Summary
Ubiquitination is a post‐translational modification implicated in a variety of cellular functions, including transcriptional regulation, protein degradation and membrane protein trafficking. Ubiquitin and the enzymes that act on it, although conserved and essential in eukaryotes, have not been well studied in parasites, despite sequencing of several parasite genomes. Several putative ubiquitin hydrolases have been identified in Plasmodium falciparum based on sequence homology alone, with no evidence of expression or function. Here we identify the first deubiquitinating enzyme in P. falciparum, PfUCH54, by its activity. We show that PfUCH54 also has deNeddylating activity, as assayed by a mammalian Nedd8‐based probe. This activity is absent from mammalian homologues of PfUCH54. Given the importance of parasitic membrane protein trafficking as well as protein degradation in the virulence of this parasite, this family of enzymes may represent a target for pharmacological intervention with this disease.
Introduction
Malaria, the disease caused by Plasmodium parasites, is a widespread and debilitating illness of utmost public health importance (Snow et al., 2005). Despite all efforts to control the morbidity and mortality associated with this parasite, a safe, efficacious vaccine has yet to be developed. Current preventive and curative drug regimens are often plagued by unpleasant side‐effects and, more importantly, limited effectiveness due to Plasmodium's efficiency in developing resistance (Schellenberg et al., 2006). Consequently, novel vaccine and drug targets are necessary for the successful control of malaria.
The key to a good therapy lies in the ability to specifically target a parasitic function with minimal effect on the host. The essential nature of ubiquitin (Ub) modifications suggests that the enzymes acting on it may be potential drug targets. Ub and some of its relatives, including Nedd8, SUMO and ISG15, are dynamic post‐translational modifiers that can be added or removed from proteins to achieve a certain localization or function. Ub is the most extensively characterized and studied modifier, involved in a variety of cellular processes including protein trafficking, protein quality control, transcriptional regulation and signalling. Its addition to target proteins is mediated by E1 Ub activating enzyme, E2 Ub conjugating enzymes and E3 Ub ligases whereas its removal is controlled by deubiquitinating proteases (DUBs), the vast majority of which are cysteine proteases.
The ubiquitin‐proteasome pathway has provided a successful therapeutic target in cancer biology. Proteasome inhibitors preferentially induce apoptosis in actively replicating cells and appear particularly effective in the treatment of multiple myeloma (Voorhees and Orlowski, 2006). There is increasing evidence that certain non‐eukaryotic pathogens, including Herpesviridae (Kattenhorn et al., 2005; Schlieker et al., 2005), the severe acute respiratory syndrome (SARS) virus (Barretto et al., 2005; Lindner et al., 2005), adenoviruses (Balakirev et al., 2002) and Chlamydia trachomatis (Misaghi et al., 2006), encode proteins capable of interfering with this pathway, presumably for immune evasive purposes.
The possibility that Plasmodium parasites have developed ways to interfere with host cellular functions is an interesting one, particularly in light of how long this parasite has had to evolve. Plasmodium has a complex system for the export of membrane proteins to the red blood cell (RBC) surface (Marti et al., 2004) and also secretes non‐membrane‐associated proteins from micronemes, rhoptries and dense granules (three types of specialized secretory organelles) into the RBC, which are essential for merozoite invasion (Binder and Kim, 2004). Whether these parasites secrete proteins capableof interfering with host ubiquitination in nucleated cells remains to be determined. As a eukaryote, Plasmodium possesses its own post‐translational modifying machinery, including a ubiquitin‐proteasome pathway. This pathway has yet to receive its due attention for therapeutic development, with the exception of a recent publication that reports toxicity of proteasome inhibitors for Plasmodium falciparum (Lindenthal et al., 2005). Although the complete sequencing of the P. falciparum genome has revealed several putative Ub C‐terminal hydrolases, there has been no evidence of expression and function for any of these gene products. The assignment of a putative deubiquitinating enzyme based solely on sequence similarity is precarious, as the enzyme may prefer to react with Ub‐like modifiers, rather than with Ub itself (Hemelaar et al., 2004; Misaghi et al., 2006). Examples of enzymes with dual specificity, for example, reactive with both Ub as well as with Nedd8, have been reported (Wada et al., 1998; Linghu et al., 2002). Here we identify a P. falciparum protein, homologous to human UCH37, which we designate PfUCH54, with apparent deubiquitinating as well as deNeddylating activity.
Results
Use of electrophilic probes to identify modified proteins
Electrophilic derivatives of Ub, SUMO, Nedd8 and ISG15 are activity‐based probes that allow identification of enzymes capable of removing these modifications from target proteins (Borodovsky et al., 2001; 2002; Hemelaar et al., 2004). These probes were equipped with epitope tags to facilitate detection, and their validation and use have been described in detail (Borodovsky et al., 2002; Hemelaar et al., 2004; Kattenhorn et al., 2005; Misaghi et al., 2006). Briefly, these probes comprise the post‐translational modifier of choice (Ub, SUMO1, Nedd8 or ISG15) with the C‐terminal glycine replaced by an electrophilic vinylmethylester (VME) or vinylmethylsulphone group. Upon attack by the target enzyme, an irreversible thioether bond is formed between enzyme and probe. These interactions can then be detected by immunoprecipitation or by immunoblotting against the epitope tag (Fig. 1).
