Abstract
Herpes simplex virus (HSV) is a human pathogen that infects epithelial cells. The cutaneous lesions, caused by the virus, spread to the nervous system creating several complications. Fusion of host membranes with the viral envelope is mandatory and mediated by a group of glycoproteins conserved in all Herpesviridae subfamilies, such as the glycoproteins B (gB), H (gH), L (gL) and D (gD). We investigated the inhibitory activity mediated by synthetic overlapping peptides spanning the entire ectodomains of gH and gL glycoproteins. We have performed a brute analysis of the complete gH/gL heterodimer in order to explore the inhibitory activity of peptides modelled on these glycoproteins against HSV‐1 infection. Twenty‐four of the gH peptides at a concentration of 150 μM reached the 50% of inhibition cut‐off. Interestingly, they are mainly located in the gH carboxy‐terminal domain. None of the gL peptides had a clear inhibiting effect. No peptide toxicity was observed by lactate dehydrogenase assay at the concentrations used in our experimental conditions. HSV‐1 therapy is based on acyclovir treatment, but some resistant strains are emerging. In this scenario, innovative approaches for HSV‐1 treatment are necessary. Our data support the direct involvement of the described domains in the process of virus penetration; therefore, these results are of relevance to the potential development of novel therapeutic compounds to prevent HSV‐1 infections. Copyright © 2017 European Peptide Society and John Wiley & Sons, Ltd.
Keywords: overlapping peptides, herpes simplex virus, infectivity inhibition, viral glycoprotein
In vitro screening of overlapping synthetic peptides derived from HSV‐1 glycoproteins for virus inhibition.
Introduction
Herpes simplex viruses (HSV) are human pathogens that infect epithelial cells 1. They are dsDNA viruses divided in three sub‐families: Alfaherpesvirinae, Betaherpesvirinae and Gammaherpesvirinae. The Alfaherpesvirinae are grouped together for their relative fast replicative cell cycle, their host range and the ability to generate latency in the sensitive ganglial nerves 2. Among this sub‐family, the important species for human infections are represented by HSV‐1, HSV‐2 and Varicella‐Zoster virus. The common HSV treatment is based on acyclovir, valacyclovir and farmcyclovir. Resistant strains are emerging; therefore, new strategies are needed to deal with these infections 3.
Therapeutic peptides have become an attractive tool in drug discovery, and the best characterized therapeutic antiviral peptide inhibitor is Enfuvirtide (fusion inhibitor) which mimics the C‐terminal repeat coil region of human immunodeficiency virus (HIV) fusion protein, gp41 4, 5, 6, 7. Specific functional domains of the proteins involved in the mechanism of cell penetration of a number of viruses have been found able to inhibit infectivity 8, 9, 10, 11, 12, 13.
HSV‐1 infection is initiated by the binding of glycoprotein C (gC) to cell surface heparan sulfate 14, 15. The HSV‐1 gB protein (gB) can also bind to heparan sulfate proteoglycans, but the binding is less efficient 16. Following attachment, gD binds to one of a number of co‐receptors on the host cell membrane, including herpesvirus entry mediator, nectin 1, nectin 2 or 3‐O‐sulfated heparan sulfate, resulting in a conformational change in gD 17, 18, 19, 20, 21, 22. The conformational change in gD is believed to trigger the formation of a fusion complex, which enables fusion to ensue. The co‐expression of gD, gB and gH‐gL heterodimer in the same cell results in cell–cell fusion, indicating that these four proteins constitute the minimal fusion apparatus 23, 24, 25, 26. Although the entry pathways of other enveloped viruses share similar patterns, most systems for which molecular details have been gathered rely on a single fusion protein 27. HSV‐1 is singular because it uses both gB and gH/gL as its core fusion machinery 28, 29. The gH/gL complex and gB only interact with each other in response to receptor binding by gD 30, 31 and this interaction is required for allowing fusion to occur 32. The tridimensional structure of gH/gL of HSV‐2 showed that the gH/gL heterodimer does not resemble any known viral fusogen, which may support different roles for gH including that of being a fusion regulator through its interaction with gB 33. A broad literature describes gH as involved in the entry process, for example, certain mutations in the transmembrane (TM) region and cytoplasmic tail affect fusion 34, 35, 36, 37, as do mutations in the region preceding the TM 38. Furthermore, peptides matching a number of regions of the gH ectodomain have been shown to interact with membranes and proposed to play a role in the fusion process, probably through regulation of gB activity and/or direct membrane interactions at the fusion site 39, 40, 41, 42, 43, 44, 45, 46, 47. In HSV‐infected cells and on mature virions, gH and gL are always found together as an heterodimer, in a stable 1 : 1 complex 33, 48 and rely on one another for proper folding, post‐translational processing and transport to the cell and virion surface 49, 50, 51, 52. HSV‐1 gH is a 838‐residue type 1 membrane glycoprotein. The ectodomain contains seven N‐glycosylation sites and eight cysteine residues forming at least two disulphide bonds between cysteines 5 and 6 (residue 554 and 589) and cysteines 7 and 8 (residues 652 and 706) 48, 53, 54; gL is a 224‐residue protein with a signal peptide but no TM region.
The crystal structure of the gH/gL complex has been solved 33 and shows an overall shape of a boot. gH can be divided in three distinct domains: (i) the N‐terminal domain, which binds gL (H1), (ii) the central helical domain (H2), and (iii) the C‐terminal β‐sandwich domain (H3).
The N‐terminal domain is located in the upper part of the gH–gL boot and consists of two subdomains, which are connected by a linker (residues Gly116‐Pro136); the first subdomain (residues Arg49‐Leu115) contains a β‐hairpin that forms a six‐stranded mixed β‐sheet with four strands coming from gL plus three short helices, while the second subdomain (residues Ala137‐Pro327) contains a six‐stranded antiparallel β‐sheet. It is interesting to note that the N‐terminal domain does not have a folded core and is the most divergent in sequence. The central domain (residues Asn332‐Phe644) is globular and mostly helical. The C‐terminal domain (residues Val645‐Pro797) is located at the toe end of the boot. It is a 10‐stranded β‐sandwich where each side is composed of a five‐stranded β‐sheet and leads to the TM region. The central and C‐terminal domains are more conserved and probably have the same fold in different gH proteins.
Similarly to the N‐terminal domain of gH, gL does not have a stable core. Only ~30% of gL residues adopt regular secondary structure, which include three helices and two β‐sheets.
Peptide‐based strategies have recently been used to study herpesvirus glycoprotein function. Several laboratories have analysed synthetic peptides homologous to regions from gC, gB and gH of different herpesviruses in order to inhibit infection 40, 44, 45, 55, 56, 57, 58, 59, 60, 61, 62.
We have previously shown that peptides modelled on gH domains with high interfacial hydrophobicity were able to inhibit HSV infectivity 45, 63, as well as peptides corresponding to helical domains 64. Recently, several studies have analysed viral glycoproteins with generalized peptide‐based approaches to dissect the whole glycoproteins in order to search for functionally important regions besides the predictable bioinformatic motifs. The mechanism that underlies the ability of some synthetic peptides to inhibit fusion is currently unclear, but some regions of these glycoproteins are exposed on the surface, and they may be involved in interactions with other proteins, or they may become exposed to the surface after the conformational rearrangements that probably take place during the fusion process; this could perhaps explain the ability of the corresponding synthetic peptides to inhibit cell fusion.
To date, few peptide molecules outside the well‐known inhibitory regions of Class 1 viral fusion proteins, the heptad repeats, should be as fusion; therefore, a brute force approach to the identification of peptide entry inhibitors may help in the dissection of HSV‐1 glycoproteins domains.
