Abstract
Tuberculosis, caused by Mycobacterium tuberculosis complex bacteria, remains one of the most pressing health problems. Despite the general trend towards reduction of the disease incidence rate, the situation remains extremely tense due to the distribution of the resistant forms. Most often, these strains emerge through the intra-host microevolution of the pathogen during treatment failure. In the present study, the focus was on three serial clinical isolates of Mycobacterium tuberculosis Beijing B0/W148 cluster from one patient with pulmonary tuberculosis, to evaluate their changes in metabolism during anti-tuberculosis therapy. Using whole genome sequencing (WGS), 9 polymorphisms were determined, which occurred in a stepwise or transient manner during treatment and were linked to the resistance (GyrA D94A; inhA t-8a) or virulence. The effect of the inhA t-8a mutation was confirmed on both proteomic and transcriptomic levels. Additionally, the amount of RpsL protein, which is a target of anti-tuberculosis drugs, was reduced. At the systemic level, profound changes in metabolism, linked to the evolution of the pathogen in the host and the effects of therapy, were documented. An overabundance of the FAS-II system proteins (HtdX, HtdY) and expression changes in the virulence factors have been observed at the RNA and protein levels.
Keywords: omics analysis, TB treatment, system analysis, Beijing B0/W148
1. Introduction
Mycobacterium tuberculosis bacteria are among the deadliest microbial pathogens in the world. Over 10 million new cases and 1.3 million lethal cases are registered annually [1]. Despite the reduction in the total tuberculosis (TB) incidence rate, the situation remains extremely tense due to the distribution of resistant forms. Among them, the multidrug-resistant- (MDR, resistance to at least isoniazid (INH) and rifampicin (RIF)) and extensively drug-resistant- (XDR, MDR phenotype with additional resistance to any fluoroquinolone (FQ) and at least one of the second-line injectable drugs capreomycin (CAP), kanamycin (KAN) or amikacin (AM)) tuberculosis pose a serious challenge for global TB control and makes successful treatment difficult or impossible [2].
Directly observed treatment (DOT) and DOT plus are the current standards of treatment strategies for the drug-susceptible and resistant forms of tuberculosis, respectively [3]. Although these protocols show high efficiency, the emergence and fixation of resistance to new antibiotics have been observed all over the world [4,5,6]. There are many reasons for this and not all of them have been fully elucidated, but it is worth mentioning that pathogen resistance develops in the patient, as the bacterium is a human obligate parasite. Treatment-driven positive selection leads to: the emergence of resistant clones within a population, their growth in a heteroresistant state and further ultimate fixation of a drug-resistant variant [7].
In recent years, multi-omics technologies have been increasingly used to understand the processes taking place in bacterial cells during the formation of resistance. For instance, the whole genome sequencing allows not only to undoubtedly distinguish mixed and unmixed infections in the patient, but also to define heterogeneity within one population [8,9,10]. The latter is often associated with the acquisition of certain drug resistance-conferring mutations, which occur independently in multiple clones and have different fitness costs, and, as a consequence, different probabilities of fixation. Apart from this, novel genetic variants may be fixed in the population by the mechanism of hitchhiking or independently [10]. A comprehensive review of these mutations has been recently published and demonstrated that most of these variants occurred in the genes, associated with the compensatory mechanisms, virulence, or relapse and survival, but their consequences remain unclear [11].
At the same time, commonly revealed mutations do not reflect all changes occurring in the cell [12]. Recently, a total of 139 genes were shown to be differentially expressed among serial isolates, while only 15 mutations were different between the strains [10]. In another work, the abundance of 46 proteins differed between resistant and sensitive strains. [13]. The in vitro experiments studying the effect of anti-tuberculosis drugs on bacteria have shown changes in the abundance of proteins related to amino acid, carbohydrate, and nucleotide metabolism [14].
In the present study, for the first time the author provided multi-omics analysis of three consecutive M. tuberculosis isolates from the same patient, to evaluate the changes in bacterial metabolism during anti-tuberculosis therapy. Based on whole genome sequencing (WGS) data, a stepwise accumulation of polymorphisms, explaining the occurrence of phenotypic resistance to fluoroquinolones and high concentrations of isoniazid, was determined. Moreover, at the proteomic and transcriptomic levels, changes were found in the loci associated with drug-resistance and virulence that only partly can be explained by mutations in the corresponding gene regions.
2. Results and Discussion
2.1. Clinical Isolates Characteristic
Three isolates from the patient with pulmonary tuberculosis have been subjected to multi-omics analysis. The first sample, RUS_B0, was isolated in 2008 before receiving TB chemotherapy. The second one, isolate 3955, was obtained one year after the treatment with high-doses of isoniazid, para-aminosalicylic acid (PASK), capreomycin, cycloserine, levofloxacin, pyrazinamide, azithromycin, clarithromycin, and amoxicillin with clavulanic acid. The 2093 sample was isolated after three years of treatment (surgery and chemotherapy according to the individual regimen).
Drug-susceptibility testing (DST) analysis has shown that the RUS_B0 isolate was MDR, while both 3955 and 2093 isolates were XDR. Phenotypic resistance to FQ evolved in the 3955 isolate after a year of treatment and was retained in the isolate 2093. Additionally, the 2093 isolate showed resistance to higher doses of isoniazid (10 mg/L).
Based on genotyping analysis, strains under study belonged to the Beijing B0/W148 cluster. Both IS6110 RFLP and 24-loci MIRU-VNTR typing have not revealed any changes in 3955 and 2093 strains relative to RUS_B0.