Figure 1.

Probe mechanisms of action. Probes were designed for the detection of enzymes capable of removing Ub and other Ub‐like proteins. An example of the Ub‐based probes is shown. Probes include a Ub moiety followed by either (A) a vinylmethylester or (B) a vinylmethylsulphone active group. Where needed, a haemagglutinin (HA) epitope tag is also included as an N‐terminal fusion. The catalytic groups act as electrophiles mimicking the C‐terminal end of the Ub gly motif and attracting DUB attack. DUBs become covalently and irreversibly attached to the probes through a thioether bond, thereby allowing for detection by anti‐HA antibodies in addition to a 10 kDa increase in size.
A search of the P. falciparum database predicted a significant number of putative C‐terminal Ub hydrolases (Table 1) inciting further investigation into existence and reactivity profiles for this family of enzymes in the parasite. The probes used in this study are all based on the mammalian sequences of Ub, Nedd8, SUMO1 and ISG15. Consequently, such probes are expected to react only if the Plasmodium‐derived modifiers are sufficiently conserved to allow cross‐reaction. A sequence alignment and consensus analysis of P. falciparum SUMO, Nedd8 and Ub modifiers with their human counterparts is shown in Fig. S1. ISG15 homologues have not been found outside of vertebrates so this alignment is not included. A SUMO‐related sequence is found in P. falciparum and shares an average of 50% amino acid sequence identity with the four human SUMO variants (48% SUMO1, 53% SUMO2, 51% SUMO3, 50% SUMO4). Likewise, a Nedd8‐like protein which is 52% identical to its human counterpart is also present. P. falciparum and human Ub, however, differ only by a single conservative amino acid replacement (E16D).
Table 1.
Putative deubiquitinating enzymes in the Plasmodium falciparum genome.
| Gene ID | Gene name |
|---|---|
| NP_703247 | Ubiquitin C‐terminal hydrolase, putative |
| NP_702688 | Ubiquitin‐specific protease, putative |
| NP_702789 | Ubiquitin C‐terminal hydrolase a, putative |
| NP_703511 | Ubiquitin C‐terminal hydrolase 2, putative |
| NP_703615 | Ubiquitin C‐terminal hydrolase, putative |
| NP_704193 | Ubiquitin C‐terminal hydrolase, putative |
| NP_701037 | Ubiquitin C‐terminal hydrolase, family 1, putative |
| NP_705079 | Ubiquitin C‐terminal Hydrolase‐like zinc finger protein |
| NP_702465 | Ubiquitin C‐terminal hydrolase, putative |
| NP_704194 | Hypothetical protein |
| NP_704848 | Hypothetical protein |
| NP_704154 | Hypothetical protein |
| NP_704195 | Hypothetical protein |
| NP_704588 | Hypothetical protein |
A P. falciparum genome database search yields a short list of putative gene products with predicted deubiquitinating activity.
Evidence of Ub and Nedd8 protease activity in P. falciparum
To determine whether P. falciparum contains proteins that have deubiquitinating, deNeddylating, deISG15ylating and deSUMOylating activity, we prepared schizont lysate from synchronized cultures. We chose schizonts, as this stage occupies the entire volume of the RBC and contains large amounts of protein. We treated the cultures with saponin to release the schizonts and to remove any unparasitized red blood cells (uRBCs). Parasites were then lysed and incubated with each of the epitope‐tagged probes. Immunoblotting revealed polypeptides reacting with Ub and Nedd8 (Fig. 2A and B). In both cases, addition of N‐ethyl‐maleimide (NEM) eliminated this reactivity, evidence of an enzymatic mechanism that involves a cysteine residue at the active site. Conversely, the SUMO and ISG15 probes failed to react with the parasite lysate (data not shown). The 50% sequence identity of P. falciparum SUMO with its mammalian counterpart could account for the absence of reactivity with the mouse‐derived probe.
Figure 2.
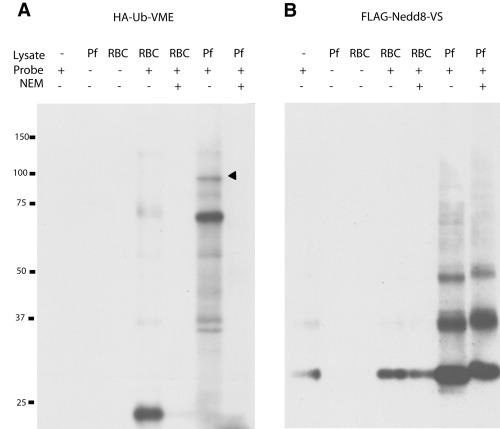
Evidence of parasite‐specific deubiquitinating and deNeddylating activity. Parasite and uninfected red blood cell (RBC) lysates were reacted with (A) HA‐Ub‐VME or (B) FLAG‐Nedd8‐VS followed by SDS‐PAGE and immunoblot with anti‐HA‐HRP or anti‐FLAG‐HRP where appropriate. Probe alone and lysate alone are shown in the left three lanes of each panel. Reaction profiles in the presence or absence of NEM are shown in the right four lanes of each panel. Numbers on the left‐hand side of (A) indicate molecular weights in kDa. The reactive polypeptide later excised for mass spectrometric analysis is indicated by the arrow. Pf, Plasmodium falciparum lysate.