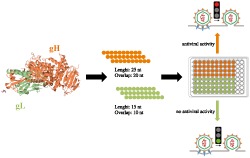
In the present study, we used a peptide scanning inhibition approach for the identification of functional domains and/or lead compounds as infectivity inhibitors. We generated overlapping peptide libraries of the complete sequences of gL and gH ectodomains. These peptides were screened for their ability to inhibit HSV‐1 infection in Vero cells at a concentration of 150 μM.
Materials and Methods
Peptide Synthesis
Peptides were prepared by standard 9‐fluorenylmethoxycarbonyl polyamine solid‐phase synthesis, using a PSSM8 multispecific peptide synthesizer (Shimadzu Corporation Biotechnology Instruments Department Kyoto JAPAN). The TGA resin (substitution 0.3 mmol g−1) was used as the solid‐phase support, and syntheses were performed on a scale of 100 μmol. All amino acids, 4 equiv. relative to resin loading, were coupled according to the TBTU/HOBT/DIEA method: 1 equiv. of Fmoc‐amino acid, 1 equiv. of TBTU, 1 equiv. of HOBT (1 M HOBT in DMF) and 2 equiv. of DIEA (2 M DIEA in DMF). The Fmoc protecting group was removed with 30% piperidine in DMF (v/v). Peptides were fully deprotected and cleaved from the resin by TFA solution (89% TFA, 5.5% thioanisole, 3.3% ethandithiol and 2.2% anisole as scavengers); the crude peptides were precipitated with ice‐cold ethyl ether, filtered, re‐dissolved in water and lyophilized. The crude peptides were purified to homogeneity by preparative reverse‐phase high‐pressure liquid chromatography on a Waters Delta Prep 3000 chromatographic system, equipped with an UV Lambda Max Mod. 481 detector. The samples were injected on a Jupiter (Phenomenex) C18 column (21.20 mm × 25 cm, 15 μm) eluted with a H2O/0.1% TFA (A) and CH3CN/0.1% TFA (B) solvent mixture. A linear gradient from 20 to 75% of B over 55 min at a flow rate of 20 ml min−1 was employed. The collected fractions were lyophilized to dryness and analysed by analytical reverse‐phase high‐pressure liquid chromatography on a Shimadzu class‐LC10 equipped with a diode array detector SPD‐M10AV using a Phenomenex C18 analytical column (10 × 250 mm, 10 μm); a linear gradient from 20 to 75% of B over 55 min at a flow rate of 1 ml min−1 was used. The identity of purified peptides was confirmed by Maldi spectrometry. All purified peptides were obtained with high yields (50–60%).
Cells and Viruses
African green monkey kidney cells (Vero) (ATCC CCL‐81) were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum. HSV‐1 (strain SC16), carrying a lacZ gene driven by the CMV IE‐1 promoter to express β‐galactosidase, was propagated on Vero cells monolayers.
Virus Entry Assays
Peptides were dissolved in DMEM without serum and used at final concentration of 150 μM. Experiments were conducted in parallel with scrambled peptides and no‐peptide controls.
To assess the effect of peptides on inhibition of HSV infectivity, a co‐treatment assay was performed in triplicate. The statistical analysis was carried out via t‐test and P‐value. The cells were incubated with peptides at 150 μM in the presence of serial dilutions of viral inoculum for 45 min at 37 °C.
After inactivation of nonpenetrated viruses by citrate buffer (pH 3.0), cell monolayers were incubated for 48 h at 37 °C in DMEM supplemented with carboxymethylcellulose. Finally, monolayers were fixed and stained with X‐gal (5‐bromo‐4‐chloro‐3‐indolyl‐β‐D‐galactopyranoside), and plaque numbers were scored. Experiments were performed in triplicate, and the percentage of inhibition was calculated with respect to no‐peptide control experiments.
The IC50 and IC90 values, peptide concentration that resulted in 50 and 90% of HSV infectivity reduction in a co‐treatment assay, respectively, were extrapolated via Graphpad Prism software from the resulting sigmoidal dose–response curve.
Toxicity
Peptide cytotoxicity was measured by a lactate dehydrogenase (LDH) assay 65 and was carried out according to manufacturer's instructions using a cytotoxicity detection kit (Roche Diagnostic SpA., Milano, Italy). Release of LDH from the cytosol to culture is a marker of cell death. The increase of LDH activity in the supernatant was related with the percentage of necrotic cells. Vero cells were cultured in a 96‐well plate at a density of 3 × 104 cells/well for 24 h, followed by treatment with investigated concentrations of each peptides for 24 h. Maximal LDH release was obtained after the treatment of control cells with 1% solution of Triton X‐100 (Sigma Chemical Company) for 10 min at room temperature. One hundred microlitres of supernatant from the top of all the wells was mixed with the prepared detection kit reagent. After 30‐min incubation, the absorbance was measured at 490 nm by Tecan spectrophotometer plate. For each experiment, at least three replicate wells were examined.
Results
To test the potential of using peptides as an antiviral agent and to identify functionally important regions of HSV‐1 membrane glycoproteins, two peptide libraries were constructed on the sequences of gH and gL ectodomains. Considering the different size of the two glycoproteins under study, we decided to prepare libraries with different overlapping lengths: a library of 25‐mer peptides extending from residues 21 to 805 (just at the beginning of the TM domain) of gH and a second library of 15‐mer peptides from residues 20 to 224 corresponding to the whole sequence of gL. The first (gH) library was made of peptides overlapping each other by 20 residues (Supplementary Table S1), while the latter was designed with a 10 residue overlap (Supplementary Table S2), and both signal peptides were excluded from our analysis. In detail, we excluded the first 25 aa from gH sequence (including the peptide signal 1–18 aa) and the first 19 aa from gL sequence. The libraries were screened for peptides with inhibitory activity by using a plaque reduction assay in which both virus and cells were exposed to the peptides prior to infection and the peptides remained present throughout the assay. The peptide efficacy was measured by a plaque reduction assay as described in the Materials and Methods. The criterion for considering a peptide of potential interest was the inhibition of at least 50% of plaque formation at a concentration of 150 μM. Figure 1 shows the gH‐overlapping peptides and their antiviral activities in the screening. Of the 148 peptides in the gH‐library, 21 peptides showed an inhibition activity more than 50% (Table 1), while the threshold was not reached by any of the gL peptides (Supplementary Table S2). The 21 gH peptides showing inhibitory activity corresponded to residues 346 to 375 (H65–H66), 381 to 405 (H72), 396 to 425 (H75–H76), 446 to 470 (H85), 456 to 485 (H87–H88), 491 to 520 (H94–H95), 506 to 530 (H97), 596 to 620 (H115), 606 to 630 (H117), 616 to 660 (H119–H123), 646 to 670 (H125), 746 to 775 (H145–H146). The locations of the inhibitory peptides on the gH crystal structure are shown in Figure 2, and we graphically subdivided their localization in four areas. It is interesting to note that some of the best inhibitors were located at the domain of interaction between gH (aa 259–323) and gL.
Figure 1.
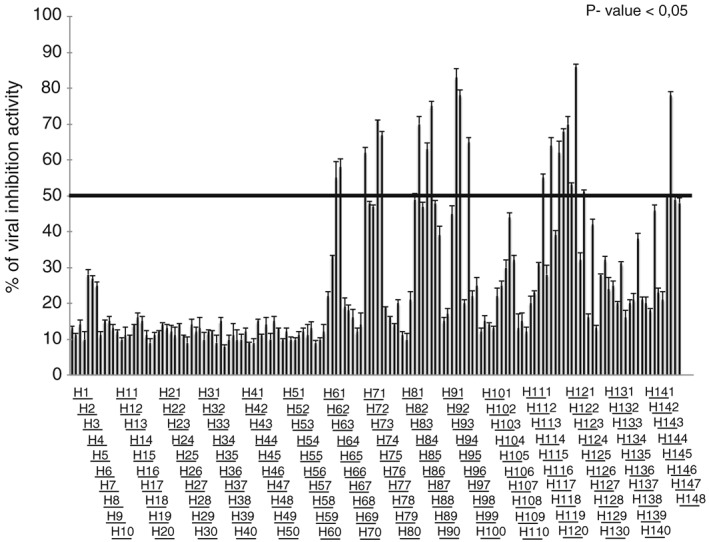
Inhibitory activity of gH peptides. The relative activity was normalized and reported as % of inhibition. The threshold lane was fixed at 50% of infection activity. The statistical analysis was carried out via P‐value < 0.05.
