Abstract
As the major etiological agent of the common cold, human rhinoviruses (HRV) cause millions of lost working and school days annually. Moreover, clinical studies proved an association between harmless upper respiratory tract infections and more severe diseases e.g. sinusitis, asthma, and chronic obstructive pulmonary disease. Both the medicinal and socio‐economic impact of HRV infections and the lack of antiviral drugs substantiate the need for intensive antiviral research. A common structural feature of the approximately 100 HRV serotypes is the icosahedrally shaped capsid formed by 60 identical copies of viral capsid proteins VP1‐4. The capsid protects the single‐stranded, positive sense RNA genome of about 7,400 bases in length. Both structural as well as nonstructural proteins produced during the viral life cycle have been identified as potential targets for blocking viral replication at the step of attachment, entry, uncoating, RNA and protein synthesis by synthetic or natural compounds. Moreover, interferon and phytoceuticals were shown to protect host cells. Most of the known inhibitors of HRV replication were discovered as a result of empirical or semi‐empirical screening in cell culture. Structure–activity relationship studies are used for hit optimization and lead structure discovery. The increasing structural insight and molecular understanding of viral proteins on the one hand and the advent of innovative computer‐assisted technologies on the other hand have facilitated a rationalized access for the discovery of small chemical entities with antirhinoviral (anti‐HRV) activity. This review will (i) summarize existing structural knowledge about HRV, (ii) focus on mechanisms of anti‐HRV agents from synthetic and natural origin, and (iii) demonstrate strategies for efficient lead structure discovery. © 2009 Wiley Periodicals, Inc. Med Res Rev, 31, No. 1, 42–92, 2010
Keywords: rhinovirus, antiviral, common cold, drug discovery
1. TAXONOMY AND HUMAN‐PATHOLOGICAL IMPACT OF HUMAN RHINOVIRUSES
A. Classification of HRV and New Findings
Human rhinoviruses (HRV) are the major cause of upper respiratory tract symptoms, the so‐called common colds in humans. Their name reflects the primary site of infection. Because HRV are nonenveloped, icosahedral viruses of small size with a diameter of about 30 nm (pico = small in Latin) that consist of an RNA genome, they were assigned to the family picornaviridae.
Currently, this virus family of the order picornavirales comprises the eight genera enterovirus, hepatovirus, cardiovirus, kobuvirus, teschovirus, erbovirus, aphthovirus, and parechovirus with 22 species and a multitude of serotypes.1 Because of high similarity in genome sequence and genome organization (Fig. 1), the former genera rhinovirus and enterovirus have been combined recently, keeping the existing name enterovirus (http://www.picornastudygroup.com/taxa/species/species.htm). An overview on the current taxonomy of picornaviruses pathogenic for humans as well as on newly proposed species of HRV is given in Table I. At present the genus enterovirus includes four approved human enterovirus (HEV) species (HEV‐A, ‐B, ‐C, and ‐D) and two approved HRV species (HRV‐A and ‐B) (http://www.picornastudygroup.com/taxa/species/-species.htm). Since 2007, the global distribution of highly divergent HRV strains was reported.2, 3, 4, 5 Based on the results of sequence, genomic, and phylogenetic analyses, it was proposed that these strains represent a new HRV species, HRV‐C.2, 3, 4, 6 In 2009, a further proposal concerning a new potential HRV‐D species was published after sequencing and analysis of all known HRV genomes.7 The approved and newly proposed species of the genus enterovirus share ≥70% homology (average amino acid identity) in the precapsid protein P1 as well as in 2C and 3CD.3, 7, 8, 9, 10
Figure 1.
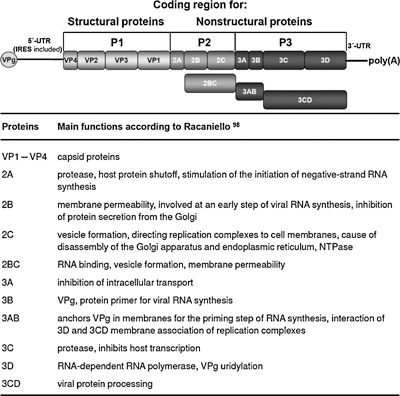
Organization of the enterovirus genome (top) and main functions of nonstructural proteins (bottom). The protein coding region of the enterovirus genome is flanked by the 5′and 3′untranslated region (UTR). A small virus protein (VPg) is covalently linked to the 5′UTR containing the internal ribosome entry site (IRES). The 3′UTR has a poly(A) tail like cellular messenger RNAs.
Table I.