To ensure that the reactive bands were not RBC‐derived, we included uRBC controls wherein we reacted uRBC lysate with the Ub and Nedd8 probes. Blotting with anti‐haemagglutinin (HA) or anti‐FLAG antibodies confirmed that the majority of the strongly reactive material was indeed of parasitic origin.
Identification of Ub‐VME‐reactive proteins
In order to further characterize these reactive polypeptides, we performed the reaction with HA‐Ub‐VME on a preparative scale. Parasite lysate was reacted with probe, followed by an anti‐HA immunoprecipitation and SDS‐PAGE. In this instance we opted for unsaponized parasites as the starting material in order to maximize the chance of detecting both internal as well as possibly secreted material that might react with the probe. As a negative control, we included an equal number of uRBCs treated in parallel. The preparative gel was silver‐stained, and polypeptides specific to the schizont lysate were excised, trypsinized and subjected to tandem mass spectrometry (MS/MS) (Fig. S2). The corresponding area in the uRBC lane was also excised and analysed to ensure parasite specificity.
Of particular interest was a polypeptide running at approximately 100 kDa, the peptides of which were absent from the uRBC control lane and which matched a protein (Pf11_0177 or NP_701037) annotated as a putative C‐terminal Ub hydrolase. The quality of the mass spectra obtained and the extent of sequence coverage allowed robust and unambiguous identification (Fig. 3). Although the gene designation suggests DUB activity, as determined by sequence similarity with known mammalian UCH‐type enzymes, these data are the first evidence that this gene product is both expressed and functional. For this reason, the identified protein was named PfUCH54, for P. falciparum Ub C‐terminal hydrolase with a molecular mass of 54 kDa. The discrepancy in predicted versus observed size of the protein (54 kDa versus ∼100 kDa) can possibly be explained by aggregation in the course of electrophoresis. It has been established that consecutive repetitions of a single amino acid, such as glutamine or alanine, can lead to protein aggregations (Ravikumar et al., 2002). We believe that the homopolymeric region of asparagines embedded in the PfUCH54 sequence may cause the observed aggregation. Peptides matching PfUCH54 were also recovered from 65 kDa and 40 kDa regions of the gel. The 65 kDa mass is consistent with a 54 kDa protein with a 10 kDa probe attachment, while the 40 kDa fragment most likely represents a degradation product. Mass spectral quality for these polypeptides was inferior to that of the 100 kDa band, hence, the latter data are presented in Fig. 3.
Figure 3.

Identification of PfUCH54 by mass spectrometry. P. falciparum and uninfected erythrocyte lysates were reacted with HA‐Ub‐VME probe on a preparative scale followed by SDS‐PAGE. Parasite‐specific reactive proteins were extracted and subjected to tandem mass spectrometry. Identified polypeptides (indicated in pink) were screened against the P. falciparum genome database and matched a 465‐amino‐acid protein annotated as a putative C‐terminal Ub hydrolase. Mass spectrometry properties of the identified peptides are shown. MH+ indicates the protonated mass of each peptide; %Mass represents the per cent of the total protein mass occupied by each peptide; AA indicates the span of amino acids covered by each peptide; and %AA identifies the per cent of amino acid coverage. Black arrows indicate potential alternative internal start sites as determined by additional translation products seen in Fig. 5.
When a blast database homology search was performed (use of BLOSUM62 matrix, gap costs for existence 11 and for extension 1; low‐complexity filter), the human proteasome‐associated DUB, UCH37, was identified as a homologue (e‐value of 1 × 10−30) despite its significantly smaller size. A sequence alignment of these two proteins, in addition to two other well‐characterized human DUBs, UCH‐L1 and UCH‐L3, revealed only moderate sequence identity but conservation of all active site residues, consistent with the observed DUB activity of the parasitic protein (Fig. 4A). The predicted active residues for PfUCH54 are Cys145, His220 and Asp235. The insertion of a homopolymeric asparagine stretch in the parasite sequence is common to many expressed proteins (Pizzi and Frontali, 2001) in P. falciparum. Although the function of these low‐complexity homopolymeric regions is not known, they may serve as sites of interaction with other proteins or may expand the parasite's antigenic repertoire, thereby contributing to immune evasion (Schofield, 1991). As the crystal structure of UCH‐L3 has been solved (Johnston et al., 1997; Misaghi et al., 2005) and as the structure of UCH37, the closest PfUCH54 homologue, is predicted to be very similar (Kelley et al., 2000), we prepared a threaded model of PfUCH54 based on that of UCH‐L3 (Fig. 4B). The predicted structure of the PfUCH54 insertions were calculated using ROSETTA (Bystroff and Shao, 2002) and manually inserted into the final model (Guex and Peitsch, 1997). We were encouraged to see that the three major insertions in the parasitic protein sequence (shown in green in the model) are unlikely to interfere with the predicted architecture of the catalytic site (active cysteine residue shown in blue), as was confirmed by experiment (see Fig. 5). If these homopolymeric amino acid stretches serve a function, it is likely one not directly related to the protein's enzymatic activity.
