Abstract
The intrinsic antibiotic resistance of Stenotrophomonas maltophilia, along with its ability to form biofilm both on abiotic surfaces and host tissues, dramatically affects the efficacy of the antibiotic therapy. In this work, 85 S. maltophilia strains isolated in several hospital of central Italy and from several clinical settings were evaluated for their genetic relatedness (by pulsed-field gel electrophoresis, PFGE), biofilm formation (by microtiter plate assay), and planktonic antibiotic resistance (by Kirby–Bauer disk diffusion technique). The S. maltophilia population showed a high genetic heterogeneity: 64 different PFGE types were identified, equally distributed in cystic fibrosis (CF) and non-CF strains, and some consisted of multiple strains. Most of the strains (88.2%) were able to form biofilm, although non-CF strains were significantly more efficient than CF strains. CF strains produced lower biofilm amounts than non-CF strains, both those from respiratory tracts and blood. Non-CF PFGE types 3 and 27 consisted of strong-producers only. Cotrimoxazole and levofloxacin were the most effective antibiotics, being active respectively against 81.2% and 72.9% of strains. CF strains were significantly more resistant to piperacillin/tazobactam compared to non-CF strains (90% versus 53.3%), regardless of sample type. Among respiratory strains, cotrimoxazole was more active against non-CF than CF strains (susceptibility rates: 86.7% versus 75%). The multidrug resistant phenotype was significantly more prevalent in CF than non-CF strains (90% versus 66.7%). Overall, the multidrug-resistance level was negatively associated with efficiency in biofilm formation. Our results showed, for the first time, that in S. maltophilia both classical planktonic drug resistance and the ability of biofilm formation might favor its dissemination in the hospital setting. Biofilm formation might in fact act as a survival mechanism for susceptible bacteria, suggesting that clinical isolates should be routinely assayed for biofilm formation in diagnostic laboratories.
Keywords: Stenotrophomonas maltophilia, biofilm formation, antibiotic resistance
1. Introduction
Among the “emerging” pathogens recognized in the recent years, Stenotrophomonas maltophilia plays a significant role in colonization and infection in hospital, and less often, community settings. This opportunistic pathogen has, in fact, been implicated in a variety of nosocomial infections, especially in intensive care unit patients (such as ventilator-associated pneumonia and sepsis), life-threatening diseases in immunocompromised patients with hematological malignancies and cancers, and respiratory tract infections in patients with chronic lung diseases [1,2].
S. maltophilia commonly causes pneumonia, bacteremia, sepsis, and wound infections, and less commonly, urinary tract infections, endocarditis, soft tissue infections, meningitis, osteochondritis, peritonitis, and ophthalmic infections [1,2]. Although it is not considered a highly virulent pathogen, S. maltophilia has been associated with high crude mortality, ranging from 25% to 75% in the case of pneumonia and from 14% to 69% in the case of bacteremia [2].
Although Pseudomonas aeruginosa is the most prevalent pathogen in cystic fibrosis (CF) patients, S. maltophilia is being increasingly isolated from CF airways, due to its ability to evade many antipseudomonal antibiotics [3,4,5,6]. In this clinical setting, the microorganism can account for perseverant colonization and chronic infection, although the clinical relevance in these patients is yet unclear. In fact, despite some studies have defined this microorganism as a colonizer, others demonstrated that its presence should not be ignored in some CF patients as S. maltophilia is associated with an increased risk of pulmonary exacerbations, the deterioration in pulmonary function, the need for lung transplantation, and death [3,4,5,6].
The biofilm-forming ability of S. maltophilia has increasingly been accepted as an important virulence trait. This microorganism can form biofilms both on abiotic surfaces and host tissues, dramatically enhancing the resistance to therapeutically important antibiotics, including aminoglycosides, fluoroquinolones, and tetracycline [7,8,9,10]. Therefore, biofilm formation might play a relevant role in the persistence of S. maltophilia infection in hospital settings, especially in CF patients where it complicates the therapeutic management of bronchial colonization- and infection [11,12,13].
However, biofilm formation is not the only reason for antimicrobial treatment failure. In fact, a distinctive feature of S. maltophilia strains is their resistance to a wide range of antibiotics, which makes these infections difficult to treat [14,15]. Nonetheless, contrarily to P. aeruginosa, treatment protocols are not yet standardized for S. maltophilia, and limited data are available about susceptibility profiles, highlighting the importance for studying the susceptibility patterns of this microorganism.
The connection between biofilm formation and planktonic antibiotic resistance is of considerable interest to biomedical researchers, although studies performed in this regard over the past two decades have yielded conflicting results, and therefore, inconsistent information [16,17,18,19,20,21]. Particularly, although some studies found a positive relationship between biofilm formation and antibiotic resistance in Staphylococcus aureus [16], Acinetobacter baumannii [18], and Staphylococcus epidermidis [19], in others an opposite trend was observed for A. baumannii [20,21].
To the best of our knowledge, a rigorous investigation in this regard has not been carried out for S. maltophilia. Therefore, in this work, we assessed the antimicrobial resistance of planktonic cells, the efficiency of biofilm formation, and the clonal relatedness in S. maltophilia. Such evaluations were extended to a larger collection of strains isolated from several clinical samples, both from CF and non-CF patients, to evaluate a possible relationship among biofilm formation ability, clinical source, and antibiotic resistance pattern. With this aim, 85 S. maltophilia strains were evaluated for their genetic relatedness by pulsed-field gel electrophoresis, antibiotic resistance by Kirby–Bauer disk diffusion technique, and capability for biofilm development in 96-well microtiter plates.
Overall, our findings highlighted that: (i) S. maltophilia enhances its persistence—both environmental and into the host—by a smart balance between antibiotic resistance and biofilm formation; and (ii) biofilm formation ability should be evaluated along with antimicrobial susceptibility testing to improve the efficacy of the treatment against biofilm-related infections.
2. Results
2.1. The S. maltophilia Population Shows a High Genetic Heterogeneity
The genetic diversity and the clonal relatedness of the tested strains were assessed by PFGE analysis, and results are shown in Table 1.
Table 1.
Clonal relatedness, antibiotic-resistance, and biofilm formation of 85 S. maltophilia strains tested in the present study. Strains were genotyped by PFGE analysis. Susceptibility tests were performed using Kirby–Bauer disk diffusion agar (SXT, cotrimoxazole; LVX, levofloxacin; CPX, ciprofloxacin; TZP: Piperacillin/tazobactam; MER, meropenem) or broth microdilution technique (CHL, chloramphenicol) and interpreted according to CLSI guidelines [22]. Biofilm formation was assessed by spectrophotometric assay after crystal violet assay and the results were categorized according to Stepanovic et al. [23].