Table 1.
Active peptide details. Inhibitory gH peptides at 150 μM with the respective sequences, % of inhibition and protein position and the respective IC50 and IC90. The highest peptide concentration used is 250 μM; therefore, Graphpad Prism results above this concentration for IC90 calculations were reported as >250 μM
| P.I. | Sequences | % inhibition at 150 μM | Position | IC50 | IC90 |
|---|---|---|---|---|---|
| H65 | PEEGTNYAQFLSRAYAEFFSGDAGA | 55 | 325–350 | 145 ± 5 | >250 |
| H66 | NYAQFLSRAYAEFFSGDAGAEQGPR | 58 | 330–355 | 147 ± 3 | >250 |
| H75 | AAHANGAVCLSDLLGFLAHSRALAG | 67 | 375–400 | 118 ± 6 | >250 |
| H76 | GAVCLSDLLGFLAHSRALAGLAARG | 71 | 380–405 | 124 ± 4 | >250 |
| H87 | LVAEILEREQSLALHALGYQLAFVL | 63 | 435–460 | 120 ± 6 | >250 |
| H88 | LEREQSLALHALGYQLAFVLDSPSA | 75 | 440–465 | 116 ± 2 | >250 |
| H94 | PSAAHLIDALYAEFLGGRVLTTPVV | 83 | 470–495 | 85 ± 8 | 180 ± 7 |
| H95 | LIDALYAEFLGGRVLTTPVVHRALF | 78 | 475–500 | 104 ± 2 | 200 ± 11 |
| H97 | GGRVLTTPVVHRALFYASAVLRQPF | 65 | 485–510 | 138 ± 3 | >250 |
| H115 | DLDESVFILDALAQATRSETPVEVL | 55 | 575–600 | 146 ± 1 | >250 |
| H117 | ALAQATRSETPVEVLAQQTHGLAST | 64 | 485–610 | 121 ± 7 | >250 |
| H119 | PVEVLAQQTHGLASTLTRWAHYNAL | 62 | 595–620 | 124 ± 4 | >250 |
| H120 | AQQTHGLASTLTRWAHYNALIRAFV | 68 | 600–625 | 117 ± 9 | >250 |
| H121 | GLASTLTRWAHYNALIRAFVPEASH | 70 | 605–630 | 111 ± 2 | >250 |
| H122 | LTRWAHYNALIRAFVPEASHRCGGQ | 53 | 610–635 | 148 ± 3 | >250 |
| H123 | HYNALIRAFVPEASHRCGGQSANVE | 86 | 615–640 | 76 ± 5 | 165 ± 8 |
| H125 | PEASHRCGGQSANVEPRILVPITHN | 51 | 625–650 | 148 ± 6 | >250 |
| H145 | VLLVDTDNTQQQIAAGPTEGAPSVF | 50 | 725–750 | 150 ± 2 | >250 |
| H146 | TDNTQQQIAAGPTEGAPSVFSSDVP | 78 | 730–755 | 102 ± 7 | 225 ± 7 |
Figure 2.
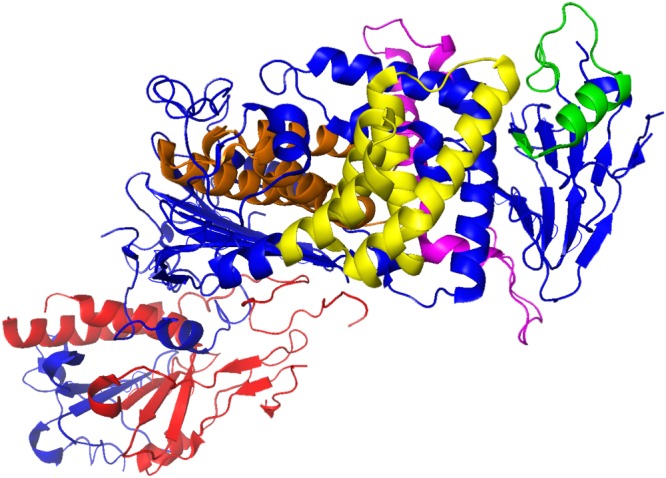
Localization of antiviral gH region in the heterodimer gH/gL. The ribbon structure of HSV‐1 gH/gL (PDB: 3M1C) and the locations of antiviral gH regions. In blue is reported gH structure, while in red gL one. Antiviral gH regions are indicated in orange, yellow, pink and green.
Previous works using a physico‐chemical algorithm, the Wimley–White Interfacial Hydrophobicity Scale (WWIHS), in combination with other structural data allowed us to predict regions in gH potentially involved in membrane interactions during the entry and fusion process, and some of them were found to possess HSV antiviral activity in dose‐dependent inhibition assays 66.
In the present study, we have extended these studies applying a more generalized peptide‐based approach to scan the entire gH ectodomain and its accompanying glycoprotein gL. The chosen threshold for including into the category of putative inhibitory peptides was dictated by the general high concentration needed for exerting an antiviral activity of HSV‐derived peptides. Previous works have shown that concentrations of peptide ranging from 10 to over 150 μM are needed to inhibit HSV infectivity. Nevertheless, the results obtained prove that peptide‐based strategies targeting regions of the proteins with predictable structures or motifs represent a valid option when searching functional regions of a protein; in fact, our whole protein scan confirmed as regions of interest several domains which were already found to posses some antiviral activity based on their structural characteristics. Peptides H85, H87 and H88, spanning the region of gH from 446 to 485, partially overlap the helical peptide previously named H‐HR1 (aa 444–479) able to reduce infectivity in a consistent manner, albeit at the high concentration of 250 μM 64, 67. Peptides H94, H95, H97 and H119, H120, H121, H122 and H123 overlap peptides gH493–537 and gH626–644, which were found able to inhibit HSV entry with approximately 60% of inhibition at 250 μM. Hydropathy analysis based on the hydrophobicity‐at‐interface scale proposed by Wimley and White 66 enabled the detection of these two domains of HSV gH able to induce rapid membrane fusion and inhibit viral entry. A shorter peptide gH493–512, corresponding only to the N‐terminus of gH493–537 and almost completely overlapping H94, H95 and H97 of the present work, was able to inhibit HSV entry even more effectively 45. Finally, peptides H145 and H146 at the extreme C‐terminus of the gH ectodomain (the TM helical domain starts at aa 804) have been previously individuated as being part of a larger pre‐TM domain with the ability of inhibiting virus infectivity, and a peptide previously named pTM6 44 exactly corresponds to peptide H146 of the present study. Nevertheless, the remaining peptides have been newly identified and presented an inhibitory activity above the set threshold. These are peptides H65–H66, H75–H76, H115, H117 and H125, which were found to exhibit inhibitory effects against HSV‐1 plaque formation (Table 1).
In order to define if chemical as well as conformational properties could represent common and shareable characteristics of active peptide, we performed bioinformatics analysis based on Pepcalc (http://www.pepcalc.com) and Pep‐Fold (http://bioserv.rpbs.univ-paris-diderot.fr/) databases, respectively (Figures 3 and 4). Chemical and physical properties highlight shared characteristic for some peptides. In detail, we reported in Figure 3 the 21 peptides subdivided for their isoelectric point grouped as acid (pH from 0 to 6), neutral (pH from 6 to 8) and basic (pH from 8 to 14). In second instance, the peptides were clustered based on their charge at pH 7. As shown in Figure 3, we found that some peptides are represented in specific clusters like H65, H145 and H146 for the first group as well as for H75, H95 and H123 in the second. Interestingly, most new inhibiting peptides are present in the acid (H65, H66, H115, H117) and basic (H125, H76) groups, while only one peptide (H75) is present in the neutral group. Moreover, we performed a primary and secondary structure analysis on the inhibiting gH peptides (Figure 4). Peptides clustered together by a sequence homology point of view (Figure 4A), and the clusters are directly associated with inhibition activity (Figure 4B). For instance, the cluster 5 had a percentage of viral inhibition close to 80%, while the clusters 3 and 6 just above 50%. Secondary structure bioinformatics peptide analysis is reported in Figure 4C, where we observed that in the cluster 5, a different distribution of helical and coil structure with low presence of extended regions is present compared to the others.