Currently Approved as well as Newly Proposed Human‐Pathogenic Picornavirus Genera, Species, and Serotypes
| Genus | Species | Serotypes | Main clinical manifestations |
|---|---|---|---|
| Enterovirus | Human enterovirus A | Coxsackievirus A (CVA) 2–8, 10, 12, 14, 16, enterovirus (EV) 71, 76, 89–91 | Herpangina (CVA), meningitis, poliomyelitis, acute flaccid paralysis (AFP), gastroenteritis, exanthema, respiratory disease, hand‐foot‐and‐mouth disease (CVA10, CVA16, EV71) |
| Human enterovirus B | CVA9, CVB1‐6, echovirus 1–7, 9, 11–21, 24–27, 29–33, EV69, 73–75, 77–88, 97, 100, 101 | Meningitis, encephalitis, minor febrile illness, respiratory disease, pericarditis, myocarditis, diarrhea, AFP, hepatitis, acute hemorrhagic conjunctivitis (CVA24) | |
| Human enterovirus C | CVA1, 11, 13, 17, 19–22, 24, EV96, poliovirus (PV) 1–3 | Poliomyelitis (PV1‐3), epidemic myalgia, Guillain‐Barré syndrome, vomiting and diarrhea, AFP | |
| Human enterovirus D | EV68 and 70 | Pneumonia and bronchiolitis, acute hemorrhagic conjunctivitis (EV70), meningoencephalitis (EV70) | |
| Human rhinovirus A | Human rhinovirus (HRV) 1A, 1B, 2, 7, 9–13, 15, 16, 18–25, 28–34, 36, 38–41, 43, 44, 46, 47, 49–51, 53–68, 71, 73–78, 80–82, 85, 88–90, 94, 96, 98, 100, Hanks | Common cold | |
| Human rhinovirus B | HRV3‐6, 14, 17, 26, 27, 35, 37, 42, 48, 52, 69, 70, 72, 79, 83, 84, 86, 91‐93, 97, 99 | Common cold | |
| Human rhinovirus Ca | Have yet to be cultivated and/or assessed for immunological cross‐reactivity | Acute lower respiratory tract infections | |
| Human rhinovirus Db | HRV8c, 45c, 95c | Common cold | |
| Hepatovirus | Hepatitis A virus | Hepatitis | |
| Parechovirus | Human parechovirus 1 and 2 (previously classified as echovirus 22 and 23), 3–6 | Mild gastrointestinal or respiratory illness | |
| Kobuvirus | Aichi virus | Gastroenteritis |
Different antigenic properties provide the basis for a further division of species into serotypes (Table I). About 100 rhinovirus serotypes are currently known. According to the currently approved taxonomy, most of them (75 and HRV Hanks) belong to HRV‐A and 25 of them to HRV‐B.11, 12 The genome of all known HRV‐A and ‐B serotypes as well as of several field isolates of HRV‐A, ‐B, and ‐C has been sequenced completely.3, 7, 13, 14, 15, 16, 17, 18, 19, 20 Viruses classified as HRV‐C could not be grown in cell culture until now.
Phylogenetic analyses have been performed with partial sequences,12, 21 as well as with the whole genome.3, 7 The most recent and comprehensive analysis of all known HRV genomes revealed that (i) HRV‐A and HRV‐C share a common ancestor, which is a sister group to HRV‐B, (ii) HRV‐C represents a third species, and (iii) a basal divergence within HRV‐A of three distinct strains that led to the proposal of a fourth species HRV‐D.
B. Association of HRV With Upper and Lower Respiratory Tract Infections
HRV‐A and ‐B most often induce a mild, usually self‐limited upper respiratory illness in humans characterized by nasal stuffiness and discharge, sneezing, sore throat, and cough. The conventional term is common cold. The common cold is a heterogeneous group of diseases caused by numerous viruses that belong to several different families e.g. rhinoviruses, coronaviruses, enteroviruses, and adenoviruses.22 But, HRV represent the most common etiological agent worldwide. A large number of distinct strains circulate each year.23 Moreover, in a family or even in a single specimen, multiple HRV serotypes were detected simultaneously.24, 25, 26 By using RT‐PCR and culture, it was shown that HRV induce 22–50% of upper respiratory tract infections in adults as well as children.27, 28, 29, 30, 31 Higher incidence has been described from September to November,32, 33, 34 and from April to May.33, 34 In some years and perhaps some geographical areas, spring was a more important time for rhinovirus transmission.33, 34 Although overall rates of respiratory illness are lower in summer, rhinoviruses are the most frequently isolated at this time of year.34 The incidence is inversely proportional to age.25, 35 By age 2 years, 91% of the children have antibodies against rhinoviruses.36 In addition to common cold, HRV are also involved in acute otitis media in children.36, 37 Moreover, data supporting a causative association with more severe lower respiratory tract infections of infants, elderly persons, and immunocompromised patients have been accumulated.38, 39, 40, 41, 42, 43, 44, 45 Studies of childhood and adult asthma have shown that HRV infections can also trigger exacerbations in patients with asthma,46, 47, 48, 49 chronic obstructive pulmonary disease,50, 51, 52, 53, 54, 55 and cystic fibrosis.56, 57, 58 The recently discovered novel rhinovirus genotype HRV‐C was associated with community outbreaks of influenza‐like, acute upper respiratory infections and severe low respiratory tract infections of infants e.g. febrile wheeze, bronchiolytis, and asthma exacerbations, which peaked in fall and winter.2, 3, 4, 5, 6, 26, 59, 60 In addition, the presence of HRV‐C in the middle ear in patients with acute otitis media was demonstrated.37
C. Transmission of HRV
HRV spread occurs by means of virus‐contaminated respiratory secretions that contain a high virus concentration.61, 62, 63 Besides direct hand‐to‐hand transmission, small‐ and large‐particle aerosol transmission of rhinoviruses has been shown.61, 64, 65 Children are important “vectors” for HRV transmission to family members.25 Moreover, studies with natural HRV‐infected adults provided evidence that daily activities of infected people can lead to contamination of environmental surfaces with HRV e.g. light switches, telephone dial buttons and handsets, and virus transfer to fingers of healthy individuals for infection.63, 66 Because viral contamination of the hands plays an important role in transmission of HRV from person‐to‐person, interruption of this step of virus transmission presents a potential target for intervention. This was experimentally proved by treatment of hands by iodine67, 68 or salicylic and pyroglutamic acid.69
D. The Pathogenesis of HRV Infections
Observations from experimentally induced infections in normal adult volunteers helped to understand the pathogenesis of HRV infections.70, 71, 72, 73, 74, 75 The 50% human infectious dose of rhinovirus is low and the infection rate between 70 and 80%. After the deposition of HRV on nasal or conjunctival mucosa, viruses are transported to the posterior nasopharynx by mucociliary action of epithelial cells. Specific receptors on epithelial cells in the adenoid area are used for binding and entry. Already 8–10 hr after intranasal inoculation, infectious virus can be detected.76 Virus shedding peaks on the second day after infection and decreases rapidly thereafter.75 But, small amounts of viruses were discovered in nasal secretions for up to 3 weeks after infection. Virus and/or viral RNA were demonstrated in the upper70 as well as lower respiratory tract.45, 72, 77, 78 Using in situ hybridization, Arruda et al. (1995) detected viral RNA in a low number of ciliated cells in nasal biopsies. In the nasopharynx, a small portion of virus‐positive ciliated as well as nonciliated cells was positive for viral RNA. In 2000 Papadopoulos et al. provided evidence that HRV may also lytically infect human bronchial epithelial cells in cell culture as well as in experimentally infected volunteers and induce the production of interleukin‐6, ‐8, and ‐16. In agreement with these results, HRV RNA was detected in 24–45% of children and 10–18% of adults with pneumonia.79, 80, 81, 82 Taken together, the results of natural cold studies as well as of experimental infection in human volunteers clearly demonstrate that HRV are able to replicate in the upper as well as in the lower airways.
