Abstract
Loop‐mediated isothermal amplification (LAMP) assays are used to detect diverse pathogens. Initially, LAMP amplicons were detected using electrophoresis; later, real‐time monitoring based on turbidity was developed to overcome the problem of contamination with environmental DNA. Recently, real‐time monitoring of fluorescence signals using a quenching primer and probe has improved the reliability of amplification signals. Here, methods of detecting LAMP amplicons are reviewed.
Keywords: detection, LAMP, QPrimer, QProbe, turbidity
Abbreviations
- CoV
coronavirus
- LAMP
loop‐mediated isothermal amplification
- MERS
Middle East respiratory syndrome
- Qprimer
quenching primer
- Qprobe
quenching probe
- RSV
respiratory syncytial virus
- RT
reverse transcription
1. INTRODUCTION
The loop‐mediated isothermal amplification (LAMP) method was developed by Notomi et al. 1 and was patented by Eiken Chemical Co., Ltd (patent no. JP2000283862, WO2002024902A1). In LAMP assays, DNA is amplified by four specific primers (F3, FIP, BIP, and B3) at 60–65°C within 1 hr, although the speed of amplification can be accelerated by adding two loop primers (LF and LB).2 Thus, LAMP is rapid and user‐friendly; it can reduce the time required to increase and decrease the incubation temperature, facilitating the choice of incubation device, that is, LAMP can be performed using a simple isothermal device, such as heat block and water bath, and the amplification ends faster than general PCRs. The principles of LAMP are described on the manufacturer's website (http://loopamp.eiken.co.jp/e/lamp/principle.html) and as an animation (http://loopamp.eiken.co.jp/e/lamp/anim.html). LAMP assays can also be used to detect RNA by adding a reverse transcription step (RT‐LAMP).1 LAMP uses four or six primers, which assures high sequence specificity. The primers are also tolerant of point mutation(s) other than mismatches at the 3′ end of the primer and in the BIP primer.3, 4, 5, 6 Therefore, LAMP enables detection of pathogen genes. LAMP assays have been developed to detect gene of bacteria,7, 8, 9, 10, 11, 12, 13 parasites,14, 15, 16 protozoa,14, 17, 18 and viruses,19, 20, 21, 22 including RNA viruses such as West Nile virus,23 dengue virus,24 rubella virus,25, 26 mumps virus,27 respiratory syncytial virus (RSV),28, 29 influenza viruses,30, 31, 32 severe acute respiratory syndrome coronavirus,33 and Middle East respiratory syndrome‐coronavirus (MERS‐CoV).5, 6, 34, 35
2. TRANSITION OF LAMP DETECTION METHODS
2.1. Detection by electrophoresis
LAMP amplicons can be visualized by electrophoresis in an agarose gel. The four basic LAMP primers are constructed based on six specific regions of the target sequence and form a unique dumbbell‐like structure, both ends of which have stem–loop structures that each comprise three specific regions. The dumbbell‐like structure is extended in a bumper‐to‐bumper manner; therefore, the amplicons are ladder‐like in appearance because they contain various numbers of dumbbell‐like structures (Figure 1). The specificity of amplification can be confirmed by incorporating restriction enzyme sites. In our RT‐LAMP for RSV,28 the subtype A amplicon is cleaved by NlaIII and that of subtype B by XbaI. If the amplification succeeds, depending on the number of used enzymes and recognition sites in targeted sequences, several bands can be seen after digestion (Figure 1). Thus, detecting LAMP amplicons by electrophoresis and restriction enzymes can confirm the specificity of amplification. However, electrophoresis is performed under open conditions, leading to contamination of the laboratory with amplified DNA. The high sensitivity of genetic diagnostic methods increases the likelihood of false positives.36, 37 LAMP produces a larger quantity of DNA than conventional PCR (11 vs. 0.2 μg in a 25 μl reaction),38 suggesting that LAMP is more likely to result in contamination of the laboratory. To prevent this, separate reagent mixing and electrophoresis rooms with a one‐way workflow are needed.39 Furthermore, training of workers is needed, and the number of tests performed daily by one person should be limited to prevent contamination; however, this is time consuming and costly. The optimum means of avoiding laboratory contamination by amplified DNA is performing detection under closed conditions, that is, without opening the reaction tube.