2.2. Intra-host Genome Evolution of M. tuberculosis Serial Isolates
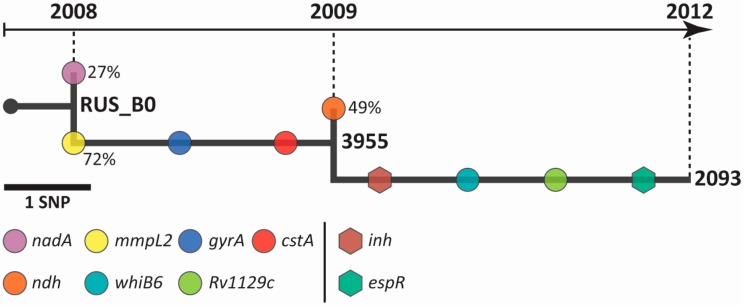
Three isolates of M. tuberculosis from a single patient, collected during different time points of treatment, were sequenced at a median depth of 383x coverage. Allele frequency of > 25% was taken as a threshold for sub-populations differentiation. In comparison to H37Rv, nine single nucleotide polymorphisms (SNPs) were variable among selected serial isolates, and seven of them were fixed in the last sample (Figure 1, Table S1).
Figure 1.
M. tuberculosis genome evolution within the patient. Sample collection time points are indicated by the numbers above the arrow. Circles and hexagons represent mutations in genes and promoter regions, respectively. Variant allele frequency is shown by the numbers beside circles.
The initial isolate RUS_B0 consisted of two populations carrying specific polymorphisms. The dominant population (72% of reads) carried synonymous SNP in the mmpL2 gene (pos. 598098c-t; I300I), while the minor population had a substitution P287S in the NadA protein (pos. 1795614c-t).
On the second time point (12 weeks, isolate 3955), the predominant population has been ultimately fixed (all reads had a mutation in mmpl2) in the sample, as well as additional polymorphisms appeared: 3427440g-t (P66P) and 7582a-c (D94A) in the cstA and gyrA gene, respectively. As in the first case, the isolate 3955 consisted of two populations. One of them (49% of reads) had a 2102714g-a mutation in the ndh gene, leading to the T110I substitution. Of note, another substitution in this codon, T110A, was previously described as associated with isoniazid resistance, but it was found only in addition to the ahpC mutation [15]. Although the presence of this mutation did not affect the isoniazid resistance of the 3955 isolate, it was assumed that it was caused by the high-doses of INH used during the treatment.
It was also observed that the sub-population with a mutation in the ndh gene has not been fixed in the patient and in the third time point, isolate 2093, all reads corresponded to the reference. Nevertheless, a novel mutation (inhA t-8a), associated with resistance to INH, has emerged in this isolate. In addition to the drug resistance-associated mutations, isolate 2093 carried SNPs in Rv1129c (1253465c-t; S357N) and whiB6 (4338344a-g; W60R) genes, as well as in the promoter region of the espR gene (c-90t).
2.3. Analysis of Drug Resistance Genes on Multi-omics Level
To evaluate the changes in bacterial metabolism during anti-tuberculosis therapy, multi-omics analysis of the serial strains was performed. The transcriptomic analysis resulted in the identification and quantification of 3889, 3894, and 3855 transcripts (TPM ≥ 1) in the RUS_B0, 3955, and 2093 strains, respectively (Table S2). According to the proteome analysis, 1941, 1728, and 1989 proteins were identified for the RUS_B0, 3955, and 2093 strains, respectively (Table S3). For quantitative proteomic analysis, 1358 proteins identified in all three strains were used. Over- and underrepresented proteins for each pair of strains are presented in Table S4.
According to DST results, all strains had the same following drug resistance-associated mutations: STR (RpsL: K43R), INH (KatG: S315T), RIF (RpoB: S450L), EMB (EmbB: Q497R), ETH (ethA: 110delA), KAN, CAP, AM (rrs: a1401g), PZA (pncA: t-11c) (Table 1). On the transcriptomic and proteomic levels, differences in the rpoB, pncA, embB genes and related proteins, as well as in the rrs gene, were not observed between the strains. Proteomic analysis revealed that in 2093 samples, abundance of RpsL and seven other ribosome proteins (Rv0055, Rv0714, Rv0720, Rv0722, Rv0979A, Rv1298, and Rv2412) was lower compared to the parental strains. This finding may be related to the fact that ribosomal proteins can be targeted by anti-tuberculosis drugs [16], and, particularly, by pyrazinamide, which was used for the treatment.
Table 1.
Results of phenotypic and genetic drug susceptibility for serial strains.
| Drug* (Resistance Associated Gene/Protein) | Clinical Isolates | ||
| RUS_B0 | 3955 | 2093 | |
| STR (Rpsl) | R (K43R) | R (K43R) | R (K43R) |
| INH (KatG) (inhA) |
R (S315T) | R (S315T) | R (S315T) R+** (t-8a) |
| RIF (RpoB) | R (S450L) | R (S450L) | R (S450L) |
| EMB (EmbB) | R (Q497R) | R (Q497R) | R (Q497R) |
| ETH (ethA) | R (110delA) | R (110delA) | R (110delA) |
| FQ (GyrA) | S | R (D94A) | R (D94A) |
| KAN, CAP, AM (rrs) | R (a1401g) | R (a1401g) | R (a1401g) |
| PZA (pncA) | R (t-11c) | R (t-11c) | R (t-11c) |
*STR - streptomycin, INH - isoniazid, RIF - rifampicin, EMB - ethambutol, ETH - ethionamide, FQ - fluoroquinolone, KAN - kanamycin, CAP - capreomycin, AM- amikacin, PZA - pyrazinamide; R - resistant, S - susceptible. **high-dose resistance (MIC=10mg/L).
For the ETH resistance-associated gene ethA carrying a frameshift mutation in the coding sequence, a reduction in gene expression during the treatment was found. Previously, the author has shown an increased level of ethA transcripts in the RUS_B0 (almost eight times) compared to the H37Rv. Its open reading frame contains numerous frame-shifts and stop codons precluding protein synthesis. The author hypothesized that MymA (Rv3083) can partly substitute the function of the inactivated gene [17]. In the present study, an up-regulation of mymA gene in 3955 strain compared to RUS_B0 was detected.