HRV infection triggers vasodilation and increased vascular permeability in the nasal mucosa, leading to nasal obstruction and rhinorrhoea. The mechanism is still incompletely understood because no histopathological changes were observed in nasal biopsy specimens from infected persons.75 This led to the suggestion that clinical symptoms are primarily caused by the inflammatory response of the host to the virus infection and not by the cytopathic effect (CPE) of HRV. Results of immunological investigations suggest a modest correlation between the concentrations of IL‐6 and IL‐8 in nasal secretions and the severity of symptoms in upper and lower HRV‐induced respiratory tract disease.78, 83 On day 2–4 after virus challenge, IL‐6 and IL‐8 concentrations were significantly greater in nasal secretions from experimentally infected symptomatic subjects than in those from infected asymptomatic or sham‐challenged subjects. IL‐8 has been proposed as a mediator of neutrophile infiltrations that are observed during symptomatic infections.
In experimental rhinovirus infection the onset of symptoms e.g. nasal stuffiness and discharge, sneezing, and cough was observed 10–12 hr after intranasal inoculation of the virus.76 In contrast to rhinovirus infections in adults, fever is found in 15% of children with upper respiratory tract infections.84 Other symptoms in children and adults may be hoarseness, headache, malaise, and lethargy. Sometimes viral infection is accompanied by bacterial complication, leading for instance to acute otitis media in about 20% of infected children, sinusitis, and pneumonia.79, 80, 81, 85, 86
Experimental infection was also used to study the causation between rhinovirus infection and asthma as well as COPD exacerbations.78, 87, 88, 89, 90, 91, 92, 93 It was shown that HRV infection enhances airway reactivity and predisposes allergic patients to develop late asthmatic reactions.88, 91 Rhinoviral colds were associated with an increase in histamine responsiveness that was accompanied by a bronchial mucosal lymphocytic and eosinophilic infiltrate.89 In a recent study, an increased HRV‐induced clinical illness severity in asthmatic compared with normal subjects was demonstrated.93 Strong relationships were shown between virus load, lower airway virus‐induced inflammation, and asthma exacerbation severity. The results of this study also indicated that augmented Th2 or impaired Th1 or IL‐10 immunity are likely important mechanisms. Mallia et al. provided evidence that low dose experimental rhinovirus infection in patients with COPD induces symptoms and lung function changes typical of an acute exacerbation of COPD.92 Viral replication and increased pro‐inflammatory cytokine response were associated with symptomatic colds, increases in lower respiratory tract symptoms and reductions in forced expiratory volume in 1s or peak expiratory flow rate.
E. Requirements for Anti‐HRV Agents
The epidemiological data and pathology of HRV infections explain their high medical and socio‐economic impact. Millions of children and adults are taken ill with common cold every year, need medical consultations, are unable to attend school and go to work.94, 95 Direct costs include hospitalization, medical fees, and symptomatic treatment. Moreover, exacerbations are the major cause of asthma and COPD morbidity, mortality, and health care costs associated with these diseases.92, 93 To date, specific drugs that prevent or reduce rhinovirus infection are not available. Common cold can be treated only symptomatically with analgesics, decongestants, antihistamines, or antitussives and antibiotics are often wrongly prescribed.96, 97 Because of the large number of circulating HRV serotypes, treatment with specific antiviral drugs is considered to be more striking than vaccination. Therefore, the search for new highly active synthetic and/or natural anti‐HRV compounds is absolutely essential and represents an important area of antiviral research. Such an anti‐HRV drug would have to be (i) with broad spectrum activity because of the high number of HRV serotypes, (ii) administered very early in infection to demonstrate a good antiviral effect because of the fast infection kinetics, (iii) very safe because of the broad application by millions of people, and (iv) directed against a highly conserved target with low risk of resistance development. Due to the very high error rates and the lack of proofreading ability in RNA polymerases of picornaviruses,98 naturally drug‐resistant variants may exist in virus populations or resistant viruses can emerge under treatment. As with HIV, another highly variable RNA virus, the risk of resistance development and/or selection of resistant virus variants could be minimized by applying combination of drugs directed against different targets. Because clinical symptoms are suggested to be primarily caused by the inflammatory response of the host to the virus infection mediated by specific cytokines, a further advantage of drug combinations could be an additional immune‐suppressive activity.
2. STRUCTURAL COMPONENTS, NONSTRUCTURAL PROTEINS, AND STAGES OF THE LIFE CYCLE OF HRV AS POTENTIAL TARGETS FOR SELECTIVE ANTIVIRAL DRUGS
The knowledge of structural components, nonstructural proteins that are necessary for viral multiplication, and stages of the viral life cycle is an essential precondition for the development of measures to prevent and treat HRV infection. The structure of HRV particles is well known. Infectious virions consist of an icosahedral protein shell (capsid) that surrounds and protects the genome, a single positive‐stranded RNA molecule of approximately 7,400 nucleotides. The organization of the enterovirus genome is shown in Figure 1. The viral genomic RNA is infectious and encodes a single, long, open reading frame flanked by untranslated regions (UTR) at the 5′ and 3′ end. A small viral protein (VPg) is covalently linked to the 5′ end. The 3′ end is polyadenylated like cellular messenger RNAs. Structural components within these UTRs e.g. the cloverleaf and the internal ribosome entry site (IRES) of the 5′UTR play an important role in RNA replication as well as protein synthesis.98 The nucleotide sequence of some regions within these structures is highly conserved among enteroviruses. Their blockade could significantly inhibit viral replication.
