RA. Prevalence and clinical symptoms of human metapneumovirus infection in hospitalized patients. J Infect Dis 2003; 188: 1571–1577. [DOI] [PubMed] [Google Scholar]
- 23. Manoha C, Espinosa S, Aho SL, Huet F, Pothier P. Epidemiological and clinical features of hMPV, RSV and RVs infections in young children. J Clin Virol 2007; 38: 221–226. [DOI] [PubMed] [Google Scholar]
- 24. Boivin G, De Serres G, Côté S, Gilca R, Abed Y, Rochette L, Bergeron MG, Déry P. Human metapneumovirus infections in hospitalized children. Emerg Infect Dis 2003; 9: 634–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Esper F, Boucher D, Weibel C, Martinello RA, Kahn JS. Human metapneumovirus infection in the United States: clinical manifestations associated with a newly emerging respiratory infection in children. Pediatrics 2003; 111: 1407–1410. [DOI] [PubMed] [Google Scholar]
- 26. Peiris JS, Tang WH, Chan KH, Khong PL, Guan Y, Lau YL, Chiu SS. Children with respiratory disease associated with metapneumovirus in Hong Kong. Emerg Infect Dis 2003; 9: 628–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Noyola DE, Alpuche‐Solís AG, Herrera‐Díaz A, Soria‐Guerra RE, Sánchez‐Alvarado J, López‐Revilla R. Human metapneumovirus infections in Mexico: epidemiological and clinical characteristics. J Med Microbiol 2005; 54: 969–974. [DOI] [PubMed] [Google Scholar]
- 28. Banerjee S, Bharaj P, Sullender W, Kabra SK, Broor S. Human metapneumovirus infections among children with acute respiratory infections seen in a large referral hospital in India. J Clin Virol 2007; 38: 70–72. [DOI] [PubMed] [Google Scholar]
- 29. Boivin G, Mackay I, Sloots TP, Madhi S, Freymuth F, Wolf D, Shemer‐Avni Y, Ludewick H, Gray GC, LeBlanc E. Global genetic diversity of human metapneumovirus fusion gene. Emerg Infect Dis 2004; 10: 1154–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sarasini A, Percivalle E, Rovida F, Campanini G, Genini F, Torsellini M, Paolucci S, Baldanti F, Marchi A, Grazia Revello M, Gerna G. Detection and pathogenicity of human metapneumovirus respiratory infection in pediatric Italian patients during a winter‐spring season. J Clin Virol 2006; 35: 59–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Agapov E, Sumino KC, Gaudreault‐Keener M, Storch GA, Holtzman MJ. Genetic variability of human metapneumovirus infection: evidence of a shift in viral genotype without a change in illness. J Infect Dis 2006; 193: 396–403. [DOI] [PubMed] [Google Scholar]
- 32. Biacchesi S, Skiadopoulos MH, Boivin G, Hanson CT, Murphy BR, Collins PL, Buchholz UJ. Genetic diversity between human metapneumovirus subgroups. Virology 2003; 315: 1–9. [DOI] [PubMed] [Google Scholar]
- 33. Hall CB, Walsh EE, Schnabel KC, Long CE, McConnochie KM, Hildreth SW, Anderson LJ. Occurrence of groups A and B of respiratory syncytial virus over 15 years: associated epidemiologic and clinical characteristics in hospitalized and ambulatory children. J Infect Dis 1990; 162: 1283–1290. [DOI] [PubMed] [Google Scholar]
- 34. Vicente D, Montes M, Cilla G, Perez‐Yarza EG, Perez‐Trallero E. Differences in clinical severity between genotype A and genotype B human metapneumovirus infection in children. Clin Infect Dis 2006; 42: e111–e113. [DOI] [PubMed] [Google Scholar]