Figure 4.
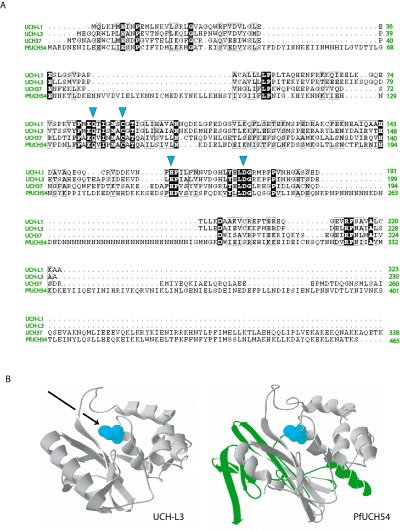
Alignment of PfUCH54 with human DUB homologues and predicted structure. A. An alignment of three human UCH‐type DUBs, UCH‐L1, UCH‐L3 and UCH37, with PfUCH54 is shown with active residues indicated by the blue arrows. B. The crystal structure of UCH‐L3 was used to model PfUCH54. Parasite‐specific insertions are shown in green. Active site cysteine residues for both enzymes are shown in blue. The arrow indicates the trajectory of Ub that is presumed for binding to UCH‐L3.
Figure 5.
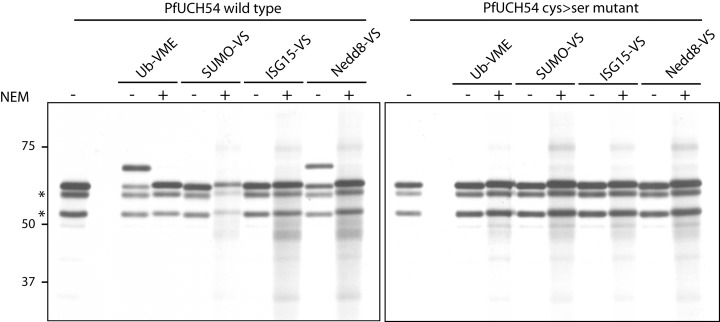
Evidence of dual Ub and Nedd8 specificity for PfUCH54. In vitro transcription/translation using HA‐tagged, wild‐type PfUCH54 (left) and the catalytically inactive cysteine to serine mutant (right) as templates was performed in the presence of 35S‐methionine. Translation products were reacted with the range of untagged probes followed by immunoprecipitation against HA. Analysis by autoradiography reveals Ub and Nedd8 activity specific to the wild‐type PfUCH54. Two unreactive truncated translation products are indicated by asterisk. Reactivity in the presence or absence of the alkylating agent NEM is shown for both the wild type and the mutant.
Parasitic DUB also has deNeddylating activity
To verify DUB activity for this protein and to probe the extent of possible deNeddylating, deSUMOylating and deISG15ylating activity, the PfUCH54 coding region (which does not contain introns) was cloned from genomic DNA and a catalytically inactive mutant was generated based on this sequence by mutating the putative active site cysteine residue to serine. Both constructs were epitope‐tagged with HA and their expression and reactivity with the probes were assessed by in vitro translation.
Although the in vitro translation product failed to react with the SUMO and ISG15 probes, modification by Ub‐VME was evident from a quantitative shift of the protein to a more slowly migrating form, consistent with covalent adduct formation with the probe (Fig. 5). Furthermore, a similar shift was also observed with the Nedd8‐vinylsulphone (VS) probe. Based on reactivity with these probes, enzymatic activity is inferred (Kattenhorn et al., 2005; Schlieker et al., 2005; Misaghi et al., 2006). Binding to both Ub‐VME and Nedd8‐VS were entirely blocked upon addition of NEM. Specificity was further substantiated by the failure of the catalytically inactive mutated protein to be modified by either probe. These data demonstrate the requirements for the cysteine residue for reactivity, a hallmark of DUB activity. In addition to the polypeptide that exhibits mobility shifts upon reaction with the probe, each sample contains two additional prominent polypeptides that do not react with the probe. Because these polypeptides are recovered via the C‐terminal epitope tag, they must correspond to N‐terminal truncations of the primary translation product. Based on the location of additional methionine residues at positions consistent with the observed size of these polypeptides, we propose that they derive from the use of alternative translation start sites (indicated in Fig. 3).
Discussion
The identification of molecules and metabolic pathways unique to P. falciparum, or sufficiently distinct from its human host to allow chemical discrimination, may yield new targets for the development of antiparasitic vaccines and therapeutic agents. Differences in the ubiquitin‐proteasome system have been successfully used for diagnosis of parasitic diseases (Telles et al., 2003). The ubiquitin‐proteasome pathway is ancient and has been conserved throughout evolution (Catic and Ploegh, 2005), and the dynamic addition and removal of Ub is central to a wide variety of eukaryotic cellular processes in organisms ranging from yeast to humans. The many effects of ubiquitination render the enzymes that act on Ub crucial to proper cellular function and survival, a possible target for development of methods to handicap pathogens. The evolutionary distance between the parasite and its host may have resulted in differences in enzyme structure sufficient to allow selective inhibition, with the core function of the enzyme remaining conserved. Interference with the ubiquitin‐proteasome pathway has been successfully targeted in cancer therapy (Voorhees and Orlowski, 2006) but has been largely overlooked within infectious disease, particularly with regard to diseases of significant public health importance, such as malaria. Indeed, interference with protein degradation in P. falciparum, by blocking proteasome function, results in growth inhibition and toxicity to the parasite (Gantt et al. 1998; Lindenthal et al., 2005) and validates this pathway as a legitimate drug target. Given the variety of molecules that mediate this branch of the ubiquitination pathway, more specific and elegant therapies might be developed by targeting individual components of the processing cascade, rather than by indiscriminately blocking the proteasome itself. The data presented here establish the expression and functionality of the enzyme PfUCH54, both as a deubiquitinating and as a deNeddylating agent and provide the first experimental evidence that these enzyme families are active in P. falciparum.