The molecular structure of HRV‐1A, HRV‐2, HRV‐3, HRV‐14, and HRV‐16 was determined by X‐ray crystallography.99, 100, 101, 102, 103, 104, 105 The results show that the viral capsid is composed of 60 protomers of each of the three outer structural proteins VP1, VP2, and VP3 and of VP4 in the interior. A star‐shaped plateau at the fivefold axis of symmetry, surrounded by a deep depression (canyon) and another smaller depression at the threefold axis were detected. Moreover, a hydrophobic pocket was found beneath the canyon floor. With exception of HRV‐14 and HRV‐3, this pocket is occupied by a fatty acid, the so‐called pocket factor. These host cell molecules have been suggested to play an important role in the viral life cycle by providing transient stability to the capsid during its movement from one host cell to another.102 The outer surface of virions contains neutralization antigenic as well as host cell binding sites. The latter allow the virus to attach to molecules of the host cell membrane (adsorption), the receptors, and to start their life cycle.106, 107, 108 Based on their receptor use, two groups of HRV can be distinguished. The majority of HRV serotypes, the major group uses intercellular adhesion molecule‐1 (ICAM‐1) as their receptor.109 The 12 viruses belonging to the minor group attach to low density lipoprotein (LDL) receptor, very‐LDL (VLDL) receptor, and LDLR‐related protein on the cells whereat multiple receptors are involved.110, 111, 112 HRV of the major group apply the canyon as attachment site for binding to ICAM‐1.113 In contrast, LDL receptors of minor group viruses bind near the tip of the five‐fold vertex.114 HRV‐87 has been shown to utilize a sialylated glycoprotein as a cellular receptor.115 Furthermore, a HRV‐89 variant as well as wild‐type HRV‐54 can use heparan sulphate proteoglycans for cell attachment in addition to ICAM‐1.116, 117 The interaction of rhinoviruses with their receptors leads to virus concentration on the cell surface. It induces the release of the pocket factor and conformational changes in the capsid and mediates viral entry via endocytosis.118, 119, 120, 121 Whereas ICAM‐1 binding directly causes uncoating,122, 123 release of the RNA genome from the capsid of LDL‐bound minor group rhinoviruses is triggered by acidification of the endosomal, pH‐dependent pathway.124, 125, 126 This detailed knowledge of the capsid structure and function as well as of the virus–receptor interaction offers a good possibility to develop antiviral drugs that interfere with the first steps of the viral life cycle, adsorption as well as uncoating.
After uncoating, rhinovirus proteins are synthesized by the translation of a single, open reading frame using cellular ribosomes. The resulting polyprotein of approximately 250 kD is cleaved by viral proteases 2Apro and 3Cpro into 11 final products (4 structural and 7 nonstrutural proteins) immediately after translation.127, 128 At first, both proteinases release themselves from the polyprotein by selfcleavage. The primary cleavage of the viral polyprotein between P1 and P2 is mediated by 2Apro. Thereafter, 3CDpro is released from the P3 precursor by autocatalytic cleavage. Next, 3Cpro and its precursor 3CDpro process proteins of the P1 (capsid proteins), P2, and P3 (nonstructural proteins) region. Interestingly, 2Apro cleaves also the eIF4GI/II component of the translation initiation factor eIF4F necessary for host cell protein synthesis,129, 130, 131 and 3Cpro and/or 3CDpro the RNA polymerase transcription factors TFIID, TFIIC, SL‐1, and UBF.98 Therefore, effective inhibition of 2Apro and 3Cpro would not only inhibit virus replication but could also prevent the shutoff of cellular protein and RNA synthesis. Moreover, the active site of proteinases is highly conserved among enterovirus serotypes.132 This high conservation in conjunction with their important role in virus multiplication predestines these enzymes as targets for antiviral therapy.
The viral RNA polymerase 3D (3Dpol) represents another very important nonstructural protein of HRV. It forms a complex with both cellular and viral proteins, the RNA replication complex.98 This enzyme synthesizes viral minus‐strand RNA and uses it as template strand for the synthesis of genomic viral RNA. VPg (3B) is the primer for negative‐ as well positive‐strand RNA synthesis. Negative‐strand, but not positive‐strand RNA synthesis, is stimulated by 2Apro. Further viral accessory proteins include 2B, 2C, 2BC, and 3AB. Besides 3Dpol these proteins can play an important role in inhibition of viral RNA synthesis by antiviral compounds.
In summary, the knowledge of the structure of the viral capsid, proteases, and polymerase and their important function in the viral life cycle predestine these proteins as potential anti‐HRV targets.
A. Experimental Approaches for Anti‐HRV Studies
HRV grow in several human and some primate cells expressing the minor group LDL receptors and/or the major group receptor ICAM‐1. Human cells susceptible to HRV infections include embryonic kidney, amnion, diploid fibroblasts from embryonic lung, tonsil, liver, intestine, and skin, adult fibroblast lines from aorta and gingival and the KB, HEP‐2, and HeLa continuous cell lines.133 But, the susceptibility of HeLa cells and human fibroblasts to virus infection may vary.32 HRV multiplication also occurs in primary human airway fibroblasts134 and differentiated bronchial epithelium.77, 78, 135 The proportion of infectible epithelial cells was shown to be between 3 and 10%.77, 135 But, enhanced levels of viral production were detected in poorly differentiated in comparison to differentiated epithelial cells.136 The degree of viral infection correlated with IL‐6 and IL‐8 induction in these cells
Virus growth causes a typical CPE characterized by ballooning, refractiveness, granularity, and shrinkage of infected cells. The HRV‐induced CPE, infectious virus titers, viral protein expression, and RNA synthesis can be chosen as parameters to evaluate the anti‐HRV activity of compounds in cell‐culture based assays. There are several methods for antiviral screening against HRV. The plaque reduction assay has been traditionally performed and accepted as the “gold standard” in antiviral testing.137, 138, 139 However, this test is laborious, time consuming, and the evaluation is subjective. Therefore, it is not suited for the routine antiviral testing. It was more and more replaced by methods based on quantification of protection from virus‐induced CPE after drug treatment. So, the CPE in sample‐treated and untreated cells has been compared by light microscopy.137, 140, 141 But this evaluation is also subjective. Another more objective approach is the spectrophotometric quantification of CPE results in neutral red or crystal violet uptake assays,12, 141, 142, 143, 144 and the tetrazolium dye reduction method.132, 145 It allows an excellent and rapid antiviral screening of large numbers of compounds using small amounts of extracts, natural, or synthetic compounds. Active samples can be scheduled for additional testing using other assays e.g. virus yield or plaque reduction assays, and for studies on the mechanism of action.