Starting from the second time point, the studied isolate 3955 acquired a D94A substitution in GyrA, which was also found in the 2093 isolate (Figure 1, Table 1). This mutation linked with resistance to FQ and, thus, correlates with the DST results. According to the RNA-seq and proteomics data, the levels of the gyrA transcript and correspondent protein were the same in all the strains. However, another gyrase subunit, GyrB, was overrepresented in the 2093 isolate.
The isoniazid-resistant strains carrying the S315T substitution in the KatG have acquired an additional mutation in the promoter region of the inhA gene (isolate 2093) (Figure 1, Table 1). Recently, it was shown that combination of KatG S315T and inhA c-15t is associated with high-level resistance (MIC ≥ 19.2 mg/L) to INH [18]. According to our study, the isolate 2093, harboring KatG S315T and inhA t-8a, was also resistant to the high drug concentrations (MIC=10 mg/L). In turn, the whole transcriptome analysis of the mabA – inhA operon revealed an increased level (7-fold) of corresponding gene transcripts in the 2093 isolate compared to the RUS_B0. It is known that INH resistance is associated with hyperproduction of the target InhA protein [19]. However, at the proteomic level, only an overabundance of MabA protein was found (changes in InhA were not detected). As mentioned above, the 2093 strain was resistant to higher concentrations of isoniazid than the RUS_B0 and 3955. InhA and MabA are both functionally and structurally related. Thus, MabA activity is also inhibited by isoniazid [20]. All of this may be evidence of high resistance of the bacterial cells.
Previously, significant differences in the levels of the katG and hspX transcripts in the INH-resistant and susceptible strains have been shown [13]. According to our data, the level of katG transcript was the same, while the expression of the hspX was higher in the 2093 and 3955 strains, compared to the RUS_B0. The abundance of KatG protein was the same after one year of therapy, but increased in the 2093 strain, which is consistent with the data, previously obtained for the INH-resistant Beijing isolates [13]. Regarding the HspX protein, its increased representation in the 3955 strain compared to RUS_B0, which went down in the strain 2093, was observed. Previously, it was proposed that HspX can be used as a marker of the disease, including its latent form [21,22]. A reduced representation of this protein in the resistant strain casts doubts on its usefulness.
2.4. Non-Specific Bacterial Response to Anti-tuberculosis Therapy
A number of studies have shown that mutations, acquired during treatment, do not reflect all metabolic changes occurring in the cell [10,12]. In our study, an increased level of the Rv2971 protein in the 2093 strain compared to the RUS_B0 and 3955 was determined. This protein is an oxidoreductase that is necessary for the growth of mycobacteria and has a potential role in detoxification of toxic metabolites. Previous studies have shown that this enzyme was differentially represented between INHr and INHs strains [23,24] and for STR [25]. In the present study, higher transcript levels of Rv2971 in the 2093, compared to other strains, were detected. Taken together, these may reflect the strain’s resistance to high-doses of isoniazid.
Previously, the author documented an increased abundance of proteins involved in lipid metabolism, in particular, those responsible for the biosynthesis of long-chain fatty acids, in strains recovered after anti-TB treatment [26]. Another study has identified differences in the abundance of Rv0241 (HtdX) and Rv3389 (HtdY) that belongs to the FAS-II system, and Rv0242c (FabG4) related to FAS-I in the INH-resistant and susceptible strains [13]. Here, the increased representation of both FAS-II system proteins in the 2093 strain, as compared to the RUS_B0, is demonstrated. However, no changes in the abundance of the FabG4 fatty acid synthase were detected. Increased representation of the Rv0469 (UmaA) protein, which is responsible for the synthesis of mycolic acids, may facilitate the formation of the cell membrane. This can prevent penetration of the anti-TB drugs, lowering their intracellular concentration and, thus, establishing favorable conditions for the survival of the pathogen and subsequent development of drug resistance. It has been frequently suggested that enzymes of the lipid metabolism play an important role in the positive selection of mycobacteria [11], and fatty acids can be considered as virulence factors of these pathogens [17,26].
2.5. Variability in the Virulence Factors Representation
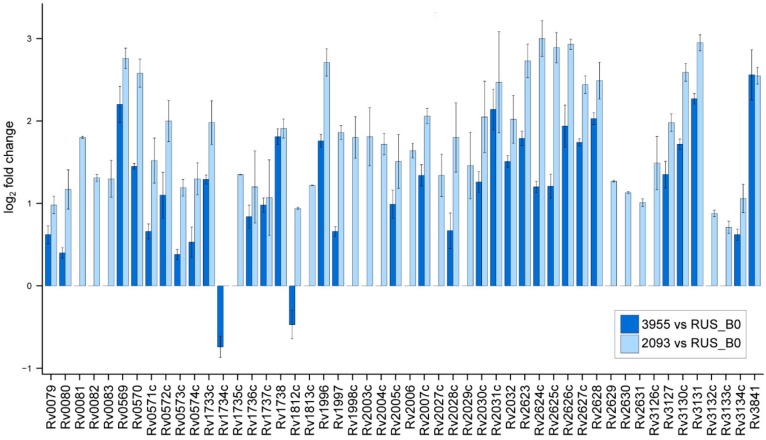
It is well known that the DosR system is involved in the response of mycobacteria to the action of the host cell immune system and to the changing environmental conditions [27,28]. The author has previously shown that under normal growth conditions the DosR regulon proteins were poorly represented in the Beijing B0/W148 cluster strains compared to the H37Rv. This is correlated with an increased abundance of the same proteins and corresponding transcripts in cluster strains under stress conditions [17]. In the present study, a statistically significant increase in the transcription levels of the DosR regulon genes on the background of anti-tuberculosis therapy was observed (Figure 2). The level of all DosR regulon genes has increased in the 3955 and 2093 compared to the RUS_B0 strain, except for the Rv1734c and Rv1812c genes, of which transcription levels were lower in the 3955 strain.