Figure 3.
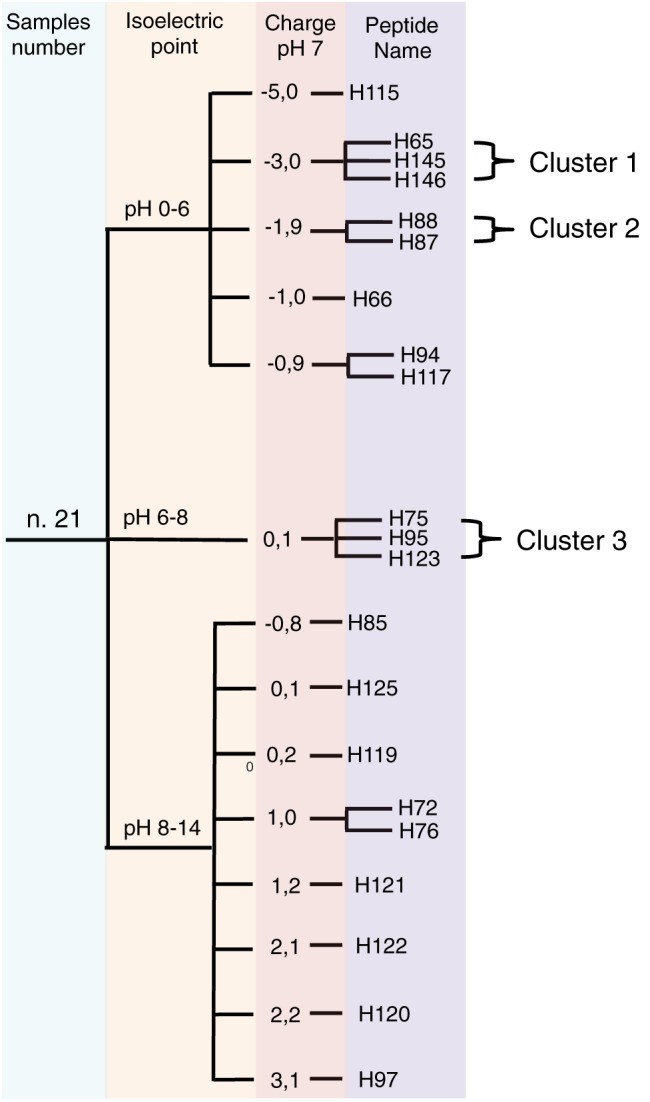
Chemical clusterization of gH active peptides. Tree clusterization of peptides based on isoelectic point and charge at pH 7.0.
Figure 4.
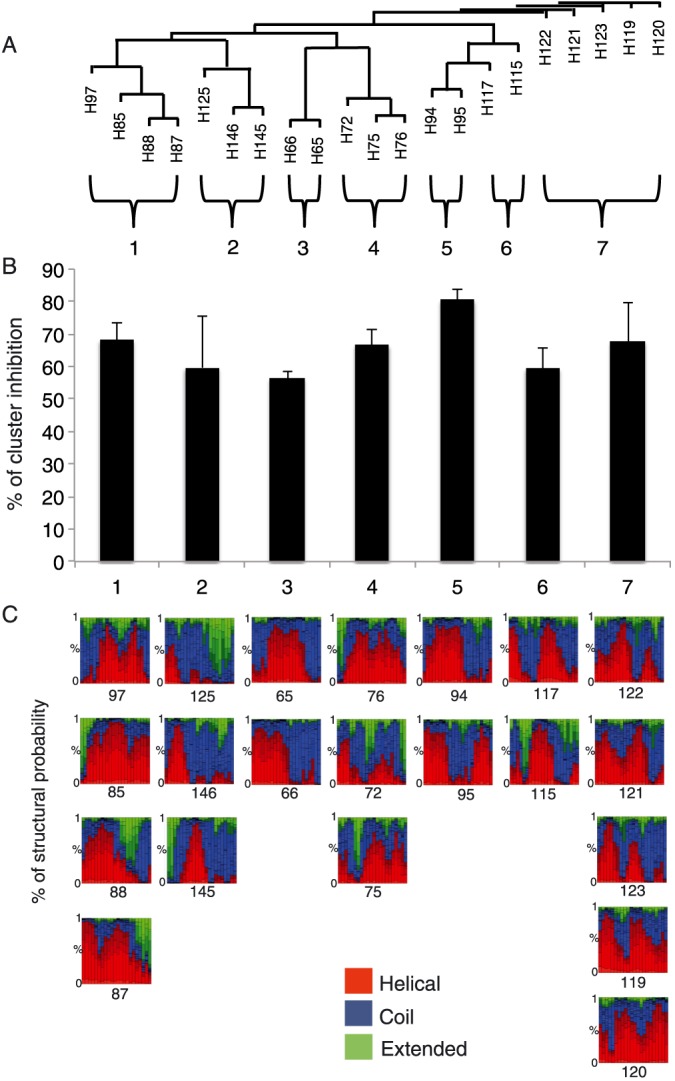
Sequence homology clusterization of gH active peptides A. Sequence homology tree based of active gH peptides. B. Inhibition activity recorded based on sequence homology clusterization. C Structural alphabet (SA) prediction profile. The probabilities, at each position of the sequence, of the 27 SA letters are sorted from helical (red), coil (blue) to extended (green). Note: SA letters are 4 residue length; therefore, the profile is of the size of the amino acid sequence minus 3.
Finally, to confirm that these peptides did not exert toxic effect on Vero cells, monolayers were exposed to a range of concentrations (10, 50, 150, 200 μM) of each inhibitory peptide for 24 h, and cell viability was assayed by an LDH assay. No statistical difference was observed between the viability of control (untreated) cells and that of cells exposed to the peptides (data not shown); thus, none of the peptides exhibited cytotoxic effects at the concentrations tested.
Discussion
Most of the viral peptide entry inhibitors described so far were intentionally designed in order to inhibit viral entry by competing with intermolecular interactions using amino acid specific sequences mostly derived from viral fusion proteins. However, an activity without a specific molecular target can be envisaged for many entry inhibitor peptides. These peptides, albeit being derived from viral surface proteins, do not belong to well‐known HR domains, but are often scattered in exposed regions of surface proteins or in domains that may only become exposed following conformational modifications triggered by binding of viruses to receptors or by environmental changes inside endosomes. The current understanding of the mechanisms of action of these peptides finds a hallmark in their propensity for membrane binding due to their interfacial hydrophobicity or amphipaticity 44, 45, 64, 68. In view of this characteristic, peptides could hamper virus entry by establishing direct physical interactions at hydrophobic interfaces on membranes or fusion proteins. Essentially, peptides can bind membranes, membrane proteins or virus particles, thereby exert a steric hindering effect on the early stages of viral penetration such as viral binding and/or membrane fusion. Sometimes, they can interfere with fusion protein modifications by premature initiation of the fusion proteins, which become unable to promote fusion once the peptides are bound to the viral envelope membrane. In this study, we have described an alternative approach for identifying functionally important regions of HSV‐1 gH and gL, showing for the first time the HSV‐1 inhibition activity of unreported peptides derived from gH protein. The approach involved the synthesis of two libraries of overlapping peptides that encompassed the entire ectodomains of gH and gL.