Figure 1.
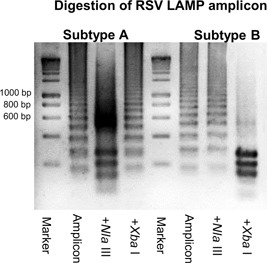
RSV RT‐LAMP was performed as described previously.28 The amplicons were purified using a QIAquick PCR purification kit and treated with NlaIII and XbaI at 37°C for 1 hr. The resulting fragments were separated by electrophoresis on 3% agarose gels and visualized using ethidium bromide staining and ultraviolet light
2.2. Detection by turbidity
Mori et al. 38 developed a turbidity‐based method for amplicon detection in a closed system, based on precipitation of magnesium pyrophosphate. The pyrophosphate ion is released from deoxynucleotide triphosphate as a by‐product of DNA polymerization by DNA polymerase, and precipitates with magnesium ion in the reaction buffer.40, 41 Because LAMP produces a large quantity of DNA, the pyrophosphate ion concentration exceeds the threshold for precipitation, resulting in the formation of a visible magnesium precipitate in an amplicon concentration‐dependent manner. By contrast, PCR yields a pyrophosphate concentration insufficient for magnesium pyrophosphate precipitation.38 The precipitate is visible at the end of amplification (Figure 2a) and the turbidity can be monitored in real‐time using a turbidimeter (Figure 2c).42 A fluorescence detection method using the fluorescent dye calcein has also been developed.43 Calcein, known as fluorexon, is quenched by manganese ions, of which it is deprived by pyrophosphate. Therefore, it fluoresces when accompanied by pyrophosphate ion production during the LAMP reaction and is visible to the naked eye under ultraviolet light (Figure 2b). The signal can be monitored using a real‐time fluorescence monitor. Turbidity monitoring enables the detection of targets under closed conditions without electrophoresis, as well as LAMP in the field and at point of care using an isothermal incubator, such as a heat block and water bath. LAMP kits for in vitro diagnosis are commercially available in Japan for whooping cough, tuberculosis, Mycoplasma pneumonia, Legionella pneumophila, influenza, and severe acute respiratory syndrome.
Figure 2.
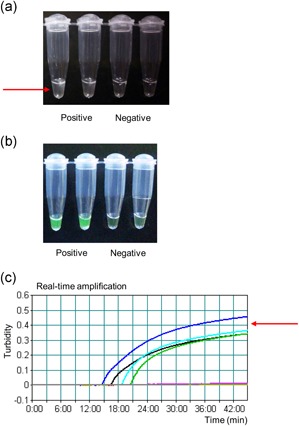
Detection of LAMP amplicons by turbidity. (a) MERS‐CoV RT‐LAMP was performed as described previously.5 A muddy white color indicates precipitation of magnesium pyrophosphate. Left two wells, positive; right two wells, negative. (b) Amplification was performed using calcein and visualized under ultraviolet light. Green fluorescence indicates positivity. (c) RSV RT‐LAMP was performed as described previously,28 and turbidity was monitored in real time. The signal is drawn as a power curve
However, the turbidity signal is derived from a by‐product of DNA amplification; it is not caused by a primer's specific reaction directly. Consequently, it is not possible to completely exclude the possibility of monitoring nonspecific amplification; salt accumulation accompanying the LAMP reaction can be induced even by a nonprimer signal or primer dimers. Thus, even if host‐derived DNA fragments or primer dimers form DNA amplification nonspecifically, the increased turbidity that accompanies this process can be monitored as a positive signal. Although rare, the possibility of nonprimer signals in turbidity monitoring cannot be excluded. Hence, validation of the integrity of the primer set is quite important to develop a LAMP system with turbidity monitoring. The primers for LAMP can be designed using online software (PrimerExplorer v. 4 or 5, Fujitsu, Tokyo, Japan; http://primerexplorer.jp/e/). However, the search results do not guarantee primer specificity. Candidates should be screened by strict validation tests to check whether they ever show nonspecific reactions in the presence of host and/or carrier nucleotides and the primer set itself. The primer set can be used for further validations only after it has been shown that it never causes nonspecific DNA amplification.