| Strain ID | Isolation Site a | PFGE Type | Susceptibility (S) or Resistance (R) to the Following Antibiotics: | Resistance Phenotype b | Biofilm Formation | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SXT | CHL | LVX | CPX | TZP | MER | Mean Value (OD492) | Class c | ||||
| SM31 | sputum | 1.1 | S | R | S | S | R | R | MDR | <0.096 | NP |
| SM10 | sputum | 2.1 | S | R | S | R | R | R | XDR | 0.633 | S |
| SM172 | sputum | 3.1 | S | R | S | R | R | R | XDR | 0.432 | S |
| SM176 | sputum | 3.1 | S | R | S | R | S | R | MDR | 0.615 | S |
| SM186 | blood | 3.1 | S | S | S | S | S | S | non-MDR | 0.575 | S |
| SM185 | blood | 3.1 | S | S | R | R | R | R | MDR | 2.282 | S |
| SM143 | sputum | 3.1 | S | R | S | R | R | R | XDR | 2.750 | S |
| SM27 | sputum | 3.1 | S | R | S | R | R | R | XDR | 0.455 | S |
| SM173 | sputum | 4.1 | S | S | S | R | R | R | MDR | 0.574 | S |
| SM139 | CF | 5.1 | S | R | S | R | S | R | MDR | 1.269 | S |
| SM115 | CF | 6.1 | S | S | S | R | R | R | MDR | 0.329 | M |
| SM119 | CF | 7.1 | S | R | S | R | R | R | XDR | 0.236 | M |
| SM108 | CF | 8.1 | R | S | S | R | R | R | XDR | 0.165 | M |
| SM190 | CF | 9.1 | R | R | R | R | R | R | PDR | 0.263 | M |
| SM192 | CF | 9.1 | R | R | R | R | R | R | PDR | 0.446 | S |
| SM193 | CF | 9.1 | R | S | S | R | R | R | XDR | 0.239 | M |
| SM194 | CF | 9.1 | S | S | R | R | R | R | MDR | 0.184 | W |
| SM195 | CF | 9.1 | S | R | R | R | R | R | XDR | 0.195 | M |
| SM134 | CF | 9.1 | S | S | R | R | R | R | XDR | 0.547 | S |
| SM135 | CF | 9.1 | S | S | R | R | R | R | MDR | 0.583 | S |
| SM136 | CF | 9.1 | R | R | R | R | R | R | PDR | 0.541 | S |
| SM106 | CF | 9.1 | S | S | S | R | R | R | MDR | 0.495 | S |
| SM110 | CF | 9.1 | R | R | R | R | R | R | PDR | 0.475 | S |
| SM171 | sputum | 10.1 | S | S | S | R | R | S | non-MDR | 1.099 | S |
| SM123 | CF | 11.1 | S | S | S | R | R | R | MDR | 0.608 | S |
| SM118 | CF | 12.1 | S | S | S | R | S | R | non-MDR | 0.755 | S |
| SM116 | CF | 13.1 | S | R | S | R | R | R | MDR | 0.293 | M |
| SM138 | CF | 14.1 | S | R | S | R | R | R | XDR | 0.391 | S |
| SM109 | CF | 15.1 | S | R | S | S | R | R | MDR | 1.035 | S |
| SM117 | CF | 16.1 | S | R | S | R | R | R | XDR | <0.096 | NP |
| SM113 | CF | 17.1 | R | S | S | R | S | S | non-MDR | <0.096 | NP |
| SM142 | CF | 18.1 | S | R | S | R | R | R | XDR | 1.545 | S |
| SM47 | sputum | 19.1 | S | R | S | S | S | R | non-MDR | 0.100 | W |
| SM157 | CF | 20.1 | S | S | S | S | S | S | non-MDR | <0.096 | NP |
| SM30 | sputum | 21.1 | S | S | S | R | S | R | non-MDR | 0.604 | S |
| SM175 | sputum | 21.1 | S | R | S | R | R | R | XDR | 0.979 | S |
| SM49 | vaginal swab | 22.1 | S | R | S | S | S | S | non-MDR | 0.746 | S |
| SM130 | CF | 23.1 | S | S | S | R | R | R | MDR | 0.546 | S |
| SM184 | blood | 24.1 | S | R | S | R | R | R | XDR | <0.096 | NP |
| SM42 | pharynx swab | 25.1 | S | S | S | R | R | R | MDR | 0.323 | M |
| SM159 | CF | 26.1 | R | S | S | S | R | R | MDR | 0.370 | M |
| SM4 | urine | 27.1 | R | R | R | R | S | R | XDR | 0.732 | S |
| SM8 | urine | 27.1 | R | S | R | R | S | R | MDR | 1.767 | S |
| SM24 | sputum | 27.1 | R | S | R | R | S | R | MDR | 0.629 | S |
| SM37 | blood | 27.1 | R | S | R | R | S | R | MDR | 0.653 | S |
| SM39 | genital swab | 27.1 | R | R | R | R | R | R | PDR | 0.459 | S |
| SM51 | blood | 28.1 | S | R | S | S | S | S | non-MDR | 1.066 | S |
| SM191 | CF | 29.1 | S | R | S | R | R | R | XDR | 0.337 | M |
| SM144 | CF | 31.1 | S | R | S | R | R | R | XDR | 0.590 | S |
| SM174 | sputum | 32.1 | S | S | S | R | R | R | MDR | 0.532 | S |
| SM150 | CF | 33.1 | S | S | S | R | R | R | MDR | 0.391 | S |
| SM180 | blood | 34.1 | S | R | S | R | S | R | MDR | 1.014 | S |
| SM181 | blood | 35.1 | S | R | S | R | R | S | MDR | <0.096 | NP |
| SM137 | CF | 36.1 | S | S | S | R | R | R | MDR | 1.123 | S |
| SM140 | CF | 37.1 | S | R | S | R | R | R | XDR | 0.233 | M |
| SM32 | sputum | 38.1 | S | S | S | S | S | S | non-MDR | 0.509 | S |
| SM21 | rectal swab | 39.1 | S | R | R | R | R | R | XDR | 0.320 | M |
| SM36 | sputum | 40.1 | S | S | S | R | S | S | non-MDR | 3.646 | S |
| SM48 | blood | 40.1 | S | S | S | R | S | R | non-MDR | 0.299 | M |
| SM43 | sputum | 41.1 | S | S | R | S | R | R | MDR | 0.470 | S |
| SM29 | sputum | 42.1 | S | S | S | R | R | R | MDR | 1.720 | S |
| SM50 | sputum | 43.1 | S | R | S | R | R | R | XDR | 2.295 | S |
| SM183 | blood | 44.1 | S | S | S | R | R | R | MDR | 0.397 | S |
| SM107 | CF | 45.1 | S | S | S | R | R | R | MDR | 0.593 | S |
| SM182 | blood | 46.1 | S | S | S | R | S | R | non-MDR | 0.831 | S |
| SM104 | CF | 47.1 | S | R | R | R | R | R | XDR | 0.349 | M |
| SM105 | CF | 48.1 | R | R | R | R | R | R | PDR | 0.581 | S |
| SM120 | CF | 49.1 | R | R | R | R | R | R | PDR | 0.352 | M |
| SM111 | CF | 50.1 | S | S | R | R | R | R | MDR | <0.096 | NP |
| SM112 | CF | 50.1 | S | S | R | R | R | R | MDR | 0.106 | W |
| SM114 | sputum | 51.1 | S | R | S | S | R | R | MDR | <0.096 | NP |
| SM122 | CF | 52.1 | S | S | S | R | R | R | MDR | 1.300 | S |
| SM45 | sputum | 53.1 | S | S | S | R | S | R | non-MDR | 0.596 | S |
| SM170 | sputum | 53.2 | S | R | R | R | R | R | XDR | 0.521 | S |
| SM46 | blood | 54.1 | S | R | S | R | S | R | MDR | 0.888 | S |
| SM5 | sputum | 55.1 | R | R | R | R | R | R | PDR | <0.096 | NP |
| SM177 | sputum | 56.1 | S | R | S | R | R | R | XDR | 0.354 | M |
| SM6 | sputum | 57.1 | S | S | S | S | S | R | non-MDR | 2.517 | S |
| SM38 | sputum | 58.1 | S | S | S | S | S | R | non-MDR | 0.896 | S |
| SM156 | CF | 59.1 | S | S | S | R | R | R | MDR | 0.276 | M |
| SM7 | pharynx swab | 60.1 | S | R | S | S | S | R | non-MDR | 1.111 | S |
| SM124 | CF | 61.1 | S | S | S | S | R | R | non-MDR | <0.096 | NP |
| SM14 | sputum | 62.1 | S | S | S | S | S | S | non-MDR | 0.847 | S |
| SM40 | pharynx swab | 63.1 | S | R | S | R | R | R | XDR | 0.476 | S |
| SM103 | CF | 64.1 | S | S | S | R | R | R | MDR | 0.400 | S |
a Isolation site: CF, isolated from the airways of cystic fibrosis patients. b Resistance phenotype, according to Magiorakos et al. [24]: MDR, multidrug-resistant; XDR: Extensively drug-resistant; PDR, pandrug-resistant; non-MDR: Non multidrug-resistant. c Biofilm classes, according to Stepanovic et al. [23]: No biofilm producer (NP; OD492 ≤ ODc; OD492 ≤ 0.096), weak biofilm producer (W; ODc < OD492 ≤ 2 × ODc; 0.096 < OD492 ≤ 0.192), moderate biofilm producer (M; 2 × ODc < OD492 ≤ 4 × ODc; 0.192 < OD492 ≤ 0.384), and strong biofilm producer (S; 4 × ODc < OD492; 0.384 < OD492), where ODc = mean OD492 of control wells + 3 SDs.