Figure 2.
DosR regulon expression. Bars indicate fold changes in gene expression in 3955 (blue bars) and 2093 (light blue bars) strains compared to the RUS_B0 strain. Error bars depict standard error of the mean from three independent experiments.
A mutation in the promoter region of espR was only detected in the 2093 strain. It has led to a decrease in the protein abundance and transcript level (albeit, not statistically significant). The virulence phenotype of bacteria is often implemented through protein hyperproduction in vivo [29], which is mediated by some intermediate activators. This also explains the low representation of the EspR and EspC proteins in the analyzed strain.
In the present study, a low abundance of the PtpA protein after the anti-TB therapy in the 2093 strain was observed. According to the transcriptome analysis, expression of the ptpA was higher in the 2093 strain. Concordantly, representation of the bifunctional protein Rv2228c, which is associated with PtpA, was higher. At the same time, the level of the Rv2228c transcript was higher in the 3955 strain, compared to the RUS_B0. It is known that PtpA (Rv2234) and PtpB (Rv0153c) are secreted proteins, which play an important role in the pathogen interaction with the host cell [30] and have orthologs among other pathogenic pro- and eukaryotes [31]. Rv2228c is presumably involved in bacterial replication, as it is only authenticated RNase HI in M. tuberculosis.
3. Materials and Methods
3.1. Mycobacterium Tuberculosis Strains and Growth Conditions
Three strains (RUS_B0, 3955, 2093) of Mycobacterium tuberculosis Beijing B0/W148 cluster from the same patient were used. Strains were grown in liquid Middlebrook 7H9 medium with Oleic Albumin Dextrose Catalase (OADC) supplement (Becton Dickinson, Franklin Lakes, USA). The cultures (90 mL) were grown in three biological replicates per strain in a cell culture flask (T175, Eppendorf, Germany) in a horizontal position. M. tuberculosis strains were incubated at 37°C for 12 days with constant shaking (5 rpm) until OD600 was reached to 0.4 [32]. The bacterial suspension from each flask was immediately aliquoted in a sterile 50 mL (for transcriptome and proteome analysis) and 15 mL (for WGS) tubes at room temperature (RT).
For WGS, an aliquot (10 mL) was centrifuged at 3200g for 10 min (RT) and the pellet was stored at -20 °C. Samples (40 mL) for transcriptomic analysis were centrifuged at 3200g for 10 min (RT) and cells pellet was frozen in liquid nitrogen and stored at −80 °C until use. For proteomic analysis, cells (40 mL of culture) were harvested by centrifugation at 3500g, 4°C for 5 min and washed with Tris-HCl and 2% Triton-X100 (pH 7.5–8). Cells were precipitated by centrifugation at 4500g, 4°C for 15 min. The pellet was frozen in liquid nitrogen and stored at −80 °C until required.
The susceptibility testing was done using the BACTEC MGIT 960 Culture system (Becton Dickinson) following the manufacturer’s protocol.
3.2. Genomic Analysis
Genomic DNA was isolated from the strains using standard extraction methods [33]. To verify that the strains belong to a Beijing B0/W148 cluster, the PCR assay was performed as described previously [34]. Spoligotyping, IS6110-RFLP and 24-MIRU-VNTR typing were performed as described in [35], [33], and [36], respectively.
Whole genome sequencing of the strains was performed on Illumina HiSeq 2500 Sequencing Platform according to the manufacturer’s instructions. Raw data were deposited in the NCBI Sequence Read Archive under accession number PRJNA421323. The circular genome of RUS_B0 strains was assembled and deposited in the NCBI earlier (CP030093.1) [37]. WGS reads were aligned to the M. tuberculosis H37Rv (NC_000962.3) and RUS_B0 (CP030093.1) genome sequences using Bowtie 2 [38]. SAMtools (v.0.1.18) [39], FreeBayes (v.1.1.0) [40] and Snippy (v.4.3.6) [41] were used for variant calling [39,42]. For FreeBayes and Snippy SNPs with a minimum mapping quality of 20, minimum coverage of 10 and alternate fraction of 0.9 were taken.
A comprehensive list of drug-resistance mutations to first- and second-line drugs were used to determine genetically resistant strains [43]. The identification of IS6110 integration sites was carried out using ISMapper pipeline [44].
3.3. Transcriptomic Analysis
3.3.1. RNA Extraction
Total RNA was isolated from all M. tuberculosis cultures as previously described [32]. In brief, bacteria were re-suspended in 1 mL Trizol (Invitrogen) and added to a 2-mL Lysing Matrix B (MP Bio, USA). Cells were disrupted by bead-beating twice for 1 min with a 2-min interval on ice. The suspension was then transferred to a new tube, where chloroform extraction was performed. RNA was precipitated by adding 0.7 volume of isopropanol and washed with 70% ethanol, air-dried and re-suspended in 100 µL DEPC-treated water.
DNase treatment was carried out with TURBO DNA-free kit (Thermo Fisher Scientific) in volumes of 100 µL and further with the RNase-Free DNase Set (Qiagen, Germany) according to the manufacturer’s protocol. RNA cleanup was performed with the RNeasy Mini Kit (Qiagen) according to the RNA Cleanup protocol and stored at −70°C until further use. DNA contamination was evaluated by PCR, using primers for amplification of the IS6110 fragment (PolyTub, Lytech, Russia). The concentration and quality of the total extracted RNA were checked by using the Quant-it RiboGreen RNA assay (Thermo Fisher Scientific) and the RNA 6000 pico chip (Agilent Technologies), respectively.