3. DETECTION USING FLUORESCENT PRIMERS/PROBES
Labeling primers with a fluorescent dye ensures detection of only primer‐derived signals. Takayama et al. developed fluorescence RT‐LAMP using a quenching primer (QPrimer)44 to detect influenza virus and RSV,45 and Nakauchi et al. developed QPrimer RT‐LAMP for rhinoviruses.46 In this system, a cytosine or guanine residue at the 5′ end of the loop primer is labeled with BODIPY (boron‐dipyrromethene) dye and the labeled and unlabeled loop primers are mixed at a 1:10 ratio. Upon annealing to its target sequence, QPrimer fluorescence is quenched by the paired guanine or cytosine residue (Figure 3). Therefore, the positive signal can be detected as a reverse sigmoid curve using an isothermal fluorescence reader, such as a real‐time PCR instrument (Figure 3). In addition, use of QPrimers reduces the LAMP detection time by several minutes relative to turbidity detection, and positive signals are typically detected within 20 min.45 Although QPrimer can resolve concerns regarding detection of primer‐derived signals directly, nonspecific DNA extension at the annealed 3′ end and/or primer dimers can result in false positives, so strict screening for primer integrity is still necessary. To overcome this, LAMP using a quenching probe (QProbe)44 was developed.6 In QProbe LAMP, the 3′ end of one of the loop primers is labeled with BODIPY dye (Figure 3); the QProbe must be designed to be several nucleotides longer than the original loop primer toward the 3′ end. Table 1 shows the sequences of QProbes used in QProbe RT‐LAMP for MERS‐CoV.6 The sequences in bold are extended relative to the loop primer, and are responsible for the specific reaction, because the sequence exists only in the target sequence and the specific amplicon. Although this makes QProbes more difficult to design than QPrimers, the labeled dye physically blocks DNA extension at the 3′ end, preventing nonspecific DNA amplification by a labeled primer and enhancing the accuracy of the signal.
Figure 3.
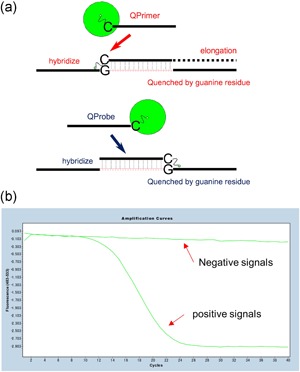
Schematic diagrams of a QPrimer and QProbe. (a) The cytosine or guanine residue is labeled with BODIPY. The QPrimer and QProbe fluoresce ordinarily. Upon annealing to their target sequence, their complementary guanine or cytosine residue quenches the fluorescence. (b) The signal can be detected as a reverse sigmoid curve using a fluorescence meter
Table 1.
Sequences of the QProbes used to detect MERS‐CoV
| For N | Sequence (5′–3′) |
|---|---|
| LB primer | GGAACCCTAACAATGATTCAGCT |
| Qprobe | GGAACCCTAACAATGATTCAGCTATTGTTACAC |
| For ORF1a | Sequence (5′–3′) |
| LB primer | GGTCACTCAAATTGCTAACATG |
| Qprobe | GGTCACTCAAATTGCTAACATGTTCTTGGAACAGAC |
The sequences in bold are extended relative to the loop primer.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
Fluorescence detection requires a fluorometer, which prevents naked‐eye endpoint detection. However, portable isothermal fluorometers enable fluorescence detection in the field. For example, the ESEQuant TS2 (Qiagen, https://www.qiagen.com/jp/products/custom-solutions/automation/ese-instruments/esequant-ts2/) is a 12 well isothermal tube scanner that has a built‐in battery pack, and so can be used outdoors. Furthermore, a lyophilized RT‐LAMP reaction mixture in a 12 well tube reduces the time required for MERS‐CoV detection.6 In other words, the lyophilized reagent contains all necessary components for RT‐LAMP, and all it takes is just to add extracted RNA and water, and it can reduce the time for regent preparation. Thus, lyophilized reagents and portable fluorescence LAMP devices enable its use in field surveillance as an accurate diagnostic tool.