A total of 64 different PFGE types were identified among the 85 S. maltophilia strains, with 30 and 34 different PFGE profiles observed among 40 CF and 45 non-CF strains, respectively. These findings indicated that the genetic heterogeneity—calculated as (number of pulsotypes/number of strains tested) x 100—is comparable in both CF and non-CF strains (75% versus 75.5%, respectively).
Six PFGE types—specifically two and four, respectively, among CF and non-CF strains—were represented by multiple strains. Among CF strains, PFGE type 9 consisted of 10 strains, followed by PFGE type 50, comprised of two strains. Among non-CF strains, PFGE type 3 was the most prevalent (six strains), followed by PFGE type 27 (five strains), and PFGE types 21 and 40 (two strains each). No PFGE types shared by CF and non-CF strains were found.
2.2. Antibiotic Resistance Is More Prevalent in CF Strains
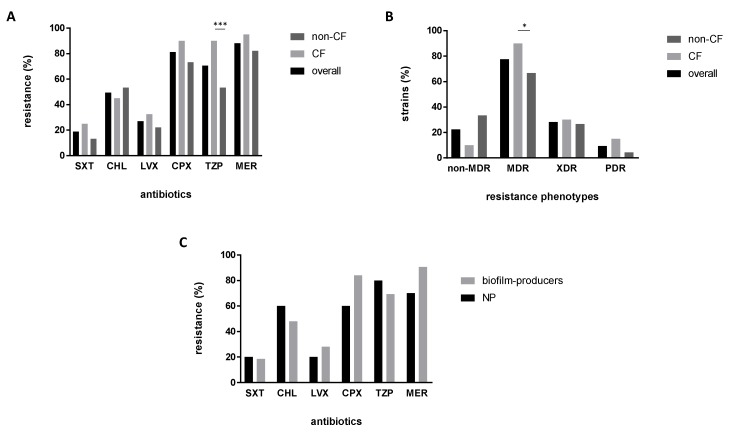
The in vitro activity of six antibiotics against S. maltophilia strains was measured by disk diffusion agar or broth microdilution, and results are shown in detail in Table 1 and summarized in Figure 1. Considering the strains as a whole, cotrimoxazole and levofloxacin were the most effective antibiotics, being active respectively against 81.2% and 72.9% of strains. Contrarily, the higher resistance rates were observed for meropenem (88.2%), followed by ciprofloxacin (81.2%), piperacillin/tazobactam (70.6%), and chloramphenicol (49.4%) (Figure 1A).
Figure 1.
Distribution of antibiotic resistance rates and profiles. S. maltophilia strains were evaluated for in vitro susceptibility to six antibiotics: SXT, cotrimoxazole; CHL, chloramphenicol; LVX, levofloxacin; CPX, ciprofloxacin; TZP, piperacillin/tazobactam; MER, meropenem. The frequencies of (A) antibiotic resistances, and (B) multiresistance profiles (non-MDR, MDR, XDR, and PDR) were calculated for cystic fibrosis (CF) (n = 40), non-CF (n = 45), and overall (n = 85) strains. (C) Resistance rates were calculated according to the capability to form biofilm: Non-producers (NP; n = 10) or biofilm-producers (n = 75; gathering strong, moderate, and weak-producers). Significance level at Fisher’s exact test: * p < 0.05; *** p < 0.001. Resistance profiles according to Magiorakos et al. [24]: MDR, multidrug-resistant strains; XDR, extensively drug-resistant strains; PDR, pandrug-resistant strains.
The strains isolated from CF airways exhibited significantly higher resistance rates to piperacillin/tazobactam compared to those isolated from non-CF patients (90% versus 53.3%, respectively; p < 0.001) (Figure 1A); the same trend was confirmed after stratification based on sample type (90% versus 62% and 36.4%, respectively, for respiratory from CF patients, respiratory from non-CF patients, and blood from non-CF patients; p < 0.01) (data not shown). Cotrimoxazole showed comparable activity in both CF and non-CF strains; however, when only respiratory strains were considered, the antibiotic was significantly more active against non-CF than CF strains (susceptibility rates: 86.7% versus 75%, respectively; p < 0.05) (Figure 1A).
The antibiotic resistance patterns revealed that most strains (66 out of 85, 77.6%) show multidrug resistance, with extensively drug-resistant (XDR) and pandrug-resistant (PDR) phenotypes, respectively, seen for 28.2% and 9.4% of the strains.
The multidrug-resistant (MDR) phenotype was observed in a proportion significantly higher in CF strains compared with non-CF strains (90% versus 66.7%, respectively; p < 0.05) (Figure 1B). CF strains also showed higher XDR and PDR prevalence compared with non-CF strains, although not at a statistically significant extent (XDR: 30% versus 26.7%, respectively; PDR: 15% versus 4.4%, respectively).
No relationship between specific PFGE types and antibiotic resistance was found.
2.3. Non-CF Strains Are Significantly More Efficient Than CF Strains in Forming Biofilm
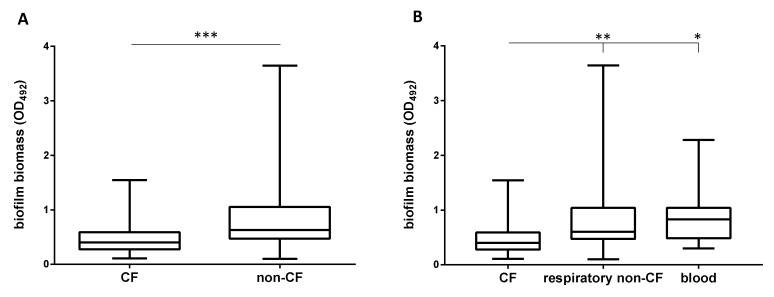
The biofilm forming ability was assessed on polystyrene using the microtiter plate method, and results are shown in detail in Table 1 and summarized in Figure 2 and Figure 3. Most of the strains (75 out of 85, 88.2%) were able to form biofilms on polystyrene, although to different extents, as indicated by OD492 values (range: 0.100–3.646; mean ± SD: 0.745 ± 0.644; coefficient of variation: 86.4%) (Table 1). The prevalence of strains able to form biofilm was comparable both in CF and non-CF strains (87.5 versus 88.9%, respectively), although the median amount of biofilm formed by non-CF strains was significantly higher than that produced by CF strains (OD492, median: 0.631 versus 0.400, respectively; p < 0.001) (Figure 2A). Biofilm formation was related to the source of isolation and patient type since CF strains produced lower biofilm biomass levels compared to those isolated from non-CF patients both from airways (OD492, median: 0.400 versus 0.604, respectively; p < 0.01) and blood (OD492, median: 0.400 versus 0.831, respectively; p < 0.05), whereas no significant differences were found among non-CF strains regardless of the sample type considered (Figure 2B).
Figure 2.