3.3.2. RNA-seq and Analysis
Total RNA (1 - 2.5 µg) was used for library preparation. Ribosomal RNA was removed from the total RNA and libraries were prepared using the ScriptSeq Complete kit (Epicentre/Illumina, Madison, USA), according to the manufacturer’s protocol. Subsequently, RNA cleanup was performed with the Agencourt RNA Clean XP kit (Beckman Coulter, Brea, USA). The library underwent a final cleanup using the Agencourt AMPure XP system (Beckman Coulter) after which the libraries’ size distribution and quality were assessed using a high sensitivity DNA chip (Agilent Technologies). Libraries were subsequently quantified by Quant-iT DNA Assay Kit, High Sensitivity (Thermo Fisher Scientific). Finally, equimolar quantities of all libraries (12 pM) were sequenced by a high throughput run on the Illumina HiSeq using 2×125 bp paired-end reads and a 5% Phix spike-in control. Before loading the cBot system, the libraries were incubated at 98°C for 2 minutes and then cooled on ice to improve the hybridization of the GC-rich sequences. In total, 114 and 110 million paired reads were obtained corresponding to 14 and 13 billion nucleotide bases for H37Rv and RUS_B0, respectively. The dataset of RNA-Seq analysis was deposited to the NCBI with the project name PRJNA421323.
Adaptors were trimmed with the Trimmomatic v0.33 tool [45]. Quality control on raw reads was carried out with FASTQC v0.11.5 [46]. The Kallisto v0.46.0 [47] software was used for the reads mapping and abundance estimation. Differential expression analysis was performed using edgeR v3.26.8 [48] package, integrated in the Degust v4.1.1 [49] web-tool. Only genes with count per million (CPM) ≥ 1 were analyzed further. Genes were filtered based on false discovery rate cutoff (FDR) ≤ 0.05 and minimum expression fold change (FC) ≥ 2. The plots were generated using the ggplot2 package in R.
3.4. Proteomic Analysis
3.4.1. Protein Extraction and Trypsin Digestion
The bacterial pellet was lysed and protein was extracted as described previously [17]. Protein concentration was measured by the Bradford method [50] using the Bradford Protein Assay Kit (Bio-Rad, Hercules, USA).
Proteolytic in-gel digestion was performed in three biological and two technical replicates: 1) sample, fractionated into 6 parts; 2) total load sample as described previously [51]. Peptides were cleaned using C18 Sep-Pak columns (Waters, Milford, USA) [17].
3.4.2. LC-MS/MS Analysis
Peptides were separated with high-performance liquid chromatography (HPLC, Ultimate 3000 Nano LC System, Thermo Fisher Scientific,) in a 15-cm long C18 column with a diameter of 75 μm (Acclaim® PepMap™ RSLC, Thermo Fisher Scientific). The peptides were eluted with a gradient from 5% to 35% of buffer B (80% acetonitrile, 0.1% formic acid) over 115 min at a flow rate of 0.3 μl/min. Total run time including 15 min to reach 99% buffer B, flushing 10 min with 99% buffer B and 15 min re-equilibration to buffer A (0.1% formic acid) amounted to 65 min. Further analysis was performed with a Q Exactive HF mass spectrometer (Q ExactiveTM HF Hybrid Quadrupole-OrbitrapTM Mass spectrometer, Thermo Fisher Scientific). Mass spectra were acquired at a resolution of 60,000 (MS) and 15,000 (MS/MS) in a range of 400-1500 m/z (MS) and 200-2000 m/z (MS/MS). An isolation threshold of 67,000 was determined for precursor selection, and up to the top 25 precursors were chosen for fragmentation with high-energy collisional dissociation (HCD) at 25 V and 100 ms activation time. Precursors with a charged state of +1 were rejected and all measured precursors were excluded from measurement for 20 s. From 2 to 4 technical runs were analyzed for each sample.
3.4.3. Protein Identification and Quantitation
Raw data was captured from the mass spectrometer and converted to MGF (Mascot Generic Format) files using ProteoWizard with the following parameters: peakPicking true 2, msLevel 2, zeroSamples removeExtra [52]. For thorough protein identification, the generated peak lists were searched with the MASCOT (v 2.5.1, Matrix Science Ltd, UK) and X! Tandem (VENGEANCE, 2015.12.15, The Global Proteome Machine Organization) search engines. Database-searching parameters were as follows: tryptic hydrolysis, no more than one missed site, the precursor and fragment mass tolerance were set at 20 ppm and 50 ppm, respectively. Oxidation of methionine was set as a possible modification, carbamidomethylation of cysteine as a fixed. For X! Tandem, parameters that allowed a quick check for protein N-terminal residue acetylation, peptide N-terminal glutamine ammonia loss or peptide N-terminal glutamic acid water loss were selected. Resulting files were submitted to the Scaffold 4 software (v 4.2.1, Proteome Software, Inc, USA) for validation and further analysis. For protein identification, the proteomic databases for the RUS_B0 (RefSeq: CP030093.1) and reference strain H37Rv (RefSeq: NC_000962.3) genomes were used. Additionally, in proteome searching database peptides containing strain-specific single amino acid polymorphisms were added according to the approach described earlier [51]. The local false discovery rate scoring algorithm with standard experiment-wide protein grouping was used. A 1% FDR threshold was applied to search results from individual datasets. For all detected proteins, functional categories (TubercuList v 2.6 (http://tuberculist.epfl.ch/)) and subcellular localizations (PSORTdb v 3.0 (http://db.psort.org/)) were established.
For label-free quantitation, raw MS data files (.wiff files) were imported and processed in Progenesis LC-MS software v.4.1 (Nonlinear Dynamics, Newcastle, UK). The results of peptide quantitation were normalized using an iterative median-based normalization as implemented in the Progenesis software. Differences in the abundance of a protein between the three biological replicates of all strains were evaluated using a two-sided unpaired Student’s T-test. P-values < 0.05 were considered statistically significant. Adjusted p-values for multiple tests (q-values) were generated using the Benjamini–Hochberg method [53].