Brute‐force scanning approaches have proved to be of interest for analysing proteins of several viruses; in fact, potential antiviral peptides against hepatitis C virus (HCV) have been discovered by screening 441 overlapping peptides (18‐mers) covering the entire HCV polyprotein 69, 70. Eleven peptides with strong antiviral activity were identified. The most active inhibitory peptide, called C5A, showed significant hydrophobic and membrane binding propensity and was suggested to directly interact with the HCV viral envelope blocking infection when treating viruses during or prior to the initial entry stage. Interestingly, C5A was also able to inhibit HIV by disrupting the integrity of the viral envelope 71. Following this line, four proteins considered to play a role for promoting HCV entry (namely: CD81, scavenger receptor B1, claudin 1 and occluding) were analysed by an overlapping 18 amino acid peptide scanning strategy, and out of 113 peptides, two of them derived from claudin 1 showed potent inhibitory activity against HCV infectivity 72. Furthermore, HCV has also been recently analysed by screening a peptide library covering the full‐length E1 and E2 amino acids sequences where a peptide from the E2 stem domain was found to block cell entry of HCV peseudoparticles at nanomolar concentrations. This peptide is of sure potential interest because it proved to be efficient against the major HCV genotypes from 1 to 6 73. Another study assessed the activity of 95‐overlapping peptides (15‐mers) covering the entire VP1 capsid protein of Enterovirus 71, an important human pathogen which may cause severe neurological complications. In this case, the strategy proved to be also usable for searching potential peptide inhibitors in non‐enveloped viruses, with the discovery of four peptides able to inhibit EV‐71 infection by more than 80% 74. A further study described a 10‐mer overlapped peptide library synthesized by combinatorial approach where a peptide derived from the S1 subunit of the spike glycoprotein of the severe acute respiratory syndrome associated coronavirus (SARS‐CoV) could block the binding to the cellular receptor and, thus, the entry into target cells 75. Bai et al. 76 used a phage display methodology to identify a peptide (named P1) to inhibit West Nile virus (WNV) infectivity, possibly by binding to the envelope glycoprotein (E protein) necessary for membrane fusion. A modified version of P1, named P9, showed a strongly increased inhibitory activity against WNV at micrometre concentrations, and, more interestingly, entered the brain by crossing the blood brain barrier to reduce infectivity in a mouse model of brain infection. Moreover, an interesting study sparkling from the observation that HIV patients co‐infected with the common and asymptomatic GB virus C (a virus related to HCV) present lower HIV viremia suggested the analysis of overlapping peptides from the E2 protein from the GB virus C and allowed the identification of two peptides able to inhibit HIV in vitro by their competitive binding at the gp41–gp120 interface 77, 78. Also, HSV glycoproteins have been analysed by a peptide scanning strategy. A library of overlapping 15‐mer peptides encompassing the ectodomain of gB was synthesized and tested for HSV infectivity inhibition 79. Seven of the peptides inhibited infectivity by 50% or more when tested at 100‐μM concentrations.
Interestingly, many of the antiviral peptides identified by these brute force approaches overlap with many peptides discovered by analysing hydrophobicity at interface scales 68. The example of HSV gB is remarkable, because all the peptides (at least the most active) had also been previously described as membrane‐interacting 80 sequences using the WWIHS66. Our results, scanning the entire sequences of HSV‐1 gH and gL, confirm that the WWIHS is a powerful mean to identify potential antiviral peptides, but some regions of potential interest can remain underscored; therefore, a systematic analysis of the whole sequence by overlapping peptide libraries can add more detailed information on the regions involved in inhibition. In fact, we were able to detect four areas where peptides could be grouped for their antiviral activity. Some of the identified regions overlap with peptides already described which were discovered by bioinformatics tools, thereby strengthening this clear relationship between the function and the physiochemical character of peptides. In particular, the four identified inhibitory areas are mainly located in the regions of the glycoprotein named H2, while only a small area (S4) is located in the H3 region. The H2 domain of gH is mainly characterized by a bundle of helices and a few extended regions. In gH derived from Pseudorabies virus (PRV), a synthaxin like bundle (SLB) is present in this region 81, and this motif has proved to be of importance because its disruption can lead to impaired replication activity of the virus 82. Interestingly, one of our inhibitory peptides is located in the HSV‐1 gH corresponding region. Surprisingly, we did not detect any activity in inhibition assays when testing gL derived peptides. This may likely account for the negligible role of gL in membrane interactions. Nevertheless, also domain H1 of gH which is mainly devoted to interact with gL did not provide any inhibitory peptides. H1 sub‐domains clamp gL like tongs and make extensive contacts between the interacting highly complementary surfaces. In fact, the two proteins need each other to fold correctly and gL is a powerful scaffolding protein for gH. The inability of gH peptides derived from the H1 domain to function as inhibitors of infectivity can be explained by the fact that the formation of the highly stable complex between the two glycoproteins happens during the maturation and egress from the infected cell; therefore, the structure is already definitive when the heterodimer becomes expressed on the mature virion envelope. Disruption of the gH:gL interaction is not likely to happen because the whole H1 domain would result in an unfolded structure in absence of gL. The four areas of active peptides are depicted in Figure 5 where the filled surface of the protein is shown. It is of interest to note that inhibitory areas S2, S3 and S4 are mainly occupying regions of gH domains H2 and H3 and are exposed to the external surface of the protein, while only area S1 is partially buried inside and only partially exposed to the outside. Therefore, this observation is consistent with the minor inhibitory activity of peptides belonging to area S1. Furthermore, inhibitory area S4 precedes the C‐terminal TM domain of gH and is part of the C‐teminal H3 ectodomain mainly composed of β‐sheets which constitute a large patch of hydrophobic residues able to interact with the cell membrane during the fusion process or modifying the curvature of the viral envelope.
Figure 5.
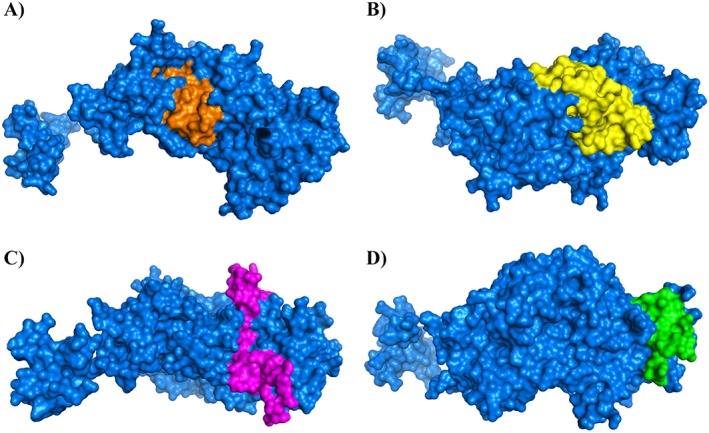
Filled surface representation of gH and the areas hosting inhibitory peptides. The target protein (gH) is rendered as a filled surface (light blue), and the locations of the four areas (from the N‐terminal to the C‐terminal of the protein) are shown. S1 is depicted in orange, S2 is in yellow, while S3 is in pink, and S4 is in green. gL is not shown, and the protein is rotated to show the best rendering for each of the inhibitory areas. Images generated from files from the PDB.
Membrane fusion is an important step in enveloped virus entry into host cells. The present study on the antiviral activity of HSV‐1 derived peptides provides a large set of data obtained with overlapping peptides covering the whole ectodomains of gH and gL. These peptides may be useful for probing gH activity during membrane fusion, but their major use could be as direct antiviral molecules. Acting on early stages, the discovered peptides can be used as microbicide for topical administration following further analysis and appropriate modifications for improving their viability and efficiency. As a matter of fact, considering their molecular weight, these peptides can be categorized as mid‐size drugs, which are, nowadays, receiving considerable attention in medicinal chemistry.