The dual deubiquitinating and deNeddylating activity of PfUCH54 is reminiscent of UCH‐L3 dual reactivity (Wada et al., 1998) and has been reported for other enzymes as well (Linghu et al., 2002; Misaghi et al., 2005), suggesting that this dual activity may be a widespread, conserved and significant phenomenon. Although the alignment of UCH‐L3 with PfUCH54 does not show a high level of identity between the two proteins, putative active site amino acids reside within highly conserved sequence blocks. The dual specificity observed for PfUCH54 does not allow us to state with confidence whether its primary function is to target Ub‐modified or Nedd8‐modified proteins. Nedd8's role, as far as is known, is restricted to the modification of cullins, which are part of the SCF‐type Ub ligases involved in cell cycle control (Chiba and Tanaka, 2004). It is possible that PfUCH54 may contribute to regulation of both Ub and Nedd8 conjugation in P. falciparum. The Nedd8 reactivity of PfUCH54 is of particular interest given that mammalian and parasitic Nedd8 share only 52% identity. Nonetheless, there are three distinct blocks of conserved sequence (aa 5–11, 42–50, 71–76; see Fig. S1), which suggests that these regions of the protein are the critical components for proper folding and function.
Non‐eukaryotic pathogens such as viruses and bacteria in some cases express proteins that can catalyse deubiquitination, presumably as part of immune evasion strategies. These proteins are either assembled or secreted into the host cell's cytoplasm where they interfere with the Ub pathway. A catalytically active truncation of UL36, the major tegument protein of herpes simplex virus‐1 (HSV‐1), is able to cleave polyubiquitin chains (Kattenhorn et al., 2005) and a related activity is conserved across the Herpesviridae (Schlieker et al., 2005). Likewise, C. trachomatis encodes two recently identified proteins with deubiquitinating activity, ChlaDUB1 and ChlaDUB2 (Misaghi et al., 2006). Homologues to these enzymes are not present in non‐pathogenic Chlamydia spp., an observation that supports the idea that these proteins may contribute to pathogenicity. Additional examples of proteins with deubiquitinating activity are also encoded by the SARS virus (Barretto et al., 2005; Lindner et al., 2005) as well as adenoviruses (Balakirev et al., 2002).
As a major aspect of P. falciparum pathogenesis is mediated by the parasite's ability to display its own proteins on the surfaces of host erythrocytes, the mechanisms surrounding secretion have been well studied (Sargeant et al., 2006). In order for a parasite protein to be secreted, it must contain a specific N‐terminal signal sequence, otherwise known as a PEXEL domain (Binder and Kim, 2004; Marti et al., 2004). PfUCH54 lacks such a sequence, suggesting that its function is intraparasitic rather than host‐targeted. In addition, asparagine repeats which are commonly found throughout the P. falciparum genome are most commonly associated with parasite‐internal proteins (Singh et al., 2004). The presence of an asparagine‐rich region in PfUCH54 and the presence of a strongly HA‐Ub‐VME‐reactive band at 54 kDa in saponized parasites (Fig. 2A) suggest intraparasitic localization, a conclusion that would require further support by direct immunohistochemical analysis.
The significance and necessity of PfUCH54 and other Ub‐reactive proteins in P. falciparum remain to be determined. Efforts to assess parasite viability in the absence of these molecules are underway in the form of knockout and mutation studies in live parasites. Unlike in other organism, there seem to be few transcriptional changes in P. falciparum following exposure to stimuli such as antifolates (Ganesan et al., 2003). This rigidity in transcription suggests that post‐transcriptional regulation, including protein turnover, is likely to play a major role in parasite gene expression and parasite viability. By targeting specific DUBs, we hope to open new avenues in the development of antimalarial therapeutics.
Experimental procedures
Parasite culturing and purification
Plasmodium falciparum parasites (3D7 strain) were maintained in culture as previously described (Trager and Jensen, 1976). Mature schizonts were isolated from synchronous cultures with a parasitaemia of 4% by two methods. For one sample, the infected RBC pellet was simply washed twice with PBS before storage at −80°C. In the other sample, the infected RBC pellet was lysed in an incubation of 0.15% saponin to release the mature schizonts before subsequent washing in PBS and storage at −80°C.