The activity of potential antiviral drugs has to be approved in vivo. Because of the high degree of species‐specific variations in ICAM‐1 preventing infection by major group HRV, practical animal models have been absent for a long time. Chimpanzees were infected with several HRV serotypes but without developing clinical signs.133 HRV do not induce infection in rabbits, guinea pigs, and weanling mice injected with HRV by different routes. One minor group HRV, serotype 2, was adapted to grow in mouse fibroblasts and used in a mouse model of rhinovirus infection in which growth could be demonstrated.146 Based on the fact that the LDL receptor family is highly conserved between human and mouse, Newcomb et al. examined whether HRV‐1B, another minor group virus, may infect mouse airways in 6‐ to 8‐week‐old female C57BL/6 mice.147 The authors demonstrated that this HRV serotype replicates and induces airway inflammation in vivo. These results strongly correspond to those of Bartlett et al. who established three novel mouse models of rhinovirus infection in BALB7c mice.148 In the first model, 6‐week‐old BALB/c mice were infected with HRV‐1B. In the second model, transgenic BALB/c mice, expressing a mouse‐human ICAM‐1 chimera, were inoculated with the major group HRV‐16. Rhinovirus‐induced exacerbation of allergic airway inflammation is mimicked in the third model.
Due to the lack of a small‐animal model for HRV infection until 2008, the experimental human challenge model has to be used to approve effects of potential antiviral drugs under controlled conditions in preclinical studies. Volunteers were experimentally inoculated with various serotypes e.g. HRV‐4, HRV‐9, HRV23, HRV‐29, and HRV‐39 to examine the efficacy of potential antiviral drugs under standardized conditions.149, 150, 151, 152, 153, 154 Examples and results of these studies with capsid‐binders, protease, and RNA synthesis inhibitors, as well as interferons are described in the following sections.
3. SYNTHETIC INHIBITORS OF HRV REPLICATION
Antiviral agents that inhibit virus attachment, capsid uncoating, protein and RNA synthesis of picornaviruses are the best studied,95, 155, 156, 157 and will be in the focus of this section.
A. Options to Prevent Virus Attachment and/or Uncoating
Inhibition of virus attachment and/or uncoating interrupts the viral life cycle at its beginning and prevents HRV infection. Options to prevent these early steps of the viral life cycle include (i) virus neutralization by HRV‐specific antibodies, (ii) receptor blockade by antibodies directed against the cellular receptors ICAM‐1 or LDL, (iii) by soluble receptor molecules, or (iv) by compounds interacting with the viral capsid.
Because of the high number of serotypes circulating often in parallel, application of HRV‐specific antibodies is thought to be no promising approach for prevention or therapy of rhinovirus infection.
In contrast, antibodies directed against the cellular receptor or soluble receptor molecules of major or minor group HRV could inhibit 90 and 10% of HRV serotypes, respectively. Therefore, the strategy to prevent virus–receptor interaction by receptor antibodies or soluble receptor molecules has been extensively evaluated in vitro as well as in vivo. The antiviral activity of ICAM‐ and LDL‐specific antibodies was confirmed in cell culture.158, 159 Furthermore, the prophylactic effectiveness and safety of intranasally administered rhinovirus murine ICAM‐1 antibody was assessed in two double‐blind, placebo‐controlled, randomized studies of volunteers experimentally inoculated with HRV‐39.160 In the result, no toxicity related to antibody application was recognized. The higher dosage of 1 mg/subject of rhinovirus murine receptor antibody did not reduce overall infection or illness rates, but was associated with a 1–2 day delay in the onset of virus shedding and cold symptoms. Viral titers and nasal symptoms were significantly reduced on the second day after challenge. In summary, the monoclonal antibody to the cellular ICAM‐1 was demonstrated to be not effective enough. A new strategy was the creation of multivalent Fab fusion proteins against ICAM‐1. A new molecule, named CFY196 demonstrated a better avidity and in vitro potency against HRV over conventional MAbs.161 CFY196 is under development as nasal spray with the name of ColdSol.