Biofilm formation according to patient and sample types. Biofilm biomass, assessed by spectrophotometric assay after crystal violet assay, was stratified according to (A) patients with (CF; n = 40) or without (non-CF; n = 45) cystic fibrosis, and (B) sample type (CF, n = 40; respiratory non-CF, n = 29; blood, n = 11). Results are shown as box and whiskers: the ends of the whiskers represent the minima and the maxima of all the data; the box always extends from the 25th to 75th percentiles, while the line in the middle of the box is plotted at the median. Significance level from Mann–Whitney test: * p < 0.05; ** p < 0.01; *** p < 0.001.
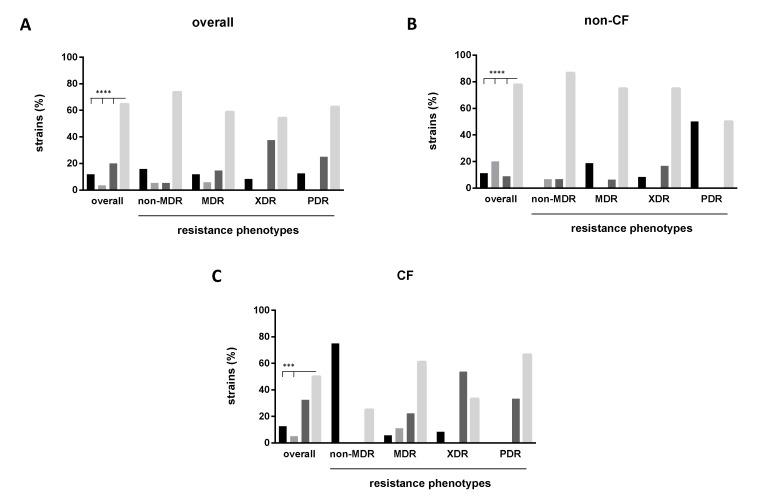
Figure 3.
Biofilm formation and multidrug-resistant phenotypes. Biofilm formation was assessed by spectrophotometric assay after crystal violet assay and categorized according to Stepanovic et al. [23]: Non-producers, weak-producers, moderate-producers, and strong-producers (left-to-right, in each series of histograms). Susceptibility tests were performed using Kirby-Bauer disk diffusion agar. Resistance profiles according to Magiorakos et al. [24]: MDR, multidrug-resistant strains; XDR, extensively drug-resistant strains; PDR, pandrug-resistant strains. Significance level at Fisher’s exact test: *** p < 0.001; **** p < 0.0001.
Categorization of biofilm formation according to Stepanovic et al. [23] showed that, overall, strong-producers were significantly more prevalent compared with other biofilm classes (64.7% versus 20%, 3.5%, and 11.8%, respectively for strong, moderate, weak, and non-producers; p < 0.0001) (Figure 3A). The same trend was observed both among non-CF (77.8%; p < 0.0001 versus other classes) (Figure 3B) and CF (50%; p < 0.0003 versus other classes except for moderate-producers) strains (Figure 3C). Further, among CF strains, the proportion of moderate-producers was higher compared to that observed for weak-producers (32.5 versus 2.5%, respectively; p < 0.01). The frequency of strong-producer strains was higher among the non-CF group than CF one (77.8% versus 50%, respectively; p < 0.05). Particularly, most of the strains from blood were strong-producers (six out of nine, 66.6%), suggesting that biofilm formation might play a role in S. maltophilia‘s tissue invasion.
Non-CF PFGE types 3 (n = six strains) and 27 (n = five strains) consisted of strong-producers only, whereas CF PFGE type 9 consisted mostly of strong-producers (six out of 10, 60%), along with moderate (three out of 10, 30%) and weak-producers (one out of 10, 10%) strains (Table 1).
2.4. Biofilm Formation and Antibiotic Resistance Are Inversely Related
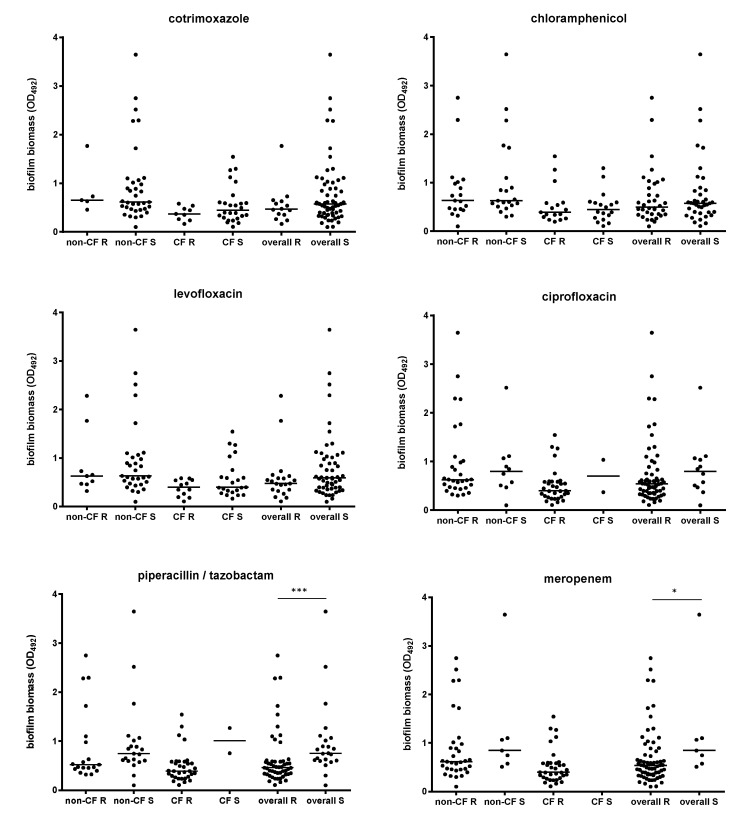
Trying to find a relationship between biofilm formation and planktonic resistance to antibiotics, we evaluated the variations in the biofilm biomass formed on the bases of both susceptibility/resistance and multi-resistance profiles. Considering the strains as a whole, those susceptible to piperacillin/tazobactam or meropenem produced significantly more biofilm than resistant counterparts (OD492, median; piperacillin/tazobactam: 0.755 versus 0.464, p < 0.001; meropenem: 0.847 versus 0.536, p < 0.05; respectively for susceptible and resistant strains) (Figure 4).
Figure 4.
Biofilm formation according to antibiotic resistance and patient type. Biofilm biomass formation, spectrophotometrically assessed by crystal violet assay, was stratified on each antibiotic tested—according to susceptibility (S) or resistance (R)—and patient type. Results are shown as scatter plots, with horizontal lines indicating the median values. Significance level at Mann–Whitney test: * p < 0.05; *** p < 0.0001.