Supporting information
Table S1. gH peptide names with the respective sequences.
Table S2. gL peptide names with the respective sequences.
Franci, G. , Falanga, A. , Zannella, C. , Folliero, V. , Martora, F. , Galdiero, M. , Galdiero, S. , Morelli, G. , and Galdiero, M. (2017) Infectivity inhibition by overlapping synthetic peptides derived from the gH/gL heterodimer of herpes simplex virus type 1. J. Pept. Sci., 23: 311–319. doi: 10.1002/psc.2979.
References
- 1. Srivastava R, Dervillez X, Khan AA, Chentoufi AA, Chilukuri S, Shukr N, Fazli Y, Ong NN, Afifi RE, Osorio N, Geertsema R, Nesburn AB, Wechsler SL, BenMohamed L. The herpes simplex virus latency‐associated transcript gene is associated with a broader repertoire of virus‐specific exhausted CD8+ T cells retained within the trigeminal ganglia of latently infected HLA transgenic rabbits. J. Virol. 2016; 90: 3913–3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aranda AM, Epstein AL. Herpes simplex virus type 1 latency and reactivation: an update. Med. Sci. (Paris) 2015; 31: 506–514. [DOI] [PubMed] [Google Scholar]
- 3. Piret J, Boivin G. Resistance of herpes simplex viruses to nucleoside analogues: mechanisms, prevalence, and management. Antimicrob. Agents Chemother. 2011; 55: 459–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Matthews T, Salgo M, Greenberg M, Chung J, DeMasi R, Bolognesi D. Enfuvirtide: the first therapy to inhibit the entry of HIV‐1 into host CD4 lymphocytes. Nat. Rev. Drug Discov. 2004; 3: 215–225. [DOI] [PubMed] [Google Scholar]
- 5. Kilby JM, Hopkins S, Venetta TM, DiMassimo B, Cloud GA, Lee JY, Alldredge L, Hunter E, Lambert D, Bolognesi D, Matthews T, Johnson MR, Nowak MA, Shaw GM, Saag MS. Potent suppression of HIV‐1 replication in humans by T‐20, a peptide inhibitor of gp41‐mediated virus entry. Nat. Med. 1998; 4: 1302–1307. [DOI] [PubMed] [Google Scholar]
- 6. Brauer F, Schmidt K, Zahn RC, Richter C, Radeke HH, Schmitz JE, von Laer D, Egerer L. A rationally engineered anti‐HIV peptide fusion inhibitor with greatly reduced immunogenicity. Antimicrob. Agents Chemother. 2013; 57: 679–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kaushik‐Basu N, Basu A, Harris D. Peptide inhibition of HIV‐1: current status and future potential. BioDrugs 2008; 22: 161–175. [DOI] [PubMed] [Google Scholar]
- 8. Lok SM, Costin JM, Hrobowski YM, Hoffmann AR, Rowe DK, Kukkaro P, Holdaway H, Chipman P, Fontaine KA, Holbrook MR, Garry RF, Kostyuchenko V, Wimley WC, Isern S, Rossmann MG, Michael SF. Release of dengue virus genome induced by a peptide inhibitor. PLoS One 2012; 7: .e50995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schmidt AG, Yang PL, Harrison SC. Peptide inhibitors of dengue‐virus entry target a late‐stage fusion intermediate. PLoS Pathog. 2010; 6: .e1000851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Porotto M, Doctor L, Carta P, Fornabaio M, Greengard O, Kellogg GE, Moscona A. Inhibition of hendra virus fusion. J. Virol. 2006; 80: 9837–9849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yi HA, Fochtman BC, Rizzo RC, Jacobs A. Inhibition of HIV entry by targeting the envelope transmembrane subunit gp41. Curr. HIV Res. 2016; 14: 283–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Krepstakies M, Lucifora J, Nagel CH, Zeisel MB, Holstermann B, Hohenberg H, Kowalski I, Gutsmann T, Baumert TF, Brandenburg K, Hauber J, Protzer U. A new class of synthetic peptide inhibitors blocks attachment and entry of human pathogenic viruses. J. Infect. Dis. 2012; 205: 1654–1664. [DOI] [PubMed] [Google Scholar]
- 13. Skalickova S, Heger Z, Krejcova L, Pekarik V, Bastl K, Janda J, Kostolansky F, Vareckova E, Zitka O, Adam V, Kizek R. Perspective use of antiviral peptides against influenza virus. Viruses 2015; 7: 5428–5442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shukla D, Spear PG. Herpesviruses and heparan sulfate: an intimate relationship in aid of viral entry. J. Clin. Invest. 2001; 108: 503–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Spear PG, Eisenberg RJ, Cohen GH. Three classes of cell surface receptors for alphaherpesvirus entry. Virology 2000; 275: 1–8. [DOI] [PubMed] [Google Scholar]
- 16. Herold BC, Visalli RJ, Susmarski N, Brandt CR, Spear PG. Glycoprotein C‐independent binding of herpes simplex virus to cells requires cell surface heparan sulphate and glycoprotein B. J. Gen. Virol. 1994; 75(Pt 6): 1211–1222. [DOI] [PubMed] [Google Scholar]
- 17. Cocchi F, Fusco D, Menotti L, Gianni T, Eisenberg RJ, Cohen GH, Campadelli‐Fiume G. The soluble ectodomain of herpes simplex virus gD contains a membrane‐proximal pro‐fusion domain and suffices to mediate virus entry. Proc. Natl. Acad. Sci. U. S. A. 2004; 101: 7445–7450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Geraghty RJ, Krummenacher C, Cohen GH, Eisenberg RJ, Spear PG. Entry of alphaherpesviruses mediated by poliovirus receptor‐related protein 1 and poliovirus receptor. Science 1998; 280: 1618–1620. [DOI] [PubMed] [Google Scholar]
- 19. Montgomery RI, Warner MS, Lum BJ, Spear PG. Herpes simplex virus‐1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell 1996; 87: 427–436. [DOI] [PubMed] [Google Scholar]
- 20. Krummenacher C, Supekar VM, Whitbeck JC, Lazear E, Connolly SA, Eisenberg RJ, Cohen GH, Wiley DC, Carfi A. Structure of unliganded HSV gD reveals a mechanism for receptor‐mediated activation of virus entry. EMBO J. 2005; 24: 4144–4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Milne RS, Hanna SL, Rux AH, Willis SH, Cohen GH, Eisenberg RJ. Function of herpes simplex virus type 1 gD mutants with different receptor‐binding affinities in virus entry and fusion. J. Virol. 2003; 77: 8962–8972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Atanasiu D, Cairns TM, Whitbeck JC, Saw WT, Rao S, Eisenberg RJ, Cohen GH. Regulation of herpes simplex virus gB‐induced cell–cell fusion by mutant forms of gH/gL in the absence of gD and cellular receptors. MBio 2013; 4: e00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Turner A, Bruun B, Minson T, Browne H. Glycoproteins gB, gD, and gHgL of herpes simplex virus type 1 are necessary and sufficient to mediate membrane fusion in a Cos cell transfection system. J. Virol. 1998; 72: 873–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Muggeridge MI. Characterization of cell–cell fusion mediated by herpes simplex virus 2 glycoproteins gB, gD, gH and gL in transfected cells. J. Gen. Virol. 2000; 81: 2017–2027. [DOI] [PubMed] [Google Scholar]
- 25. Pertel PE, Fridberg A, Parish ML, Spear PG. Cell fusion induced by herpes simplex virus glycoproteins gB, gD, and gH‐gL requires a gD receptor but not necessarily heparan sulfate. Virology 2001; 279: 313–324. [DOI] [PubMed] [Google Scholar]