Antagonism of virus–receptor interaction was considered as another promising way to prevent HRV attachment to host cells. Soluble forms of fully or truncated ICAM‐1,162, 163, 164 and LDL or VLDL‐receptor concatemers165, 166, 167 exhibited antiviral activity against major and minor group HRV, respectively, in cell culture. Soluble forms of ICAM‐1 compete with receptor binding sites on the virus capsid, hinder an early infection event such as entry or uncoating, or directly inactivate HRV due to the formation of empty capsids.163, 168, 169, 170 A soluble LDL receptor fragment neutralized viral infectivity by aggregation.171 Concatemers of the third ligand binding module of the VLDL‐receptor did not lead to viral aggregation but blocked the receptor binding sites and possibly inhibited viral uncoating by cross‐linking the viral capsid subunits via multi‐module binding.166 The antiviral activity of a truncated, soluble form of ICAM‐1 was proved in HRV‐16 infected chimpanzee.172 In randomized, double‐blind, placebo‐controlled trials, the safety and efficacy of intranasal administration of tremacamra, a soluble ICAM‐1 in experimental HRV‐39‐induced colds in humans, was shown.173 No further development was reported for these agents.95
A further option to prevent virus attachment was described for low‐molecular‐weight compounds, the so‐called capsid‐binding agents, which enter the small hydrophobic pocket within viral capsid protein 1 beneath the ICAM‐binding canyon of HRV.174, 175 Zhang et al. showed that drug may integrate into mature viruses by diffusion as well as into progeny viruses during assembly.176 When HRV‐14 and HRV‐16 were grown in the presence of pleconaril, a higher occupancy occurred than when the drug was introduced into the already‐assembled viruses. In doing so, capsid‐binders induce conformational changes of the canyon of HRV‐3 and HRV‐14, hinder virus‐receptor interactions, and prevent attachment to host cells.105, 175, 177, 178, 179 In addition, uncoating of both HRV serotypes was shown to be inhibited as a result of a potential loss of flexibility of the viral capsid after drug binding. In contrast to HRV‐3 and HRV‐14, capsid‐binding compounds did not prevent attachment of HRV‐1A. Results from X‐ray studies showed that drug binding into the hydrophobic pocket of HRV‐1A replaces the pocket factor but induces only very small conformational changes.180 Therefore, Kim et al. suggested that the observed conformational changes are too small to affect receptor binding. But, capsid‐binding compounds prevented attachment of HRV‐16 possessing a pocket factor like HRV‐1A without distinct deformation of the pocket.102, 176, 181 Further results from comparative antiviral studies with different capsid‐binding compounds and HRV, representative for the major and minor group, did not reveal a correlation between inhibition of adsorption and receptor grouping or antiviral grouping.182 The reasons for the difference in the mode of action of capsid‐binding compounds related to attachment inhibition are not fully understood until now. Taken together, inhibition of RNA uncoating was found for all investigated serotypes after drug binding independent of receptor grouping whereas prevention of virus attachment was found to be an additional mode of action for individual viruses and/or drugs.
Till now, various potent compounds belonging to diverse chemical classes have been described as uncoating inhibitors. Just to give an impression of diversity, the structures of disoxaril and pleconaril,12, 183, 184, 185 pirodavir and the oxime ether,14, 141, 142, 186, 187 the isoxazole derivate compound,19, 143, 188 the imidazole derivative SCH 38057,189, 190, 191 the chalcone Ro 09‐0881,192, 193 4′,6 dichloroflavan and isoflavan,137, 194 the pyridine derivative MDL 20,957,195, 196 and the phenoxybenzene MDL‐860197, 198 that exhibit a potent anti‐HRV activity (Table II) are shown as examples in Figure 2. They inhibit most of HRV serotypes and a couple of them also affect enteroviruses, however, with varying susceptibility. Based on variability of susceptibility to capsid‐binders of different length, HRV serotypes were classified into two different groups, A and B.140 Several of the given examples of compounds were also clinically tested.
Table II.
Antiviral Target, 50% Inhibitory Concentration (IC50) Against Rhinoviruses and Assays Used for the Determination of Antiviral Activity of Examples of Highly Active Synthetic Compounds Described in This Review (Chemical Structures are Presented in Figs. 2–4)
| Antiviral activity in vitro | |||||
|---|---|---|---|---|---|
| Antiviral target | Synthetic compound |
 (µM) (µM) |
Assay | HRV tested | Citation |
| Capsid binder | Disoxaril | 0.01–18.1 | Plaque reduction assay | 35 serotypes | 139 |
| Pleconaril | 0.03–17.5; seven completely resistant HRV‐B | Cell protection assay with crystal violet | All serotypes | 12 | |
| Pirodavir | 0.003–39.0; four completely resistant HRV | Cell protection assay with MTT | All serotypes | 187 | |
| Oxime ether 14 | 0.002–0.02 | Cell protection assay | 16 serotypes | 141 | |
| Compound 19 | 0.01 | Cell protection assay with crystal violet | HRV‐2 | 188 | |
| SCH 38057 | 20.4–29.2 | Plaque reduction assay | 6 serotypes | 191 | |
| Ro 09‐0881 | 0.01–9.1 | Plaque reduction assay | 12 serotypes | 193 | |
| BW863C | 0.007 | Plaque reduction assay | HRV‐1B | 194 | |
| Isoflavan | ∼0.48 | Plaque reduction assay | HRV‐1B | 137 | |
| MDL 20,957 | 0.02 | Plaque reduction assay | 32 serotypes | 196 | |
| MDL‐860 | 3.25a | Virus yield reduction assay | 90 serotypes | 198 | |
| 2A and 3C Proteinase | Homophthalimides | 4.2 | Virus yield reduction assay | HRV‐2 and HRV‐14 | 215 |
| 3C Proteinase | Rupintrivir | 0.003–0.08 | Cell protection assay with XTT | 48 serotypes | 227 |
| Compound 1 | 0.014–0.12 | Cell protection assay with XTT | 35 serotypes | 231 | |
| RNA synthesis | 2‐Furylmercury chloride | 0.02–2.5; 4 completely resistant HRV | Not described | 17 serotypes | 235 |
| Ribavirin | 73.8 and 123.0 102.5 and 159.8 | Cell protection assay Plaque reduction assay | HRV‐2 and HRV‐14, resp. | 239 | |
| Enviroxime | 0.17–1.0 | Plaque reduction assay | 15 serotypes | 244 | |
| 0.03–0.2 | Cell protection assay with crystal violet | 12 serotypes | 245 | ||
a1 µg/mL reduced the virus yield of 72 serotypes by at least 1.0 log10, 12 serotypes were intermediately inhibited, and 6 not.
Figure 2.
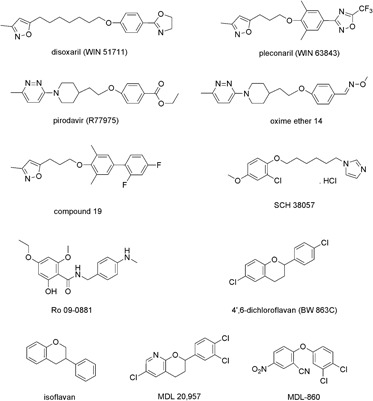
Chemical structures of selected capsid‐binding agents with potent anti‐HRV activity.