Although each detection method has advantages and disadvantages, overall, LAMP is a superior assay. Its high sensitivity, specificity, and rapidity are advantageous for use in clinical laboratories compared with rapid test kits based on antigen detection. Indeed, the sensitivity of LAMP is superior to that of rapid test kits based on immunochromatography.28, 29 Thus, LAMP may replace such kits in the future.
ACKNOWLEDGMENTS
I thank Mr Shohei Semba of the Eiken Chemical Co., Ltd. and Dr Ikuyo Takayama, Dr Mina Nakauchi, and Dr Tsutomu Kageyama of the Influenza Virus Research Center, National Institute of Infectious Diseases for technical support. This work was supported by a Grant‐in‐Aid (Research Program on Emerging and Re‐emerging Infectious Diseases, No. 18fk0108030j0402) from the Japan Agency for Medical Research and Development (AMED), and a Grant‐in‐Aid for Scientific Research (C: 17K10133) from the Japan Society for the Promotion of Science.
DISCLOSURE
The author declares that he has no competing interests.
Shirato K. Detecting amplicons of loop‐mediated isothermal amplification. Microbiology and Immunology. 2019;63:407‐412. 10.1111/1348-0421.12734
References
REFERENCES
- 1. Notomi T, Okayama H, Masubuchi H, et al. Loop‐mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:63e‐63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nagamine K, Hase T, Notomi T. Accelerated reaction by loop‐mediated isothermal amplification using loop primers. Mol Cell Probes. 2002;16:223‐9. [DOI] [PubMed] [Google Scholar]
- 3. Peyrefitte CN, Boubis L, Coudrier D, et al. Real‐time reverse‐transcription loop‐mediated isothermal amplification for rapid detection of rift valley Fever virus. J Clin Microbiol. 2008;46:3653‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang D. Effect of internal primer–template mismatches on loop‐mediated isothermal amplification. Biotechnol Biotechnol Equip. 2016;30:314‐18. [Google Scholar]
- 5. Shirato K, Yano T, Senba S, et al. Detection of Middle East respiratory syndrome coronavirus using reverse transcription loop‐mediated isothermal amplification (RT‐LAMP). Virol J. 2014;11:139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shirato K, Semba S, El‐Kafrawy SA, et al. Development of fluorescent reverse transcription loop‐mediated isothermal amplification (RT‐LAMP) using quenching probes for the detection of the Middle East respiratory syndrome coronavirus. J Virol Methods. 2018;258:41‐48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Adhikari BR, Pandey BD, Ghimire P, et al. Loop‐mediated isothermal amplification (LAMP) for the direct detection of human pulmonary infections with environmental (nontuberculosis) mycobacteria. Jpn J Infect Dis. 2009;62:212‐4. [PubMed] [Google Scholar]
- 8. Ueda S, Kuwabara Y. The rapid detection of salmonella from food samples by loop‐mediated isothermal amplification (LAMP). Biocontrol Sci. 2009;14:73‐6. [DOI] [PubMed] [Google Scholar]
- 9. Geojith G, Dhanasekaran S, Chandran SP, Kenneth J. Efficacy of loop mediated isothermal amplification (LAMP) assay for the laboratory identification of Mycobacterium tuberculosis isolates in a resource limited setting. J Microbiol Methods. 2011;84:71‐3. [DOI] [PubMed] [Google Scholar]
- 10. Gotoh K, Nishimura N, Ohshima Y, et al. Detection of Mycoplasma pneumoniae by loop‐mediated isothermal amplification (LAMP) assay and serology in pediatric community‐acquired pneumonia. J Infect Chemother. 2012;18:662‐7. [DOI] [PubMed] [Google Scholar]
- 11. Kamachi K, Toyoizumi‐Ajisaka H, Toda K, et al. Development and evaluation of a loop‐mediated isothermal amplification method for rapid diagnosis of Bordetella pertussis infection. J Clin Microbiol. 2006;44:1899‐902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Neonakis IK, Gitti Z, Krambovitis E, Spandidos DA. Molecular diagnostic tools in mycobacteriology. J Microbiol Methods. 2008;75:1‐11. [DOI] [PubMed] [Google Scholar]