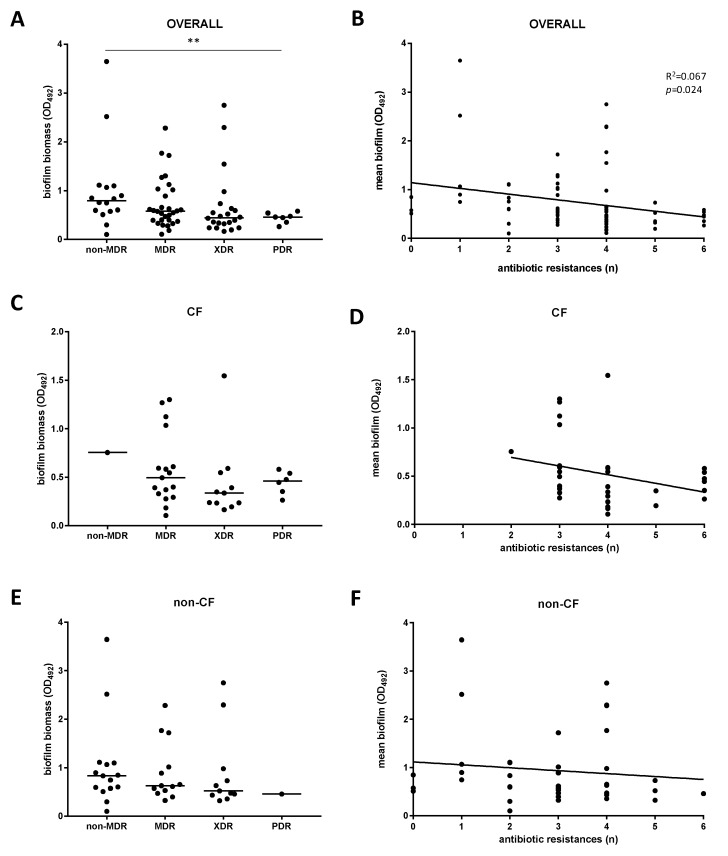
Next, stratifying the biofilm biomass formation on the resistance phenotype, we found that non-MDR strains were more efficient in forming biofilm compared to PDR strains (OD492, median: 0.793 versus 0.459, respectively; p < 0.01) (Figure 5A), whereas no significant differences were observed among CF (Figure 5C) and non-CF (Figure 5E) strains. The multidrug-resistance level—calculated as the number of antibiotic resistances showed by each strain—was negatively associated with mean biofilm formed only when strains were considered as a whole (p = 0.024, linear regression analysis) (Figure 5B). A similar, although not statistically significant, trend was observed among both CF (Figure 5D) and non-CF (Figure 5F) strains. Confirming this trend, the frequency of non-producer strains observed for non-MDR group was significantly higher compared with other groups considered (75% versus 5.6%, 8.3%, and 0%, respectively, for MDR, XDR, and PDR strains; p < 0.01) (Figure 3C). Further, both in “overall” and non-CF groups, the frequency of strong-producers was higher in non-MDR group compared with MDR, XDR, and PDR, although these differences were not statistically significant (Figure 3A,B).
Figure 5.
Biofilm formation according to the antibiotic resistance level. Biofilm formation was measured by crystal violet assay, and antibiotic resistance by disk diffusion technique. Biofilm formation was stratified according to (A,C,E) the resistance phenotype (non-MDR, MDR, XDR, PDR)— showing results as scatter plot, with the horizontal line indicating the median value—and (B,D,F) the number of resistances to each antibiotic tested. Significance level: ** p < 0.01, Mann-–Whitney test; r2: 0.067, p = 0.024, linear regression analysis.
The resistance rates exhibited by biofilm non-producer strains were comparable to those observed among biofilm-producers (weak, moderate, and strong) (Figure 1C). Non-producer strains showed both MDR (seven out of 10; 70%) and non-MDR (three out of 10; 30%) phenotypes.
3. Discussion
The present study was undertaken to investigate the clonal relatedness, antibiotic-resistance patterns, and biofilm forming ability of a population of S. maltophilia strains isolated from different hospitals in central Italy, as a representative for several clinical settings. The possible relationship among these traits was also assessed.
The persistence of bacterial infections is due to the emergence of persister cells, whose physiologically dormant state makes them able to elude antibiotic killing. The selection for persister cells occurs under conditions that include hostile host environments, a damage response being caused by sublethal concentrations of antibiotics, and bacterial biofilms [25].
Biofilms are microbial communities wrapped in a polysaccharidic matrix produced by the bacteria, which are adhered to an inert or biotic surface. Biofilms are far less susceptible to antibiotics than planktonic cells, and therefore, they have been associated with a wide range of infections in a clinical setting, from those related to exogenous devices (i.e., catheters or prosthetic joints) to chronic tissue infections, such as those occurring in the lungs of CF patients [26,27].
In this work, we found that biofilm forming ability is highly preserved in S. maltophilia and could, therefore, be accepted as an important virulence trait underlying treatment failure, recurrences, and/or persistence of colonization. Biofilm formation appeared to be related to both the site of infection and patient type. In fact, respiratory strains from CF lung were less efficient in forming biofilm with respect to those isolated from the airways of patients without CF, as indicated by differences in both median biofilm biomass and frequency of strong-producers, suggesting that S. maltophilia can successfully adapt to a highly stressful environment, such as a CF lung, by paying a “biological cost.”
Molecular analysis of XbaI-digested DNA resulted in 64 distinct PFGE types; 90.6% of those consisted of only one isolate, confirming the great flexibility of S. maltophilia to evolve, regardless of the clinical setting considered [11,28,29]. On the other hand, the evidence for PFGE types consisting of multiple strains shows that the same strain was isolated from different patients over a period, emphasizing the persistence and dissemination of S. maltophilia in the hospital environment. This extraordinary variability, besides suggesting that most strains were acquired independently rather than because of cross-transmission, could also be the consequence of the high propensity to form biofilm we found in this study. In fact, earlier observations showed that the characteristic environment created within a biofilm enhances both the proportion of hypermutable strains and the horizontal transfer of genetic material with consequent genetic rearrangement by recombination [30,31,32]. This makes S. maltophilia populations significantly complex and dynamic, being able to fluctuate rapidly under changing selective pressures.
Treatment of S. maltophilia infection is a challenge for clinicians due to its natural resistance to many antimicrobial drugs, mostly involving the study of strains isolated from CF patients [33,34]. Overall, our results confirmed the well-established multi-resistant phenotype of S. maltophilia in the hospital setting. Most strains, in fact, showed MDR phenotype (77.6%), with a frequency significantly higher in CF strains compared with non-CF strains (90% versus 66.7%, respectively).
SXT is the antibiotic of choice for treating S. maltophilia infections, although in recent years, increasing rates of resistance, ranging from 16% to 45%, were reported, especially in CF patients [9,35,36]. Considering available clinical breakpoints, our results confirmed SXT is the most active compound against all the strains tested. The overall resistance to SXT was 18.7%, although its activity was significantly higher in non-CF than CF strains (96.3% versus 75%, respectively), in agreement with previous findings [37,38].
Recently, a systematic review reported that fluoroquinolones show comparable effects on the mortality of S. maltophilia infection compared with trimethoprim-sulfamethoxazole, supporting the use of fluoroquinolones in clinical S. maltophilia infections [39], although high rates of resistance are also increasingly being reported. In the current study, the newer fluoroquinolone levofloxacin showed better activity against S. maltophilia than ciprofloxacin (susceptibility rates: 77.9% versus 18.8%, respectively), confirming previous studies [9,37,38].
We found that CF strains were significantly more resistant to piperacillin-tazobactam than non-CF strains (90% versus 53.3%, respectively). This finding might be the consequence of the airways’ concomitant colonization/infection by both S. maltophilia and P. aeruginosa observed in CF patients [35,40,41]. Consequently, the higher use of this antipseudomonal beta-lactam/beta-lactamase inhibitor combination in CF patients might have exerted a positive antibiotic pressure associated with higher levels of resistance.
To explore whether there was any correlation between biofilm formation and planktonic antibiotic resistance, first, we analyzed the biofilm biomass formed and the composition of the biofilm formation groups, with respect to resistance phenotypes. The most noteworthy information obtained from the present work, seen for the first time, was the overall negative relationship between the ability to form biofilm and the level of antibiotic resistance. A general trend was in fact seen; that is, the amount of biofilm formed was inversely correlated with the number of resistances to antibiotics. Confirming this trend, we observed that non-MDR strains were more efficient at forming biofilm compared to PDR strains; further, the frequency of non-producer strains among non-MDR strains was significantly higher compared with that shown by the multi-resistance phenotypes considered. We did not find any significant relationship between biofilm categories and resistance profiles, except for the CF population, where strains exhibiting more robust biofilm formation likely contained a larger proportion of MDR strains compared with strains from non-CF patients. This finding probably suggests that the relationship between drug susceptibility and biofilm formation is influenced by the environmental pressure at the site of infection.
Afterward, to determine whether biofilm formation is correlated with resistance to any antibiotic(s), we compared the biofilm forming capacities among strains with different resistance profiles to each of the antibiotics tested. A negative correlation between biofilm amount and resistance profile was observed in the case of piperacillin/tazobactam and meropenem, where the susceptible strains could form higher biofilm biomass amounts than resistant strains.