Studying the development of clinically effective capsid‐binders, the long road to the discovery of a clinically effective anti‐HRV drug becomes apparent. One well‐described example represents the discovery and optimization of capsid‐binders from Sterling Winthrop Pharmaceutical group, the so‐called WIN compounds.174 First inhibitors originated from juvenile hormone mimetics that demonstrated some activity against HRV‐1A. Determination of the X‐ray structure of HRV‐14 helped to understand the compounds' binding sites at the virus capsid.103 Results from subsequent X‐ray studies of HRV‐WIN compound complexes revealed the location and nature of binding sites and provided information concerning interactions within these sites.175, 179, 180 This knowledge was used for optimization and design of new compounds.199 Optimized WIN compounds, for example disoxaril and pleconaril (Fig. 2), consist of a methylisoxazol ring, a substituted phenoxy group, and a five‐membered heteroatom ring and inhibit a broad spectrum of rhinoviruses and enteroviruses (Table II).12, 183, 184, 185 In 1985, the first broad‐spectrum WIN compound disoxaril (Win 51711) was tested in clinical trials.139 The development of crystallurea in human volunteers treated with high doses as well as its low bioavailability (15%) prohibited subsequent development. Thereafter, results from SAR and QSAR analysis were used to further enhance the potency and spectrum of activity. In 1992, another compound, WIN 54954, was clinically tested. It was not effective in humans infected with HRV‐23 and HRV‐29.153 Moreover, it was rapidly metabolized and induced a reversible hepatitis. Consequently, the further clinical development was stopped. The better understanding of pharmacokinetic properties of capsid binders and synthetic chemistry efforts led to the discovery of pleconaril, an orally bioavailable, well‐tolerated capsid‐binder that inhibits most rhinovirus as well as various enterovirus serotypes.12, 183, 184, 185 In 2000, Schiff et al. published the efficacy of pleconaril in an experimentally induced coxsackievirus A infection in humans.200 In phase II placebo‐controlled, natural cold trials, the drug produced a moderate reduction of 1–1.5 day in the medium time to elevation of illness compared with placebo.201 These results were confirmed in two subsequent pivotal studies.202 Besides the moderate clinical efficacy, these studies revealed that 13% of baseline isolates were not susceptible to pleconaril and 11% developed reduced susceptibility (defined as 10‐fold increase in baseline value). In a subsequent study the relationship of pleconaril susceptibility and clinical outcomes in the treatment of common cold caused by rhinoviruses was demonstrated.203 Based on drug interaction, marginal treatment effect, and possibility of transmission of resistant viruses, the FDA did not approve the applied oral administration of pleconaril for the treatment of common cold. The molecular mechanism of drug interaction of orally given pleconaril was shown to be based on hepatic cytochrome P450 3A activation.204 To reduce adverse effects, Shering‐Plough under license of ViroPharma completed a phase II clinical trial with an intranasal formulation of pleconaril for the potential treatment of common cold in high‐risk populations in 2007. The results were not published until now.
Pyridazine analogues developed by Janssen Research Foundation represent another example for the long road to discovery of a clinically effective capsid‐binder.186 In 1992, the broad‐spectrum activity of pirodavir (Fig. 2; Table II) against rhinoviruses was published.187 In the same year the results of a randomized, double‐blind, placebo‐controlled trail to assess the therapeutic efficacy of intranasal pirodavir in natural common colds were described.205 Possibly as a result of poor water solubility and rapid hydrolysis of pirodavir, no clinical benefit was found. The problem of ester hydrolysis was resolved by the development of oxime ether analogues of pirodavir by Biota. An example is shown in Figure 2. Like pirodavir these new analogues are potent inhibitors of rhinoviruses.142 An advantage over pirodavir is their improved bioavailability. BTA‐798, an antiviral analogue with long half‐life and good oral bioavailability, was scheduled to a phase II clinical trial in 2008. The results have not yet been published. In summary, despite extensive research leading to the discovery of potent anti‐HRV capsid‐binders, no agent has been approved for prevention and/or therapy of rhinovirus‐induced diseases so far.
B. Development of Protease Inhibitors
Nearly, the same conclusion has to be drawn for protease inhibitors. Because of their pivotal role for viral polyprotein processing and the high conservation of critical amino acids,132 2Apro as well as 3Cpro represent potential anti‐HRV targets. Results from cell culture‐based assays provided evidence that inhibition of HRV replication is in principle possible. For example, processing of the HRV‐2 polyprotein was prevented by pyrrolidine dithiocarbamate treatment in virus‐infected HeLa cells.206 In contrast to other enteroviruses,207, 208, 209, 210 pretreatment of cell monolayers with different nitric oxide donors leading to S‐nitrosylation of 2Apro and 3Cpro had neither an effect on virus replication nor on HRV‐induced IL‐8 elaboration.211 The proteolytic activity of 2Apro of HRV‐14 was specifically inhibited by two elastase‐specific inhibitors,212 and an antiviral peptide representing a derivative of the caspase inhibitor zVAD.fmk.213, 214 Homophthalimides, e.g. LY353349 (Fig. 3; Table II), were described as inhibitors of 2Apro as well as 3Cpro.215 In contrast to protease 3C, no structure–activity relationship studies have been reported for HRV 2A protease. Moreover, protease 2A accomplishes only one cleavage in HRV polyprotein, while protease 3C performs all other cleavages. After elucidation of the crystal structure of 3Cpro,216 computer modeling of structural features of protease inhibitors became possible.217 Furthermore, structure‐based design was used to develop mechanism‐based inhibitors of the 3C protease with