- 13. Inoue H, Noda A, Agata K, et al. Examination of the rapid detection of legionella from bathwater samples by the loop‐mediated isothermal amplification and polymerase chain reaction methods. J Antibact Antifung Agents. 2004;32:481‐87. [Google Scholar]
- 14. Wang LX, He L, Fang R, et al. Loop‐mediated isothermal amplification (LAMP) assay for detection of Theileria sergenti infection targeting the p33 gene. Vet Parasitol. 2010;171:159‐62. [DOI] [PubMed] [Google Scholar]
- 15. Arimatsu Y, Kaewkes S, Laha T, Hong SJ, Sripa B. Rapid detection of Opisthorchis viverrini copro‐DNA using loop‐mediated isothermal amplification (LAMP). Parasitol Int. 2012;61:178‐82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nakao R, Stromdahl EY, Magona JW, et al. Development of loop‐mediated isothermal amplification (LAMP) assays for rapid detection of Ehrlichia ruminantium . BMC Microbiol. 2010;10:296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Plutzer J, Karanis P. Rapid identification of Giardia duodenalis by loop‐mediated isothermal amplification (LAMP) from faecal and environmental samples and comparative findings by PCR and real‐time PCR methods. Parasitol Res. 2009;104:1527‐33. [DOI] [PubMed] [Google Scholar]
- 18. He L, Zhou YQ, Oosthuizen MC, Zhao JL. Loop‐mediated isothermal amplification (LAMP) detection of Babesia orientalis in water buffalo (Bubalus babalis, Linnaeus, 1758) in China. Vet Parasitol. 2009;165:36‐40. [DOI] [PubMed] [Google Scholar]
- 19. Reddy AK, Balne PK, Reddy RK, Mathai A, Kaur I. Loop‐mediated isothermal amplification assay for the diagnosis of retinitis caused by herpes simplex virus‐1. Clin Microbiol Infect. 2011;17:210‐3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ihira M, Yoshikawa T, Enomoto Y, et al. Rapid diagnosis of human herpesvirus 6 infection by a novel DNA amplification method, loop‐mediated isothermal amplification. J Clin Microbiol. 2004;42:140‐5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Okamoto S, Yoshikawa T, Ihira M, et al. Rapid detection of varicella‐zoster virus infection by a loop‐mediated isothermal amplification method. J Med Virol. 2004;74:677‐82. [DOI] [PubMed] [Google Scholar]
- 22. Iwata S, Shibata Y, Kawada J, et al. Rapid detection of Epstein‐Barr virus DNA by loop‐mediated isothermal amplification method. J Clin Virol. 2006;37:128‐33. [DOI] [PubMed] [Google Scholar]
- 23. Parida M, Posadas G, Inoue S, Hasebe F, Morita K. Real‐time reverse transcription loop‐mediated isothermal amplification for rapid detection of West Nile virus. J Clin Microbiol. 2004;42:257‐63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Parida M, Horioke K, Ishida H, et al. Rapid detection and differentiation of dengue virus serotypes by a real‐time reverse transcription‐loop‐mediated isothermal amplification assay. J Clin Microbiol. 2005;43:2895‐903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mori N, Motegi Y, Shimamura Y, et al. Development of a new method for diagnosis of rubella virus infection by reverse transcription‐loop‐mediated isothermal amplification. J Clin Microbiol. 2006;44:3268‐73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Abo H, Okamoto K, Anraku M, et al. Development of an improved RT‐LAMP assay for detection of currently circulating rubella viruses. J Virol Methods. 2014;207:73‐7. [DOI] [PubMed] [Google Scholar]
- 27. Okafuji T, Yoshida N, Fujino M, et al. Rapid diagnostic method for detection of mumps virus genome by loop‐mediated isothermal amplification. J Clin Microbiol. 2005;43:1625‐31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shirato K, Nishimura H, Saijo M, et al. Diagnosis of human respiratory syncytial virus infection using reverse transcription loop‐mediated isothermal amplification. J Virol Methods. 2007;139:78‐84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ushio M, Yui I, Yoshida N, et al. Detection of respiratory syncytial virus genome by subgroups‐A, B specific reverse transcription loop‐mediated isothermal amplification (RT‐LAMP). J Med Virol. 2005;77:121‐7. [DOI] [PubMed] [Google Scholar]