Susceptible bacteria reasonably need alternative strategies which are of no use to resistant bacteria, in order to escape antibiotic treatment and support their survival within the host. In agreement with earlier works [20,21,42], overall, our findings suggest that, also in S. maltophilia, susceptible bacteria may use biofilm formation in this regard, probably because the biofilm-mediated resistance might be less expensive in terms of energy requirements than chromosomal resistance mechanisms. However, the findings that nearly all strains unable to form biofilm maintain the MDR phenotype and show antibiotic-resistance rates comparable to those observed in biofilm-forming strains, indicate that other factors might be involved and should be examined to confirm such speculation.
4. Materials and Methods
4.1. Bacterial Strains and Growth Conditions
A total of 85 consecutive, non-repetitive S. maltophilia strains were isolated between 2017 and 2018: 40 from respiratory samples collected from CF patients attending the Cystic Fibrosis Unit at “Bambino Gesù” Hospital (Rome, Italy), and 45 from different sites (29 from respiratory tract, 11 from blood, three from anogenital swabs, and two from urine) of non-CF patients attended at various hospitals of central Italy.
All strains were grown at 37 °C onto Tryptone soy agar (Oxoid SpA; Rodano, Milan, Italy) and were identified as S. maltophilia using manual (API20NE) or automated (Vitek 2) systems (bio-Mérieux, Marcy l’Etoile, France). Strains were stored in Cryobank (Copan Diagnostics, Murrieta, CA, USA) at −80 °C and were cultivated in Tryptone soy broth (Oxoid SpA) at 37 °C for 18–20 h without shaking for further analysis.
4.2. Genetic Relatedness and Cluster Analysis
Bacterial DNA was digested with the restriction enzyme XbaI as previously described, with minor modifications [11]. PFGE was carried out using the following parameters: Initial switch and final switch times were 5 and 35 s, respectively; run time was 20 h, and the temperature was 12 °C for 20 h at 6.0 V/cm; and the included angle was 120°. Isolates with identical PFGE patterns were assigned to the same PFGE type and subtype. Isolates differing by one to three bands were considered genetically related and were assigned to the same PFGE type with different PFGE subtypes. Isolates with PFGE patterns differing by more than 4 bands were considered genetically unrelated and were assigned to different PFGE types. PFGE types were analyzed with BioNumerics software for Windows (version 2.5; Applied Maths, Ghent, Belgium). Normalization of DNA banding patterns was performed with bacteriophage lambda concatemer ladder standards. The banding patterns were compared by computer-assisted analysis using the unweighted pair group method with arithmetic averages (UPGMA) and with the Dice similarity coefficient. A tolerance of 1.5% in band position was applied during DNA pattern comparisons.
4.3. Antibiotic Susceptibility Testing
Six antibiotics were tested in vitro against S. maltophilia strains using the Kirby–Bauer disc diffusion technique (meropenem, ciprofloxacin, piperacillin/tazobactam, levofloxacin, and cotrimoxazole) or the broth microdilution method (chloramphenicol) according to the Clinical and Laboratory Standards Institute (CLSI) guidelines [22]. Pseudomonas aeruginosa ATCC 27853 was used as a reference strain. Strains categorized as resistant or intermediate, as defined using clinical breakpoints as interpretive criteria provided by CLSI [22], were grouped as “resistant” for data analysis. The activities of piperacillin-tazobactam, ciprofloxacin, and meropenem—whose CLSI breakpoints were not determined for S. maltophilia—were investigated using interpretation afforded for P. aeruginosa.
Since we tested the activity of antibiotics belonging to five different classes, according to Magiorakos et al. [24], we defined a strain as: Multidrug-resistant (MDR), if non-susceptible to at least one agent in three or more antimicrobial categories; extensively drug-resistant (XDR), if non-susceptible to at least one agent in four antimicrobial categories (i.e., bacterial isolates remain susceptible to only one category); pandrug-resistant (PDR), if non-susceptible to all agents in all antimicrobial categories (i.e., no agents tested as susceptible for that organism). Strains resistant to no more than 2 antimicrobial categories were described separately and referred to as non-MDR for the correlation analyses between antibiotic resistance and biofilm formation.
4.4. Biofilm Formation Assay
The ability of each isolate to form biofilm was quantitatively assessed using the microtiter plate method and crystal violet staining, as previously described [40]. In brief, overnight growth in TSB was adjusted to an optical density measured at 550 nm of 1.00 (corresponding to 1 × 109 CFU/mL) and then diluted 1:100 in fresh TSB. Two-hundred microliters of this standardized inoculum was dispensed to each well of a sterile flat-bottom polystyrene 96-well microtiter tissue culture plate (Iwaki, Bibby srl; Milan, Italy) and incubation was at 37 °C, under an aerobic atmosphere. Control wells contained medium alone. After a 24 h incubation, non-adherent bacteria were removed by washing twice with 200 μL sterile phosphate-buffered saline pH 7.3 (PBS) (Sigma-Aldrich Italia, Milan, Italy), and biofilm amount was measured by crystal violet assay. Briefly, biofilm samples were fixed at 60 °C for 1 h, then stained for 5 min with 200 μL crystal violet. Excess stain was rinsed off with running tap water, and then the plates were air-dried. Crystal violet was extracted by exposure at room temperature for 15 min to 200 μL glacial acetic acid 33% (Sigma-Aldrich), and biofilm biomass (including adherent bacteria and EPS) was then assessed by measuring the optical density at 492 nm (OD492) (SpectraMax 190; Molecular Devices, Sunnyvale, CA, USA). Strains were classified into the following categories: No biofilm producer (OD492 ≤ ODc), weak biofilm producer (ODc < OD492 ≤ 2 × ODc), moderate biofilm producer (2 × ODc < OD492 ≤ 4 × ODc), and strong biofilm producer (4 × ODc < OD492), where ODc = mean OD492 of control (without inoculum) wells + 3 standard deviations (SDs) [23]. The reference S. maltophilia ATCC13637 strain was used as a positive control for strong biofilm formation (mean OD492: 2.629), whereas the S. epidermidis ATCC 12228 strain was used as negative control.
4.5. Statistical Analysis
Each experiment was carried out at least in triplicate and repeated on two different occasions (n ≥ 6). The Gaussian distribution of results was assessed by the D’Agostino and Pearson normality test. Differences in biofilm biomass between groups were evaluated by Mann–Whitney test, whereas differences between proportions were analyzed by Fisher’s exact test. Correlation between biofilm formation and antibiotic-resistance level was measured by linear regression analysis. All intermediate susceptibilities to the antibiotics were considered to be indicative of antibiotic resistance. Data analyses were carried out using GraphPad Software (Prism 7.0 for Windows; GraphPad software Inc.; San Diego, CA, USA), considering p-values less than 0.05 as statistically significant.
5. Conclusions
The findings from the present work indicated, for the first time in literature, that in S. maltophilia the possession of both classical drug resistance mechanisms in planktonic phase and the ability to form biofilm might contribute to the microorganism’s survival in a stressful environment, favoring its dissemination in the hospital setting.
The biofilm production probably acts as a survival mechanism for bacteria, especially in cases of susceptibility or when resistance level is not enough; bacteria that achieve resistance through biofilm production do not need to develop or maintain the mechanisms responsible for resistance of planktonic cells. Testing for biofilm formation is important in deciding the pathogenicity of clinical S. maltophilia isolates, and therefore, should be routinely performed in diagnostic laboratories. This evaluation might be paired with the antibiogram to predict possible cases of non-eradication of the pathogen and/or to apply synergic treatments facilitating antibiotic passage through the biofilm layer. However, considering the intrinsic antibiotic-resistance shown by sessile communities, our findings further indicate the poor therapeutic predictive value of the standard, planktonic-based, antibiogram and the consequent need to develop alternative, biofilm-based, antimicrobial susceptibility tests.