4. Conclusions
This is a first report of the system omics analysis of M. tuberculosis Beijing B0/W148 cluster strains isolated from one patient at different stages of TB therapy. A clear correlation between genetic and phenotypic resistance to the antibiotics was demonstrated, documenting changes in the lipid metabolism, and production of virulence factors on the proteomic and transcriptomic levels. These changes imply that the anti-tuberculosis therapy play a crucial role in the regulation of bacterial biochemical pathways.
We show that integrating various approaches can advance our knowledge of the role and significance of microevolution in M. tuberculosis infection.
Acknowledgments
We acknowledge the IBMC “Human Proteome” Core Facility for assistance with the generation of mass spectrometry data. We thank the Center for Precision Genome Editing and Genetic Technologies for Biomedicine, Federal Research and Clinical Center of Physical-Chemical Medicine of Federal Medical Biological Agency for providing the WGS platform.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-0817/9/2/131/s1. Table S1: Variable SNPs among tree clinical isolates. Table S2: List of identified and quantified transcripts for RUS_B0, 3955 and 2093 strains. Table S3: Proteins and peptides identified in RUS_B0, 3955 and 2093 strains. Table S4: Quantitative proteomic analysis of RUS_B0, 3955 and 2093 strains.
Author Contributions
Conceptualization, J.B. and E.S.; methodology, J.B., E.S., K.K., M.D. and V.V.; software, D.B., A.G., and G.A.; formal analysis, D.B., A.G., G.A., K.K., M.D. and V.V.; investigation, J.B., E.S.; resources, V.Z., E.I. and V.G.; data curation, J.B., E.S. and V.Z.; writing – original draft preparation, J.B. and E.S.; writing – review & editing, J.B, E.S. and D.B.; supervision, E.I.; project administration, V.G.; funding acquisition, J.B. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by Russian Foundation for Basic Research (project 18-34-00168).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.WHO . Weekly Epidemiological Record. Volume 95. WHO; Geneva, Switzerland: 2020. pp. 1–12. [Google Scholar]
- 2.WHO . Weekly Epidemiological Record No 45. Volume 81. WHO; Geneva, Switzerland: 2006. pp. 425–432. [Google Scholar]
- 3.WHO . Treatment of TB: Guidelines for National Programmes. WHO; Geneva, Switzerland: 1997. WHO/TB/97.220. [Google Scholar]
- 4.Calver A.D., Falmer A.A., Murray M., Strauss O.J., Streicher E.M., Hanekom M., Liversage T., Masibi M., Van Helden P.D., Warren R.M., et al. Emergence of increased resistance and extensively drug-resistant tuberculosis despite treatment adherence, South Africa. Emerg. Infect. Dis. 2010;16:264–271. doi: 10.3201/eid1602.090968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dye C. Doomsday postponed? Preventing and reversing epidemics of drug-resistant tuberculosis. Nat. Rev. Microbiol. 2009;7:81–87. doi: 10.1038/nrmicro2048. [DOI] [PubMed] [Google Scholar]
- 6.Müller B., Borrell S., Rose G., Gagneux S. The heterogeneous evolution of multidrug-resistant Mycobacterium tuberculosis. Trends Genet. 2013;29:160–169. doi: 10.1016/j.tig.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Didelot X., Walker A.S., Peto T.E., Crook D.W., Wilson D.J. Within-host evolution of bacterial pathogens. Nat Rev Microbiol. 2016;14:150–162. doi: 10.1038/nrmicro.2015.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun G., Luo T., Yang C., Dong X., Li J., Zhu Y., Zheng H., Tian W., Wang S., Barry C.E., et al. Dynamic population changes in Mycobacterium tuberculosis during acquisition and fixation of drug resistance in patients. J. Infect. Dis. 2012;206:1724–1733. doi: 10.1093/infdis/jis601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Merker M., Kohl T.A., Roetzer A., Truebe L., Richter E., Rüsch-Gerdes S., Fattorini L., Oggioni M.R., Cox H., Varaine F., et al. Whole genome sequencing reveals complex evolution patterns of multidrug-resistant Mycobacterium tuberculosis Beijing strains in patients. PLoS ONE. 2013;8:e82551. doi: 10.1371/journal.pone.0082551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eldholm V., Norheim G., von der Lippe B., Kinander W., Dahle U.R., Caugant D.A., Mannsåker T., Mengshoel A.T., Dyrhol-Riise A.M., Balloux F. Evolution of extensively drug-resistant Mycobacterium tuberculosis from a susceptible ancestor in a single patient. Genome Biol. 2014;15:490. doi: 10.1186/s13059-014-0490-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ley S.D., de Vos M., Van Rie A., Warren R.M. Deciphering Within-Host Microevolution of Mycobacterium tuberculosis through Whole-Genome Sequencing: The Phenotypic Impact and Way Forward. Microbiol. Mol. Biol. Rev. 2019;83:e00062-18. doi: 10.1128/MMBR.00062-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davies A.P., Billington O.J., McHugh T.D., Mitchison D.A., Gillespie S.H. Comparison of phenotypic and genotypic methods for pyrazinamide susceptibility testing with Mycobacterium tuberculosis. J. Clin. Microbiol. 2000;38:3686–3688. doi: 10.1128/JCM.38.10.3686-3688.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.María Nieto L.R., Mehaffy C., Dobos K.M. Comparing isogenic strains of Beijing genotype Mycobacterium tuberculosis after acquisition of Isoniazid resistance: A proteomics approach. Proteomics. 