Cloning and mutagenesis
Plasmodium falciparum genomic DNA was prepared using the Blood and Body Fluid Spin Protocol in Qiagen's QIAamp DNA Mini Kit and QIAamp DNA Blood Mini Kit Handbook according to manufacturer's specifications (Valencia, CA). Starting parasite material was prepared by incubating 8 × 107 parasitized RBCs of mixed stages with saponin (0.15%) for 10 min at room temperature. Released parasites were centrifuge‐pelleted at 500 g for 5 min and washed in chilled PBS twice. Subsequently, supernatant was removed and pellets were flash frozen and stored at −80°C.
Pf11_0177 is the intronless, open reading frame on chromosome 11 that encodes PfUCH54. Primers were designed against the 5′ (CGGAATTCATGGCGAGGGATAATGAAAACA) and 3′ (CCGCTCGAGAGATTTTGTTGCATTTTTTAATTT) regions of the open reading frame with EcoRI and XhoI linkers, respectively, for directional cloning. The sequence was PCR‐amplified with Pfu polymerase and the product was digested and ligated into the multicloning site of pcDNA3.1(+) in frame with a C‐terminal HA epitope tag. The ligation mixture was transformed to competent Escherichia coli and colonies harbouring a plasmid were picked for DNA isolation. Clones positive for an insert of the correct size were selected and their DNA sequence was verified. Correct clones were used for subsequent studies.
Site‐directed mutagenesis was performed using Stratagene's QuikChange II Site‐Directed mutagenesis Kit (La Jolla, CA). Complementary primers spanning the area immediately around the catalytic cysteine codon were designed (5′ > 3′ GTTATACCAAATGCAAGTGCTACACAAGC and 3′ > 5′ GCTTGTGTAGCACTTGCATTTGGTATAAC) to change the cysteine to a serine by mutation of a single base (indicated by the underlined nucleotides). Thirty cycles of PCR followed by DpnI digestion to remove the methylated template plasmid DNA yielded product which was transformed to competent E. coli as described above. Properly mutated clones were identified by sequence verification.
Probes
Haemagglutinin‐ or FLAG‐tagged, VME or VS Ub, Nedd8, ISG15 and SUMO electrophilic probes were previously generated and described (Borodovsky et al., 2001; 2002; Hemelaar et al., 2004). Reactions with parasite lysate were carried out for 90 min at a concentration of approximately 0.3 μg of probe for every 1 μg of parasite protein. For the immunoblot the lysed equivalent of 5 × 107 schizonts or uRBCs were loaded per lane. For the preparative gel, 4 mg of total lysed protein (derived from 12 × 1010 RBC containing 2.5 × 109 schizonts or 12 × 1010 uRBC) was reacted with 40 μg of probe.
Immunoblot
Parasite lysate was separated by gel electrophoresis and proteins were transferred to PVDF which were blocked overnight in 5% milk. Membranes were then probed with anti‐HA‐HRP (Roche, Mannheim, Germany) or anti‐FLAG‐HRP (Sigma, St Louis, MO) antibodies at a dilution of 1:1000 in PBS‐Tween for 1 h at room temperature. Following six 10 min washes in PBS‐Tween, proteins were visualized by chemiluminescence (Perkin Elmer, Boston, MA).
In vitro translation and immunoprecipitation
Template plasmid DNA of either the wild‐type or mutated gene was added at a concentration of 200–300 μg ml−1 to 30 μl of rabbit reticulocyte lysate TNT T7 Quick Coupled Translation/Transcription System mastermix (Promega, Madison, WI). Each reaction was supplemented with 1 μCi of 35S‐methionine (Perkin Elmer) and incubated at 30°C for 1 h. Each sample mixture was then reacted with 0.3 μg of untagged probe (either Ub‐VME, Nedd8‐VS, ISG15‐VS or SUMO‐VS) for 90 min at room temperature and then diluted to a 1 ml volume in NP‐40 lysis buffer [10 mM Tris (pH 7.8), 150 mM NaCl, 5 mM MgCl2, 0.5% NP‐40]. Enzyme–probe complexes were immunoprecipitated by 12CA5 mouse monoclonal anti‐HA antibody for 45 min at 4°C followed by addition of Immobilized Protein‐A agarose (Repligen, Waltham, MA) for another 45 min at 4°C. Beads were spun and washed in washing buffer [0.5% NP‐40, 50 mM Tris (pH 7.4), 150 mM NaCl, 5 mM EDTA] three times, resuspended in reducing sample buffer and the eluate was subjected to SDS‐PAGE (8% polyacrylamide). Results were analysed by autoradiography.
Modelling
A homology model of PfUCH54 was constructed based on the solved structure of UCH‐L3, and predicted secondary structure of the Plasmodium protein. Inspection of sequence alignment of PfUCH54 with UCH‐L1, UCH‐L3 and UCH37 shows regions of similarity in all four proteins that are punctuated by large (> 25 amino acids) insertions in PfUCH54. These insertions were omitted from the initial modelling as they are present in regions of UCH‐L3 that are not part of the Ub binding site or the active site of UCH‐L3.