Our results also raise questions about the mechanisms underlying the complex relationship between biofilm formation ability and antibiotic resistance, and how resistant strains achieve high levels of biofilm-specific resistance despite producing lesser amounts of biofilms. Further studies are warranted to better clarify these mechanisms, in order to develop new and more effective prophylactic and therapeutic strategies for dealing with infections caused by S. maltophilia, an opportunistic and often multi-drug resistant, nosocomial pathogen.
Acknowledgments
The authors wish to thank Sherry Lynn Jones for the language revision of the manuscript.
Author Contributions
Conceptualization, A.P., G.G., and G.D.B.; methodology, A.P. and G.D.B.; software, G.D.B.; validation, A.P. and G.D.B.; formal analysis, A.P., V.S., E.F., and G.G.; investigation, A.P.; data curation, A.P. and G.D.B.; writing—Original draft preparation, A.P. and G.D.B.; writing—Review and editing, A.P., V.S., E.F., G.G., and G.D.B.; supervision, G.D.B.; project administration, G.D.B.; funding acquisition, G.D.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by “G. d’Annunzio” University of Chieti-Pescara, “Fondo Ricerca di Ateneo, anno 2017”.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Looney W.J., Narita M., Mühlemann K. Stenotrophomonas maltophilia: An emerging opportunist human pathogen. Lancet Infect. Dis. 2009;9:312–323. doi: 10.1016/S1473-3099(09)70083-0. [DOI] [PubMed] [Google Scholar]
- 2.Brooke J.S. Stenotrophomonas maltophilia: An emerging global opportunistic pathogen. Clin. Microbiol. Rev. 2012;25:2–41. doi: 10.1128/CMR.00019-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Waters V., Yau Y., Prasad S., Lu A., Atenafu E., Crandall I., Tom S., Tullis E., Ratjen F. Stenotrophomonas maltophilia in cystic fibrosis: Serologic response and effect on lung disease. Am. J. Respir. Crit. Care Med. 2011;183:635–640. doi: 10.1164/rccm.201009-1392OC. [DOI] [PubMed] [Google Scholar]
- 4.Goss C.H., Mayer-Hamblett N., Aitken M.L., Rubenfeld G.D., Ramsey B.W. Association between Stenotrophomonas maltophilia and lung function in cystic fibrosis. Thorax. 2004;59:955–959. doi: 10.1136/thx.2003.017707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waters V., Atenafu E.G., Lu A., Yau Y., Tullis E., Ratjen F. Chronic Stenotrophomonas maltophilia infection and mortality or lung transplantation in cystic fibrosis patients. J. Cyst. Fibros. 2013;12:482–486. doi: 10.1016/j.jcf.2012.12.006. [DOI] [PubMed] [Google Scholar]
- 6.Raidt L., Idelevich E.A., Dübbers A., Küster P., Drevinek P., Peters G., Kahl B.C. Increased prevalence and resistance of important pathogens recovered from respiratory specimens of cystic fibrosis patients during a decade. Pediatr. Infect. Dis. J. 2015;34:700–705. doi: 10.1097/INF.0000000000000714. [DOI] [PubMed] [Google Scholar]
- 7.Di Bonaventura G., Prosseda G., Del Chierico F., Cannavacciuolo S., Cipriani P., Petrucca A., Superti F., Ammendolia M.G., Concato C., Fiscarelli E., et al. Molecular characterization of virulence determinants of Stenotrophomonas maltophilia strains isolated from patients affected by cystic fibrosis. Int. J. Immunopathol. Pharmacol. 2007;20:529. doi: 10.1177/039463200702000311. [DOI] [PubMed] [Google Scholar]
- 8.Di Bonaventura G., Spedicato I., D’Antonio D., Robuffo I., Piccolomini R. Biofilm formation by Stenotrophomonas maltophilia: Modulation by quinolones, trimethoprim-sulfamethoxazole, and ceftazidime. Antimicrob. Agents Chemother. 2004;48:151–160. doi: 10.1128/AAC.48.1.151-160.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun E., Liang G., Wang L., Wei W., Lei M., Song S., Han R., Wang Y., Qi W. Antimicrobial susceptibility of hospital acquired Stenotrophomonas maltophilia isolate biofilms. Braz. J. Infect. Dis. 2016;20:365–373. doi: 10.1016/j.bjid.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pompilio A., Crocetta V., Confalone P., Nicoletti M., Petrucca A., Guarnieri S., Fiscarelli E., Savini V., Piccolomini R., Di Bonaventura G. Adhesion to and biofilm formation on IB3-1 bronchial cells by Stenotrophomonas maltophilia isolates from cystic fibrosis patients. BMC Microbiol. 2010;10:102. doi: 10.1186/1471-2180-10-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pompilio A., Crocetta V., Ghosh D., Chakrabarti M., Gherardi G., Vitali L.A., Fiscarelli E., Di Bonaventura G. Stenotrophomonas maltophilia phenotypic and genotypic diversity during a 10-year colonization in the lungs of a cystic fibrosis patient. Front. Microbiol. 2016;7:1551. doi: 10.3389/fmicb.2016.01551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Abreu P.M., Farias P.G., Paiva G.S., Almeida A.M., Morais P.V. Persistence of microbial communities including Pseudomonas aeruginosa in a hospital environment: A potential health hazard. BMC Microbiol. 2014;14:118. doi: 10.1186/1471-2180-14-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Esposito A., Pompilio A., Bettua C., Crocetta V., Giacobazzi E., Fiscarelli E., Jousson O., Di Bonaventura G. Evolution of Stenotrophomonas maltophilia in cystic fibrosis lung over chronic infection: A genomic and phenotypic population study. Front. Microbiol. 2017;8:1590. doi: 10.3389/fmicb.2017.01590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanchez M.B. Antibiotic resistance in the opportunistic pathogen Stenotrophomonas maltophilia. Front. Microbiol. 2015;6:658. doi: 10.3389/fmicb.2015.00658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeon Y.D., Jeong W.Y., Kim M.H., Jung I.Y., Ahn M.Y., Ann H.W., Ahn J.Y., Han S.H., Choi J.Y., Song Y.G., et al. Risk factors for mortality in patients with Stenotrophomonas maltophilia bacteremia. Medicine. 2016;95:e4375. doi: 10.1097/MD.0000000000004375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kwon A.S., Park G.C., Ryu S.Y., Lim D.H., Lim D.Y., Choi C.H., Park Y., Lim Y. Higher biofilm formation in multidrug-resistant clinical isolates of Staphylococcus aureus. Int. J. Antimicrob. Agents. 2008;32:68–72. doi: 10.1016/j.ijantimicag.2008.02.009. [DOI] [PubMed] [Google Scholar]
- 17.Shrestha L.B., Bhattarai N.R., Khanal B. Comparative evaluation of methods for the detection of biofilm formation in coagulase-negative staphylococci and correlation with antibiogram. Infect. Drug Resist. 2018;11:607–613. doi: 10.2147/IDR.S159764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Badave G.K., Kulkarni D. Biofilm producing multidrug resistant Acinetobacter baumannii: An emerging challenge. J. Clin. Diagn. Res. 2015;9:DC08. doi: 10.7860/JCDR/2015/11014.5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sahal G., Bilkay I.S. Multi drug resistance in strong biofilm forming clinical isolates of Staphylococcus epidermidis. Braz. J. Microbiol. 2014;45:539–544. doi: 10.1590/S1517-83822014005000042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krzyściak P., Chmielarczyk A., Pobiega M., Romaniszyn D., Wójkowska-Mach J. Acinetobacter baumannii isolated from hospital-acquired infection: Biofilm production and drug susceptibility. APMIS. 2017;125:1017–1026. doi: 10.1111/apm.12739. [DOI] [PubMed] [Google Scholar]