- 26. Fan Q, Longnecker R, Connolly SA. Substitution of herpes simplex virus 1 entry glycoproteins with those of saimiriine herpesvirus 1 reveals a gD‐gH/gL functional interaction and a region within the gD profusion domain that is critical for fusion. J. Virol. 2014; 88: 6470–6482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. White JM, Delos SE, Brecher M, Schornberg K. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit. Rev. Biochem. Mol. Biol. 2008; 43: 189–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Connolly SA, Jackson JO, Jardetzky TS, Longnecker R. Fusing structure and function: a structural view of the herpesvirus entry machinery. Nat. Rev. Microbiol. 2011; 9: 369–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Eisenberg RJ, Atanasiu D, Cairns TM, Gallagher JR, Krummenacher C, Cohen GH. Herpes virus fusion and entry: a story with many characters. Viruses 2012; 4: 800–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Atanasiu D, Whitbeck JC, Cairns TM, Reilly B, Cohen GH, Eisenberg RJ. Bimolecular complementation reveals that glycoproteins gB and gH/gL of herpes simplex virus interact with each other during cell fusion. Proc. Natl. Acad. Sci. U. S. A. 2007; 104: 18718–18723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fan Q, Longnecker R, Connolly SA. A functional interaction between herpes simplex virus 1 glycoprotein gH/gL domains I and II and gD is defined by using alphaherpesvirus gH and gL chimeras. J. Virol. 2015; 89: 7159–7169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Atanasiu D, Whitbeck JC, de Leon MP, Lou H, Hannah BP, Cohen GH, Eisenberg RJ. Bimolecular complementation defines functional regions of Herpes simplex virus gB that are involved with gH/gL as a necessary step leading to cell fusion. J. Virol. 2010; 84: 3825–3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chowdary TK, Cairns TM, Atanasiu D, Cohen GH, Eisenberg RJ, Heldwein EE. Crystal structure of the conserved herpesvirus fusion regulator complex gH‐gL. Nat. Struct. Mol. Biol. 2010; 17: 882–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Browne H, Bell S, Minson T, Wilson DW. An endoplasmic reticulum‐retained herpes simplex virus glycoprotein H is absent from secreted virions: evidence for reenvelopment during egress. J. Virol. 1996; 70: 4311–4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Harman A, Browne H, Minson T. The transmembrane domain and cytoplasmic tail of herpes simplex virus type 1 glycoprotein H play a role in membrane fusion. J. Virol. 2002; 76: 10708–10716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wilson DW, Davis‐Poynter N, Minson AC. Mutations in the cytoplasmic tail of herpes simplex virus glycoprotein H suppress cell fusion by a syncytial strain. J. Virol. 1994; 68: 6985–6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kharkwal H, Furgiuele SS, Smith CG, Wilson DW. Herpes simplex virus capsid–organelle association in the absence of the large tegument protein UL36p. J. Virol. 2015; 89: 11372–11382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Galdiero M, Whiteley A, Bruun B, Bell S, Minson T, Browne H. Site‐directed and linker insertion mutagenesis of herpes simplex virus type 1 glycoprotein H. J. Virol. 1997; 71: 2163–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Galdiero S, Falanga A, Vitiello M, Browne H, Pedone C, Galdiero M. Fusogenic domains in herpes simplex virus type 1 glycoprotein H. J. Biol. Chem. 2005; 280: 28632–28643. [DOI] [PubMed] [Google Scholar]
- 40. Gianni T, Martelli PL, Casadio R, Campadelli‐Fiume G. The ectodomain of herpes simplex virus glycoprotein H contains a membrane alpha‐helix with attributes of an internal fusion peptide, positionally conserved in the herpesviridae family. J. Virol. 2005; 79: 2931–2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Galdiero S, Russo L, Falanga A, Cantisani M, Vitiello M, Fattorusso R, Malgieri G, Galdiero M, Isernia C. Structure and orientation of the gH625‐644 membrane interacting region of herpes simplex virus type 1 in a membrane mimetic system. Biochemistry 2012; 51: 3121–3128. [DOI] [PubMed] [Google Scholar]
- 42. Galdiero S, Falanga A, Vitiello M, Raiola L, Russo L, Pedone C, Isernia C, Galdiero M. The presence of a single N‐terminal histidine residue enhances the fusogenic properties of a Membranotropic peptide derived from herpes simplex virus type 1 glycoprotein H. J. Biol. Chem. 2010; 285: 17123–17136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Galdiero S, Falanga A, Vitiello M, Raiola L, Fattorusso R, Browne H, Pedone C, Isernia C, Galdiero M. Analysis of a membrane interacting region of herpes simplex virus type 1 glycoprotein H. J. Biol. Chem. 2008; 283: 29993–30009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Galdiero S, Falanga A, Vitiello M, D'Isanto M, Collins C, Orrei V, Browne H, Pedone C, Galdiero M. Evidence for a role of the membrane‐proximal region of herpes simplex virus Type 1 glycoprotein H in membrane fusion and virus inhibition. Chembiochem 2007; 8: 885–895. [DOI] [PubMed] [Google Scholar]
- 45. Galdiero S, Falanga A, Vitiello M, D'Isanto M, Cantisani M, Kampanaraki A, Benedetti E, Browne H, Galdiero M. Peptides containing membrane‐interacting motifs inhibit herpes simplex virus type 1 infectivity. Peptides 2008; 29: 1461–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Galdiero S, Falanga A, Vitiello G, Vitiello M, Pedone C, D'Errico G, Galdiero M. Role of membranotropic sequences from herpes simplex virus type I glycoproteins B and H in the fusion process. Biochim. Biophys. Acta 2010; 1798: 579–591. [DOI] [PubMed] [Google Scholar]
- 47. Falanga A, Vitiello MT, Cantisani M, Tarallo R, Guarnieri D, Mignogna E, Netti P, Pedone C, Galdiero M, Galdiero S. A peptide derived from herpes simplex virus type 1 glycoprotein H: membrane translocation and applications to the delivery of quantum dots. Nanomedicine 2011; 7: 925–934. [DOI] [PubMed] [Google Scholar]
- 48. Peng T, Ponce de Leon M, Novotny MJ, Jiang H, Lambris JD, Dubin G, Spear PG, Cohen GH, Eisenberg RJ. Structural and antigenic analysis of a truncated form of the herpes simplex virus glycoprotein gH–gL complex. J. Virol. 1998; 72: 6092–6103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cairns TM, Friedman LS, Lou H, Whitbeck JC, Shaner MS, Cohen GH, Eisenberg RJ. N‐terminal mutants of herpes simplex virus type 2 gH are transported without gL but require gL for function. J. Virol. 2007; 81: 5102–5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hutchinson L, Browne H, Wargent V, Davis‐Poynter N, Primorac S, Goldsmith K, Minson AC, Johnson DC. A novel herpes simplex virus glycoprotein, gL, forms a complex with glycoprotein H (gH) and affects normal folding and surface expression of gH. J. Virol. 1992; 66: 2240–2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Roop C, Hutchinson L, Johnson DC. A mutant herpes simplex virus type 1 unable to express glycoprotein L cannot enter cells, and its particles lack glycoprotein H. J. Virol. 1993; 67: 2285–2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rogalin HB, Heldwein EE. Interplay between the herpes simplex virus 1 gB cytodomain and the gH cytotail during cell–cell fusion. J. Virol. 2015; 89: 12262–12272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cairns TM, Landsburg DJ, Whitbeck JC, Eisenberg RJ, Cohen GH. Contribution of cysteine residues to the structure and function of herpes simplex virus gH/gL. Virology 2005; 332: 550–562. [DOI] [PubMed] [Google Scholar]