2016;16:1376–1380. doi: 10.1002/pmic.201500403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Danelishvili L., Shulzhenko N., Chinison J.J.J., Babrak L., Hu J., Morgun A., Burrows G., Bermudeza L.E. Mycobacterium tuberculosis proteome response to antituberculosis compounds reveals metabolic “escape” pathways that prolong bacterial survival. Antimicrob. Agents Chemother. 2017;61:e00430-17. doi: 10.1128/AAC.00430-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee A.S.G., Teo A.S.M., Wang S.Y. Novel mutations in ndh in isoniazid-resistant Mycobacterium tuberculosis isolates. Antimicrob. Agents Chemother. 2001;45:2157–2159. doi: 10.1128/AAC.45.7.2157-2159.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Finken M., Kirschner P., Meier A., Wrede A., Böttger E.C. Molecular basis of streptomycin resistance in Mycobacterium tuberculosis: Alterations of the ribosomal protein S12 gene and point mutations within a functional 16S ribosomal RNA pseudoknot. Mol. Microbiol. 1993;9:1239–1246. doi: 10.1111/j.1365-2958.1993.tb01253.x. [DOI] [PubMed] [Google Scholar]
- 17.Bespyatykh J., Shitikov E., Butenko I., Altukhov I., Alexeev D., Mokrousov I., Dogonadze M., Zhuravlev V., Yablonsky P., Ilina E., et al. Proteome analysis of the Mycobacterium tuberculosis Beijing B0/W148 cluster. Sci. Rep. 2016;6:28985. doi: 10.1038/srep28985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lempens P., Meehan C.J., Vandelannoote K., Fissette K., De Rijk P., Van Deun A., Rigouts L., De Jong B.C. Isoniazid resistance levels of Mycobacterium tuberculosis can largely be predicted by high-confidence resistance-conferring mutations. Sci. Rep. 2018;8:3246. doi: 10.1038/s41598-018-21378-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Banerjee A., Dubnau E., Quemard A., Balasubramanian V., Um K.S., Wilson T., Collins D., De Lisle G., Jacobs W.R. inhA, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis. Science. 1994;263:227–230. doi: 10.1126/science.8284673. [DOI] [PubMed] [Google Scholar]
- 20.Ducasse-Cabanot S., Cohen-Gonsaud M., Marrakchi H., Nguyen M., Zerbib D., Bernadou J., Daffé M., Labesse G., Quémard A.A. In Vitro Inhibition of the Mycobacterium tuberculosis-Ketoacyl-Acyl Carrier Protein Reductase MabA by Isoniazid. Antimicrob. Agents Chemother. 2004;48:242–249. doi: 10.1128/AAC.48.1.242-249.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Castro-Garza J., García-Jacobo P., Rivera-Morales L.G., Quinn F.D., Barber J., Karls R., Haas D., Helms S., Gupta T., Blumberg H., et al. Detection of anti-HspX antibodies and HspX protein in patient sera for the identification of recent latent infection by Mycobacterium tuberculosis. PLoS ONE. 2017;12:e0181714. doi: 10.1371/journal.pone.0181714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dhiman A., Haldar S., Mishra S.K., Sharma N., Bansal A., Ahmad Y., Kumar A., Sharma T.K., Tyagi J.S. Generation and application of DNA aptamers against HspX for accurate diagnosis of tuberculous meningitis. Tuberculosis. 2018;112:27–36. doi: 10.1016/j.tube.2018.07.004. [DOI] [PubMed] [Google Scholar]
- 23.Jiang X., Zhang W., Gao F., Huang Y., Lv C., Wang H. Comparison of the proteome of isoniazid-resistant and -susceptible strains of Mycobacterium tuberculosis. Microb. Drug Resist. 2006;12:231–238. doi: 10.1089/mdr.2006.12.231. [DOI] [PubMed] [Google Scholar]
- 24.Phong T.Q., Ha D.T.T., Volker U., Hammer E. Using a Label Free Quantitative Proteomics Approach to Identify Changes in Protein Abundance in Multidrug-Resistant Mycobacterium tuberculosis. Indian J. Microbiol. 2015;55:219–230. doi: 10.1007/s12088-015-0511-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sharma P., Kumar B., Gupta Y., Singhal N., Katoch V.M., Venkatesan K., Bisht D. Proteomic analysis of streptomycin resistant and sensitive clinical isolates of Mycobacterium tuberculosis. Proteome Sci. 2010;8:59. doi: 10.1186/1477-5956-8-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bespyatykh J., Shitikov E., Zgoda V., Smolyakov A., Zamachaev M., Dogonadze M., Zhuravlev V., Ilina E. Changes in metabolism of Mycobacterium tuberculosis Beijing B0/W148 cluster against the background of anti-tuberculosis therapy. FEBS J. 2017;284(Suppl. 1):347–347. [Google Scholar]
- 27.Dasgupta N., Kapur V., Singh K.K., Das T.K., Sachdeva S., Jyothisri K., Tyagi J.S. Characterization of a two-component system, devR-devS, of Mycobacterium tuberculosis. Tuber. Lung Dis. 2000;80:141–159. doi: 10.1054/tuld.2000.0240. [DOI] [PubMed] [Google Scholar]
- 28.Converse P.J., Karakousis P.C., Klinkenberg L.G., Kesavan A.K., Ly L.H., Allen S.S., Grosset J.H., Jain S.K., Lamichhane G., Manabe Y.C., et al. Role of the dosR-dosS two-component regulatory system in Mycobacterium tuberculosis virulence in three animal models. Infect. Immun. 2009;77:1230–1237. doi: 10.1128/IAI.01117-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cao G., Howard S.T., Zhang P., Hou G., Pang X. Functional analysis of the EspR binding sites upstream of espR in Mycobacterium tuberculosis. Curr. Microbiol. 2013;67:572–579. doi: 10.1007/s00284-013-0404-8. [DOI] [PubMed] [Google Scholar]