- 21.Qi L., Li H., Zhang C., Liang B., Li J., Wang L., Du X., Liu X., Qiu S., Song H. Relationship between antibiotic resistance, biofilm formation, and biofilm-specific resistance in Acinetobacter baumannii. Front. Microbiol. 2016;7:483. doi: 10.3389/fmicb.2016.00483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.CLSI . Performance Standards for Antimicrobial Susceptibility Testing. 29th ed. Clinical and Laboratory Standards Institute; Wayne, PA, USA: 2019. CLSI Supplement M100. [Google Scholar]
- 23.Stepanovic S., Vukovic D., Hola V., Di Bonaventura G., Djukic S., Cirkovic I., Ruzicka F. Quantification of biofilm in microtiter plates: Overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. APMIS. 2007;115:891–899. doi: 10.1111/j.1600-0463.2007.apm_630.x. [DOI] [PubMed] [Google Scholar]
- 24.Magiorakos A.P., Srinivasan A., Carey R.B., Carmeli Y., Falagas M.E., Giske C.G., Harbarth S., Hindler J.F., Kahlmeter G., Olsson-Liljequist B., et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012;18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 25.Harms A., Maisonneuve E., Gerdes K. Mechanisms of bacterial persistence during stress and antibiotic exposure. Science. 2016;354:aaf4268. doi: 10.1126/science.aaf4268. [DOI] [PubMed] [Google Scholar]
- 26.Ciofu O., Rojo-Molinero E., Macià M.D., Oliver A. Antibiotic treatment of biofilm infections. APMIS. 2017;125:304–319. doi: 10.1111/apm.12673. [DOI] [PubMed] [Google Scholar]
- 27.Azeredo J., Azevedo N.F., Briandet R., Cerca N., Coenye T., Costa A.R., Desvaux M., Di Bonaventura G., Hébraud M., Jaglic Z., et al. Critical review on biofilm methods. Crit. Rev. Microbiol. 2017;43:313–351. doi: 10.1080/1040841X.2016.1208146. [DOI] [PubMed] [Google Scholar]
- 28.Tanimoto K. Stenotrophomonas maltophilia strains isolated from a university hospital in Japan: Genomic variability and antibiotic resistance. J. Med. Microbiol. 2013;62:565–570. doi: 10.1099/jmm.0.051151-0. [DOI] [PubMed] [Google Scholar]
- 29.Neela V., Rankouhi S.Z., van Belkum A., Goering R.V., Awang R. Stenotrophomonas maltophilia in Malaysia: Molecular epidemiology and trimethoprim-sulfamethoxazole resistance. Int. J. Infect. Dis. 2012;16:e603–e607. doi: 10.1016/j.ijid.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 30.Prunier A.L., Malbruny B., Laurans M., Brouard J.F., Leclercq R. High rate of macrolide resistance in Staphylococcus aureus strains from patients with cystic fibrosis reveals high proportions of hypermutable strains. J. Infect. Dis. 2003;187:1709–1716. doi: 10.1086/374937. [DOI] [PubMed] [Google Scholar]
- 31.Molin S., Tolker-Nielsen T. Gene transfer occurs with enhanced efficiency in biofilms and induces enhanced stabilisation of the biofilm structure. Curr. Opin. Biotechnol. 2003;14:255–261. doi: 10.1016/S0958-1669(03)00036-3. [DOI] [PubMed] [Google Scholar]
- 32.Lee H.W., Koh Y.M., Kim J., Lee J.C., Lee Y.C., Seol S.Y., Cho D.T., Kim J. Capacity of multidrug-resistant clinical isolates of Acinetobacter baumannii to form biofilm and adhere to epithelial cell surfaces. Clin. Microbiol. Infect. 2008;14:49–54. doi: 10.1111/j.1469-0691.2007.01842.x. [DOI] [PubMed] [Google Scholar]
- 33.Safdar A., Rodriguez G., Balakrishnan M., Tarrand J., Rolston K. Changing trends in etiology of bacteremia in patients with cancer. Eur. J. Clin. Microbiol. Infect. Dis. 2006;25:522–526. doi: 10.1007/s10096-006-0173-4. [DOI] [PubMed] [Google Scholar]
- 34.Barbolla R., Catalano M., Orman B.E., Famiglietti A., Vay C., Smayevsky J., Centrón D., Piñeiro S.A. Class 1 integrons increase trimethoprim-sulfamethoxazole MICs against epidemiologically unrelated Stenotrophomonas maltophilia isolates. Antimicrob. Agents Chemother. 2004;48:666–669. doi: 10.1128/AAC.48.2.666-669.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De Dios Caballero J., del Campo R., Royuela A., Solé A., Máiz L., Olveira C., Quintana-Gallego E., de Gracia J., Cobo M., de la Pedrosa E.G., et al. Bronchopulmonary infection–colonization patterns in Spanish cystic fibrosis patients: Results from a national multicenter study. J. Cyst. Fibros. 2016;15:357–365. doi: 10.1016/j.jcf.2015.09.004. [DOI] [PubMed] [Google Scholar]
- 36.Valenza G., Tappe D., Turnwald D., Frosch M., König C., Hebestreit H., Abele-Horn M. Prevalence and antimicrobial susceptibility of microorganisms isolated from sputa of patients with cystic fibrosis. J. Cyst. Fibros. 2008;7:123–127. doi: 10.1016/j.jcf.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 37.Chang Y.T., Lin C.Y., Chen Y.H., Hsueh P.R. Update on infections caused by Stenotrophomonas maltophilia with particular attention to resistance mechanisms and therapeutic options. Front. Microbiol. 2015;6:893. doi: 10.3389/fmicb.2015.00893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Díez-Aguilar M., Ekkelenkamp M., Morosini M.I., Merino I., de Dios Caballero J., Jones M., van Westreenen M., Tunney M.M., Cantón R., Fluit A.C. Antimicrobial susceptibility of non-fermenting Gram-negative pathogens isolated from cystic fibrosis patients. Int. J. Antimicrob. Agents. 2019;53:84–88. doi: 10.1016/j.ijantimicag.2018.09.001. [DOI] [PubMed] [Google Scholar]
- 39.Ko J.H., Kang C.I., Cornejo-Juárez P., Yeh K.M., Wang C.H., Cho S.Y., Gözel M.G., Kim S.H., Hsueh P.R., Sekiya N., et al. Fluoroquinolones versus trimethoprim-sulfamethoxazole for the treatment of Stenotrophomonas maltophilia infections: A systematic review and meta-analysis. Clin. Microbiol. Infect. 2019;25:546–554. doi: 10.1016/j.cmi.2018.11.008. [DOI] [PubMed] [Google Scholar]
- 40.Pompilio A., Crocetta V., De Nicola S., Verginelli F., Fiscarelli E., Di Bonaventura G. Cooperative pathogenicity in cystic fibrosis: Stenotrophomonas maltophilia modulates Pseudomonas aeruginosa virulence in mixed biofilm. Front. Microbiol. 2015;6:951. doi: 10.3389/fmicb.2015.00951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Green H., Jones A. The microbiome and emerging pathogens in cystic fibrosis and non-cystic fibrosis bronchiectasis. Semin. Respir. Crit. Care Med. 2015;36:225–235. doi: 10.1055/s-0035-1546752. [DOI] [PubMed] [Google Scholar]
- 42.Baldassarri L., Creti R., Recchia S., Imperi M., Facinelli B., Giovanetti E., Pataracchia M., Alfarone G., Orefici G. Therapeutic failures of antibiotics used to treat macrolide-susceptible Streptococcus pyogenes infections may be due to biofilm formation. J. Clin. Microbiol. 2006;44:2721–2727. doi: 10.1128/JCM.00512-06. [DOI] [PMC free article] [PubMed] [Google Scholar]