Supporting information
Fig. S1. Alignment of human and P. falciparum Ub and Ub‐like proteins. The extent of amino acid sequence homology between human and P. falciparum (A) SUMO variants (B) Nedd8 and (C) Ub. Solid blocks indicating regions of conserved amino acids. Amino acid sequences were obtained from the GenBank database. The accession number are as follows: SUMO GI:23504543, Nedd8 GI:23615283, Ub GI:23508814. Fig. S2. Preparative silver‐stained gel for mass spectrometric analysis. Lysates derived from 12 x 1010 RBC containing 2.5 x 109 schizonts or 12 x 1010 uRBC were reacted with HA‐Ub‐VME probe. Reactive material was immunoprecipitated with anti‐HA resin, SDS‐PAGE separated, and polypeptides indicated by the arrows were excised and subjected to tandem mass spectrometry. PfUCH54‐matching peptides were found in bands indicated by the asterisks.
Supporting info item
Supporting info item
Acknowledgements
We thank Dr Greg Korbel and Dr Daniël Blom for insightful discussion as well as critical review of the manuscript. We also thank Bradley Coleman for providing genomic parasite DNA and Gijsbert Grotenbreg for assistance with ChemDraw.
References
- Balakirev, M.Y. , Jaquinod, M. , Haas, A.L. , and Chroboczek, J. (2002) Deubiquitinating function of adenovirus proteinase. J Virol 76: 6323–6331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barretto, N. , Jukneliene, D. , Ratia, K. , Chen, Z. , Mesecar, A.D. , and Baker, S.C. (2005) The papain‐like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J Virol 79: 15189–15198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder, E.M. , and Kim, K. (2004) Location, location, location: trafficking and function of secreted proteases of Toxoplasma and Plasmodium . Traffic 5: 914–924. [DOI] [PubMed] [Google Scholar]
- Borodovsky, A. , Kessler, B.M. , Casagrande, R. , Overkleeft, H.S. , Wilkinson, K.D. , and Ploegh, H.L. (2001) A novel active site‐directed probe specific for deubiquitylating enzymes reveals proteasome association of USP14. EMBO J 20: 5187–5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borodovsky, A. , Ovaa, H. , Kolli, N. , Gan‐Erdene, T. , Wilkinson, K.D. , Ploegh, H.L. , and Kessler, B.M. (2002) Chemistry‐based functional proteomics reveals novel members of the deubiquitinating enzyme. Chem Biol 9: 1149–1159. [DOI] [PubMed] [Google Scholar]
- Bystroff, C. , and Shao, Y. (2002) Fully automated abinitio protein structure prediction using I‐SITES, HMMSTR and ROSETTA. Bioinformatics 18: S54–S61. [DOI] [PubMed] [Google Scholar]
- Catic, A. , and Ploegh, H.L. (2005) Ubiquitin – conserved protein or selfish gene? Trends Biochem Sci 30: 600–604. [DOI] [PubMed] [Google Scholar]
- Chiba, T. , and Tanaka, K. (2004) Cullin‐based ubiquitin ligase and its control by NEDD8‐conjugating system. Curr Protein Pept Sci 5: 177–184. [DOI] [PubMed] [Google Scholar]
- Ganesan, K. , Jiang, L. , White, J. , and Rathod, P.K. (2003) Rigidity of the Plasmodium transcriptome revealed by a lethal antifolate. Abstract #2E. Molecular Parasitology Meeting XIV, Woods Hole, MA, 14–18 September.
- Gantt, S.M. , Myung, J.M. , Briones, M.R.S. , Li, W.D. , Corey, E.J. , Omura, S. , et al. (1998) Proteasome inhibitors block development of Plasmodium spp. Antimicrob Agents 42: 2731–2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guex, N. , and Peitsch, M.C. (1997) SWISS‐MODEL and the Swiss‐PdbViewer: an environment for comparative protein modeling. Electrophoresis 15: 2714–2723. [DOI] [PubMed] [Google Scholar]
- Hemelaar, J. , Borodovsky, A. , Kessler, B.M. , Reverter, D. , Cook, J. , Kolli, N. , et al. (2004) Specific and covalent targeting of conjugating and deconjugating enzymes of ubiquitin‐like proteins. Mol Cell Biol 24: 84–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston, S.C. , Larsen, C.N. , Cook, W.J. , Wilkinson, K.D. , and Hill, C.P. (1997) Crystal structure of a deubiquitinating enzyme (human UCH‐L3) at 1.8 angstrom resolution. EMBO J 16: 3787–3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kattenhorn, L.M. , Korbel, G.A. , Kessler, B.M. , Spooner, E. , and Ploegh, H.L. (2005) A deubiquitinating enzyme encoded by HSV‐1 belongs to a family of cysteine proteases that is conserved across the family Herpesviridae . Mol Cell 19: 547–557. [DOI] [PubMed] [Google Scholar]
- Kelley, L.A. , MacCallum, R.M. , and Sternberg, M.J.E. (2000) Enhanced genome annotation using structural profiles in the program 3D‐PSSM. J Mol Biol 299: 499–520. [DOI] [PubMed] [Google Scholar]
- Lindenthal, C. , Weich, N. , Chia, Y.S. , Heussler, V. , and Klinkert, M.Q. (2005) The proteasome inhibitor MLN‐273 blocks exoerythrocytic and erythrocytic development of Plasmodium parasites. Parasitology 131: 37–44. [DOI] [PubMed] [Google Scholar]