- 54. Stampfer SD, Heldwein EE. Stuck in the middle: structural insights into the role of the gH/gL heterodimer in herpesvirus entry. Curr. Opin. Virol. 2013; 3: 13–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gianni T, Fato R, Bergamini C, Lenaz G, Campadelli‐Fiume G. Hydrophobic alpha‐helices 1 and 2 of herpes simplex virus gH interact with lipids, and their mimetic peptides enhance virus infection and fusion. J. Virol. 2006; 80: 8190–8198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lopper M, Compton T. Coiled‐coil domains in glycoproteins B and H are involved in human cytomegalovirus membrane fusion. J. Virol. 2004; 78: 8333–8341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Okazaki K, Kida H. A synthetic peptide from a heptad repeat region of herpesvirus glycoprotein B inhibits virus replication. J. Gen. Virol. 2004; 85: 2131–2137. [DOI] [PubMed] [Google Scholar]
- 58. Vitiello M, Galdiero M, Galdiero M. Inhibition of viral‐induced membrane fusion by peptides. Protein Pept. Lett. 2009; 16: 786–793. [DOI] [PubMed] [Google Scholar]
- 59. Galdiero S, Falanga A, Tarallo R, Russo L, Galdiero E, Cantisani M, Morelli G, Galdiero M. Peptide inhibitors against herpes simplex virus infections. J. Pept. Sci. 2013; 19: 148–158. [DOI] [PubMed] [Google Scholar]
- 60. Trybala E, Olofsson S, Mardberg K, Svennerholm B, Umemoto K, Glorioso JC, Bergstrom T. Structural and functional features of the polycationic peptide required for inhibition of herpes simplex virus invasion of cells. Antivir. Res. 2004; 62: 125–134. [DOI] [PubMed] [Google Scholar]
- 61. Wang X, Chi X, Wang M. Structural characteristics and antiviral activity of multiple peptides derived from MDV glycoproteins B and H. Virol. J. 2011; 8: 190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Agelidis AM, Shukla D. Cell entry mechanisms of HSV: what we have learned in recent years. Future Virol 2015; 10: 1145–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Xu Y, Yu S, Zou JW, Hu G, Rahman NA, Othman RB, Tao X, Huang M. Identification of peptide inhibitors of enveloped viruses using support vector machine. PLoS One 2015; 10: .e0144171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Galdiero S, Vitiello M, D'Isanto M, Falanga A, Collins C, Raieta K, Pedone C, Browne H, Galdiero M. Analysis of synthetic peptides from heptad‐repeat domains of herpes simplex virus type 1 glycoproteins H and B. J. Gen. Virol. 2006; 87: 1085–1097. [DOI] [PubMed] [Google Scholar]
- 65. Vitiello M, D'Isanto M, Finamore E, Ciarcia R, Kampanaraki A, Galdiero M. Role of mitogen‐activated protein kinases in the iNOS production and cytokine secretion by Salmonella enterica serovar Typhimurium porins. Cytokine 2008; 41: 279–285. [DOI] [PubMed] [Google Scholar]
- 66. Wimley WC, White SH. Experimentally determined hydrophobicity scale for proteins at membrane interfaces. Nat. Struct. Biol. 1996; 3: 842–848. [DOI] [PubMed] [Google Scholar]
- 67. Gianni T, Menotti L, Campadelli‐Fiume G. A heptad repeat in herpes simplex virus 1 gH, located downstream of the alpha‐helix with attributes of a fusion peptide, is critical for virus entry and fusion. J. Virol. 2005; 79: 7042–7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Badani H, Garry RF, Wimley WC. Peptide entry inhibitors of enveloped viruses: the importance of interfacial hydrophobicity. Biochim. Biophys. Acta 1838; 2014: 2180–2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Cheng G, Montero A, Gastaminza P, Whitten‐Bauer C, Wieland SF, Isogawa M, Fredericksen B, Selvarajah S, Gallay PA, Ghadiri MR, Chisari FV. A virocidal amphipathic {alpha}‐helical peptide that inhibits hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. U. S. A. 2008; 105: 3088–3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. El‐Shenawy R, Tabll A, Bader El Din NG, El Abd Y, Mashaly M, Abdel Malak CA, Dawood R, El‐Awady M. Antiviral activity of virocidal peptide derived from NS5A against two different HCV genotypes: an in vitro study. J. Immunoassay Immunochem. 2015; 36: 63–79. [DOI] [PubMed] [Google Scholar]
- 71. Li GR, He LY, Liu XY, Liu AP, Huang YB, Qiu C, Zhang XY, Xu JQ, Yang W, Chen YX. Rational design of peptides with anti‐HCV/HIV activities and enhanced specificity. Chem. Biol. Drug Des. 2011; 78: 835–843. [DOI] [PubMed] [Google Scholar]
- 72. Si Y, Liu S, Liu X, Jacobs JL, Cheng M, Niu Y, Jin Q, Wang T, Yang W. A human claudin‐1‐derived peptide inhibits hepatitis C virus entry. Hepatology 2012; 56: 507–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Chi X, Niu Y, Cheng M, Liu X, Feng Y, Zheng F, Fan J, Li X, Jin Q, Zhong J, Li YP, Yang W. Identification of a potent and broad‐spectrum hepatitis C virus fusion inhibitory peptide from the E2 stem domain. Sci. Rep. 2016; 6: 25224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Tan CW, Chan YF, Sim KM, Tan EL, Poh CL. Inhibition of enterovirus 71 (EV‐71) infections by a novel antiviral peptide derived from EV‐71 capsid protein VP1. PLoS One 2012; 7: .e34589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Hu H, Li L, Kao RY, Kou B, Wang Z, Zhang L, Zhang H, Hao Z, Tsui WH, Ni A, Cui L, Fan B, Guo F, Rao S, Jiang C, Li Q, Sun M, He W, Liu G. Screening and identification of linear B‐cell epitopes and entry‐blocking peptide of severe acute respiratory syndrome (SARS)‐associated coronavirus using synthetic overlapping peptide library. J. Comb. Chem. 2005; 7: 648–656. [DOI] [PubMed] [Google Scholar]
- 76. Bai F, Town T, Pradhan D, Cox J, Ashish, Ledizet M, Anderson JF, Flavell RA, Krueger JK, Koski RA, Fikrig E. Antiviral peptides targeting the west nile virus envelope protein. J. Virol. 2007; 81: 2047–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Koedel Y, Eissmann K, Wend H, Fleckenstein B, Reil H. Peptides derived from a distinct region of GB virus C glycoprotein E2 mediate strain‐specific HIV‐1 entry inhibition. J. Virol. 2011; 85: 7037–7047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Eissmann K, Mueller S, Sticht H, Jung S, Zou P, Jiang S, Gross A, Eichler J, Fleckenstein B, Reil H. HIV‐1 fusion is blocked through binding of GB Virus C E2‐derived peptides to the HIV‐1 gp41 disulfide loop [corrected]. PLoS One 2013; 8: .e54452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Akkarawongsa R, Pocaro NE, Case G, Kolb AW, Brandt CR. Multiple peptides homologous to herpes simplex virus type 1 glycoprotein B inhibit viral infection. Antimicrob. Agents Chemother. 2009; 53: 987–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Galdiero S, Vitiello M, D'Isanto M, Falanga A, Cantisani M, Browne H, Pedone C, Galdiero M. The identification and characterization of fusogenic domains in herpes virus glycoprotein B molecules. Chembiochem 2008; 9: 758–767. [DOI] [PubMed] [Google Scholar]
- 81. Matsuura K, Himuro C, Yokoi T, Yamamoto Y, Vargo EL, Keller L. Identification of a pheromone regulating caste differentiation in termites. Proc. Natl. Acad. Sci. U. S. A. 2010; 107: 12963–12968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Bohm SW, Backovic M, Klupp BG, Rey FA, Mettenleiter TC, Fuchs W. A replication defect of pseudorabies virus induced by targeted alpha‐helix distortion in the syntaxin‐like bundle of glycoprotein H (V275P) is corrected by an adjacent compensatory mutation (V271A). J. Gen. Virol. 2015; 96: 2349–2354. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. gH peptide names with the respective sequences.
Table S2. gL peptide names with the respective sequences.