- 30. Imai M, Ninomiya A, Minekawa H, et al. Development of H5‐RT‐LAMP (loop‐mediated isothermal amplification) system for rapid diagnosis of H5 avian influenza virus infection. Vaccine. 2006;24:6679‐82. [DOI] [PubMed] [Google Scholar]
- 31. Mahony J, Chong S, Bulir D, Ruyter A, Mwawasi K, Waltho D. Multiplex loop‐mediated isothermal amplification (M‐LAMP) assay for the detection of influenza A/H1, A/H3 and influenza B can provide a specimen‐to‐result diagnosis in 40 min with single genome copy sensitivity. J Clin Virol. 2013;58:127‐31. [DOI] [PubMed] [Google Scholar]
- 32. Kubo T, Agoh M, Mai LQ, et al. Development of a reverse transcription‐loop‐mediated isothermal amplification assay for detection of pandemic (H1N1) 2009 virus as a novel molecular method for diagnosis of pandemic influenza in resource‐limited settings. J Clin Microbiol. 2010;48:728‐35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hong, TC, Mai, QL, Cuong, DV, et al. Development and evaluation of a novel loop‐mediated isothermal amplification method for rapid detection of severe acute respiratory syndrome coronavirus. J Clin Microbiol. 2004;42:1956‐61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bhadra S, Jiang YS, Kumar MR, Johnson RF, Hensley LE, Ellington AD. Real‐time sequence‐validated loop‐mediated isothermal amplification assays for detection of Middle East respiratory syndrome coronavirus (MERS‐CoV). PLoS One. 2015;10:e0123126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lee SH, Baek YH, Kim YH, Choi YK, Song MS, Ahn JY. One‐pot reverse transcriptional loop‐mediated isothermal amplification (RT‐LAMP) for detecting MERS‐CoV. Front Microbiol. 2016;7:2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lo YM, Mehal WZ, Fleming KA. False‐positive results and the polymerase chain reaction. Lancet. 1988;332:679. [DOI] [PubMed] [Google Scholar]
- 37. Kitchin PA, Szotyori Z, Fromholc C, Almond N. Avoidance of PCR false positives [corrected]. Nature. 1990;344:201. [DOI] [PubMed] [Google Scholar]
- 38. Mori Y, Nagamine K, Tomita N, Notomi T. Detection of loop‐mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem Biophys Res Commun. 2001;289:150‐4. [DOI] [PubMed] [Google Scholar]
- 39. Kwok S, Higuchi R. Avoiding false positives with PCR. Nature. 1989;339:237‐8. [DOI] [PubMed] [Google Scholar]
- 40. Saiki RK, Scharf S, Faloona F, et al. Enzymatic amplification of beta‐globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science. 1985;230:1350‐4. [DOI] [PubMed] [Google Scholar]
- 41. Compton J. Nucleic acid sequence‐based amplification. Nature. 1991;350:91‐2. [DOI] [PubMed] [Google Scholar]
- 42. Mori Y, Kitao M, Tomita N, Notomi T. Real‐time turbidimetry of LAMP reaction for quantifying template DNA. J Biochem Biophys Methods. 2004;59:145‐57. [DOI] [PubMed] [Google Scholar]
- 43. Tomita N, Mori Y, Kanda H, Notomi T. Loop‐mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nat Protoc. 2008;3:877‐82. [DOI] [PubMed] [Google Scholar]
- 44. Kurata S, Kanagawa T, Yamada K, et al. Fluorescent quenching‐based quantitative detection of specific DNA/RNA using a BODIPY(R) FL‐labeled probe or primer. Nucleic Acids Res. 2001;29:34e‐34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Takayama I, Nakauchi M, Takahashi H, et al. Development of real‐time fluorescent reverse transcription loop‐mediated isothermal amplification assay with quenching primer for influenza virus and respiratory syncytial virus. J Virol Methods. 2019;267:53‐58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nakauchi M, Takayama I, Takahashi H, et al. Development of real‐time fluorescent reverse transcription loop‐mediated isothermal amplification assays for rhinovirus detection. J Med Virol. 2019;91:1232‐1238. [DOI] [PMC free article] [PubMed] [Google Scholar]