- Lindner, H.A. , Fotouhi‐Ardakani, N. , Lytvyn, V. , Lachance, P. , Sulea, T. , and Menard, R. (2005) The papain‐like protease from the severe acute respiratory syndrome coronavirus is a deubiquitinating enzyme. J Virol 79: 15199–15208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linghu, B. , Callis, J. , and Goebl, M.G. (2002) Rub1p processing by Yuh1p is required for wild‐type levels of Rub1p conjugation to Cdc53p. Eukaryot Cell 1: 491–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marti, M. , Good, R.T. , Rug, M. , Knuepfer, E. , and Cowman, A.F. (2004) Targeting malaria virulence and remodeling proteins to the host erythrocyte. Science 306: 1930–1933. [DOI] [PubMed] [Google Scholar]
- Misaghi, S. , Galardy, P.J. , Meester, W.J.N. , Ovaa, H. , Ploegh, H.L. , and Gaudet, R. (2005) Structure of the ubiquitin hydrolase UCH‐L3 complexed with a suicide substrate. J Biol Chem 280: 1512–1520. [DOI] [PubMed] [Google Scholar]
- Misaghi, S. , Balsara, Z.B. , Catic, A. , Spooner, E. , Ploegh, H.L. , and Starnbach, M.N. (2006) Chlamydia trachomatis‐derived deubiquitinating enzymes in mammalian cells during infection. Mol Microbiol 61: 142–150. [DOI] [PubMed] [Google Scholar]
- Pizzi, E. , and Frontali, C. (2001) Low‐complexity regions in Plasmodium falciparum proteins. Genome Res 11: 218–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar, B. , Duden, R. , and Rubinsztein, D.C. (2002) Aggregate‐prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum Mol Genet 11: 1107–1117. [DOI] [PubMed] [Google Scholar]
- Sargeant, T. , Marti, M. , Caler, E. , Carlton, J. , Simpson, K. , Speed, T. , and Cowman, A. (2006) Lineage‐specific expansion of proteins exported to erythrocytes in malaria parasites. Genome Biol 7: R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellenberg, D. , Abdulla, S. , and Roper, C. (2006) Current issues for anti‐malarial drugs to control P. falciparum malaria. Curr Mol Med 6: 253–260. [DOI] [PubMed] [Google Scholar]
- Schlieker, C. , Korbel, G.A. , Kattenhorn, L.M. , and Ploegh, H.L. (2005) A deubiquitinating activity is conserved in the large tegument protein of the Herpesviridae . J Virol 79: 15582–15585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield, L. (1991) On the function of repetitive domains in protein antigens of Plasmodium and other eukaryotic parasites. Parasitol Today 7: 99–105. [DOI] [PubMed] [Google Scholar]
- Singh, G.P. , Chandra, B.R. , Bhattacharya, A. , Akhouri, R.R. , Singh, S.K. , and Sharma, A. (2004) Hyper‐expansion of asparagines correlates with an abundance of proteins with prion‐like domains in Plasmodium falciparum . Mol Biochem Parasitol 137: 307–319. [DOI] [PubMed] [Google Scholar]
- Snow, R.W. , Guerra, C.A. , Noor, A.M. , Myint, H.Y. , and Hay, S.I. (2005) The global distribution of clinical episodes of Plasmodium falciparum malaria. Nature 434: 214–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telles, S. , Abate, T. , Slezynger, T. , and Henriquez, D.A. (2003) Trypanosoma cruzi ubiquitin as an antigen in the differential diagnosis of Chagas disease and leishmaniasis. FEMS Immunol Med Microbiol 37: 23–28. [DOI] [PubMed] [Google Scholar]
- Trager, W. , and Jensen, J.B. (1976) Human malaria parasites in continuous culture. Science 193: 673–675. [DOI] [PubMed] [Google Scholar]
- Voorhees, P.M. , and Orlowski, R.Z. (2006) The proteasome and proteasome inhibitors in cancer therapy. Annu Rev Pharmacol 46: 189–213. [DOI] [PubMed] [Google Scholar]
- Wada, H. , Kito, K. , Caskey, L.S. , Yeh, E.T.H. , and Kamitani, T. (1998) Cleavage of the C‐terminus of NEDD8 by UCH‐L3. Biochem Biophys Res Comm 251: 688–692. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Alignment of human and P. falciparum Ub and Ub‐like proteins. The extent of amino acid sequence homology between human and P. falciparum (A) SUMO variants (B) Nedd8 and (C) Ub. Solid blocks indicating regions of conserved amino acids. Amino acid sequences were obtained from the GenBank database. The accession number are as follows: SUMO GI:23504543, Nedd8 GI:23615283, Ub GI:23508814. Fig. S2. Preparative silver‐stained gel for mass spectrometric analysis. Lysates derived from 12 x 1010 RBC containing 2.5 x 109 schizonts or 12 x 1010 uRBC were reacted with HA‐Ub‐VME probe. Reactive material was immunoprecipitated with anti‐HA resin, SDS‐PAGE separated, and polypeptides indicated by the arrows were excised and subjected to tandem mass spectrometry. PfUCH54‐matching peptides were found in bands indicated by the asterisks.
Supporting info item
Supporting info item