- 30.Koul A., Choidas A., Treder M., Tyagi A.K., Drlica K., Singh Y., Ullrich A. Cloning and characterization of secretory tyrosine phosphatases of Mycobacterium tuberculosis. J. Bacteriol. 2000;182:5425–5432. doi: 10.1128/JB.182.19.5425-5432.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kraeva N., Leštinová T., Ishemgulova A., Majerová K., Butenko A., Vaselek S., Bespyatykh J., Charyyeva A., Spitzová T., Kostygov A.Y., et al. LmxM.22.0250-Encoded Dual Specificity Protein/Lipid Phosphatase Impairs Leishmania mexicana Virulence In Vitro. Pathogens. 2019;8:241. doi: 10.3390/pathogens8040241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Benjak A., Sala C., Hartkoorn R.C. Whole-transcriptome sequencing for high-resolution transcriptomic analysis in Mycobacterium tuberculosis. Methods Mol. Biol. 2015;1285:17–30. doi: 10.1007/978-1-4939-2450-9_2. [DOI] [PubMed] [Google Scholar]
- 33.Van Embden J.D., Cave M.D., Crawford J.T., Dale J.W., Eisenach K.D., Gicquel B., Hermans P., Martin C., McAdam R., Shinnick T.M. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: Recommendations for a standardized methodology. J. Clin. Microbiol. 1993;31:406–409. doi: 10.1128/JCM.31.2.406-409.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mokrousov I., Vyazovaya A., Zhuravlev V., Otten T., Millet J., Jiao W.W., Shen A.D., Rastogi N., Vishnevsky B., Narvskaya O. Real-time PCR assay for rapid detection of epidemiologically and clinically significant Mycobacterium tuberculosis Beijing genotype isolates. J. Clin. Microbiol. 2014;52:1691–1693. doi: 10.1128/JCM.03193-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bespyatykh J.A., Zimenkov D.V., Shitikov E.A., Kulagina E.V., Lapa S.A., Gryadunov D.A., Ilina E.N., Govorun V.M. Spoligotyping of Mycobacterium tuberculosis complex isolates using hydrogel oligonucleotide microarrays. Infect. Genet. Evol. 2014;26:41–46. doi: 10.1016/j.meegid.2014.04.024. [DOI] [PubMed] [Google Scholar]
- 36.Supply P., Allix C., Lesjean S., Cardoso-Oelemann M., Rüsch-Gerdes S., Willery E., Savine E., de Haas P., van Deutekom H., Roring S., et al. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J. Clin. Microbiol. 2006;44:4498–4510. doi: 10.1128/JCM.01392-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bespyatykh J., Shitikov E., Andrei G., Smolyakov A., Klimina K., Veselovsky V., Malakhova M., Georgij A., Dogonadze M., Manicheva O., et al. System OMICs analysis of Mycobacterium tuberculosis Beijing B0/W148 cluster. Sci. Rep. 2019;9:1–11. doi: 10.1038/s41598-019-55896-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramirez-Gonzalez R.H., Bonnal R., Caccamo M., Maclean D. Bio-samtools: Ruby bindings for SAMtools, a library for accessing BAM files containing high-throughput sequence alignments. Source Code Biol. Med. 2012;7:6. doi: 10.1186/1751-0473-7-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Garrison E., Marth G. Haplotype-based variant detection from short-read sequencing. arXiv. 20121207.3907 [Google Scholar]
- 41.Seemann T. GitHub-Tseemann/Snippy: Rapid Haploid Variant Calling and Core Genome Alignment. [(accessed on 23 January 2020)];2015 Available online: https://github.com/tseemann/snippy.
- 42.Nurk S., Bankevich A., Antipov D., Gurevich A., Korobeynikov A., Lapidus A., Prjibelsky A., Pyshkin A., Sirotkin A., Sirotkin Y., et al. Assembling Genomes and Mini-Metagenomes from Highly Chimeric Reads. Springer; Berlin/Heidelberg, Germany: 2013. pp. 158–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schleusener V., Köser C.U., Beckert P., Niemann S., Feuerriegel S. Mycobacterium tuberculosis resistance prediction and lineage classification from genome sequencing: Comparison of automated analysis tools. Sci. Rep. 2017;7:46327. doi: 10.1038/srep46327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hawkey J., Hamidian M., Wick R.R., Edwards D.J., Billman-Jacobe H., Hall R.M., Holt K.E. ISMapper: Identifying transposase insertion sites in bacterial genomes from short read sequence data. BMC Genom. 2015;16:667. doi: 10.1186/s12864-015-1860-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bolger A.M., Lohse M., Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Andrews S. FastQC: A Quality Control Tool for High Throughput Sequence Data. [(accessed on 24 January 2020)];Babraham Bioinform. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- 47.Bray N.L., Pimentel H., Melsted P., Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 48.Robinson M.D., Mccarthy D.J., Smyth G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Powell D.R. Degust: Interactive RNA-seq Analysis. [(accessed on 20 January 2020)]; Available online: http://victorian-bioinformatics-consortium.github.io/degust/
- 50.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 51.Bespyatykh J., Smolyakov A., Guliaev A., Shitikov E., Arapidi G., Butenko I., Dogonadze M., Manicheva O., Ilina E., Zgoda V., et al. Proteogenomic analysis of Mycobacterium tuberculosis Beijing B0/W148 cluster strains. J. Proteom. 2019;192:18–26. doi: 10.1016/j.jprot.2018.07.002. [DOI] [PubMed] [Google Scholar]
- 52.Kessner D., Chambers M., Burke R., Agus D., Mallick P. ProteoWizard: Open source software for rapid proteomics tools development. Bioinformatics. 2008;24:2534–2536. doi: 10.1093/bioinformatics/btn323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Benjamini Y., Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. 1995;57:289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.