potent antiviral activity against multiple HRV serotypes.218, 219 Highly active compounds incorporate various Michael acceptor moieties, irreversibly bind to 3Cpro, and exhibit anti‐HRV‐14 activity in HeLa cells.220, 221 Structure–activity studies were performed to optimize protease inhibitors.220, 222, 223, 224, 225, 226 These efforts resulted in the identification of a highly active anti‐HRV compound, AG7088 (rupintrivir; Fig. 3; Table II) that entered clinical trials. In cell culture, AG7088 inhibited a broad spectrum of laboratory HRV as well as clinical isolates.132, 183, 227 In a single‐cycle, time‐of‐addition assay it demonstrated antiviral activity when added up to 6 hr after infection.227 Inhibition of HRV replication strongly correlated with reduction in the level of IL‐6 and IL‐8 release into cell supernatant, leading to the suggestion that this agent may not only block virus replication but also diminish symptoms.228 The pharmacokinetics and safety of rupintrivir were proved in two double‐blind, randomized, placebo‐controlled studies.229 Intranasal rupintrivir, administered as single doses of 4 and 8 mg or every 3 hr, six times per day, for 7 days, was safe and well tolerated. Three double blind, placebo‐controlled clinical trials were conducted to assess rupintrivir nasal spray (2% solution) for prevention and treatment of experimentally induced rhinovirus colds in healthy volunteers.230 Rupintrivir prophylaxis reduced the proportion of subjects with positive viral culture by 26% and viral titers but did not decrease the frequency of colds. Drug treatment led to the reduction of the mean total daily symptom score by 33%. Subjects receiving rupintrivir also demonstrated significantly lower viral titers and RNA levels than placebo‐treated subjects on days 2, 3, and 5 and on days 2 and 3, respectively. There was no influence on the proportion of subjects with positive viral culture and the frequency of colds. Clinical development was terminated because rupintrivir did not act in a subsequent natural infection study in patients.231 In parallel research efforts, an orally bioavailable inhibitor of HRV 3Cpro, i.e. (E)‐(S)‐4‐((S)‐2‐ 3‐[(5‐methyl‐isoxazole‐3‐carbonyl)‐amino]‐2‐oxo‐2H‐pyridin‐1‐yl
3‐[(5‐methyl‐isoxazole‐3‐carbonyl)‐amino]‐2‐oxo‐2H‐pyridin‐1‐yl ‐pent‐4‐ynoylamino)‐5‐((S)‐2‐oxo‐pyrrolidin‐3‐yl)‐pent‐2‐enoic acid ethyl ester (Fig. 3; Table II), was discovered.225, 231 Like rupintrivir, this compound is an irreversible inhibitor incorporating a Michael acceptor moiety that forms a covalent bond with the 3C protease active site cysteine. It demonstrated an antiviral activity against all HRV and related picornaviruses tested.231 In a phase 1 clinical study, compound 1 was shown to be safe and well tolerated. According to a publication of Patick, no further clinical development was planned for this compound.95
‐pent‐4‐ynoylamino)‐5‐((S)‐2‐oxo‐pyrrolidin‐3‐yl)‐pent‐2‐enoic acid ethyl ester (Fig. 3; Table II), was discovered.225, 231 Like rupintrivir, this compound is an irreversible inhibitor incorporating a Michael acceptor moiety that forms a covalent bond with the 3C protease active site cysteine. It demonstrated an antiviral activity against all HRV and related picornaviruses tested.231 In a phase 1 clinical study, compound 1 was shown to be safe and well tolerated. According to a publication of Patick, no further clinical development was planned for this compound.95
Figure 3.
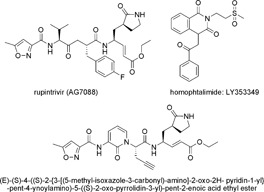
Chemical structures of most active inhibitors of 2A and 3C protease of HRV.
C. Inhibition of Viral RNA Synthesis
The blocking of viral RNA synthesis during replication represents another site for chemotherapeutic interdiction. It was shown that, rhinoviral RNA can be targeted in a sequence‐specific manner by deoxyribozymes,232 morpholino oligomers,233 and small interfering ribonucleic acids.234 The efficacy of the latter two approaches was confirmed in cell culture. In addition, 2‐furylmercury chloride (Fig. 4; Table II),235 flavonoids for example 3‐methylquercetin (Fig. 5; Table III),236 and pyrrolidine dithiocarbamate (Fig. 4) interfered with rhinoviral RNA synthesis and inhibited HRV replication in cell culture‐based assays.206, 237 The nucleoside analog ribavirin (Fig. 4; Table II) that inhibits a broad spectrum of RNA as well as DNA viruses acts also against HRV‐2 in HeLa cells.238, 239 The cellular inosine monophosphate dehydrogenase that controls de novo synthesis of purine nucleosides represents the principal target in the mode of action of ribavirin.240 Moreover, when ribavirin is incorporated into picornavirus RNA, it pairs equally well with either uracil or cytosine inducing mutations that can be lethal to RNA viruses.241 Further identified mechanisms of action for ribavirin include inhibition of genomic RNA capping, enhancement of host T‐cell‐mediated immunity against viral infections through helping to switch the host T‐cell phenotype from type 2 to type 1.242 Another compound with potent anti‐HRV activity in vitro is enviroxime (Fig. 4; Table II), a benzimidazole derivative.243, 244 It inhibits viral plus strand RNA synthesis.245 In particular the 3A protein, which is involved in the initiation of plus strand RNA synthesis, was implicated as likely target of drug activity.246, 247 However, results from another study suggest that enviroxime targets a complex of proteins and/or cellular factors and that the exact mechanism remains to be studied.248 Although there was a statistically significant reduction in clinical score in a prophylactic study with HRV‐9‐infected volunteers,152 enviroxime failed in experimentally induced HRV‐4 and HRV‐39 infection,149, 151 and in clinical studies,248, 249, 250 because of poor bioavailability and side effects. In an attempt to overcome the marked hydrophobicity, water insolubility, and toxicity, Wyde et al. incorporated enviroxime into liposomes and then tested the anti‐HRV activity and toxicity of the liposome‐incorporated enviroxime in cell culture.251 The liposome preparation of enviroxime inhibited HRV‐1A and HRV13 as effective as the parent compound and was 10‐ to ≥50‐fold less toxic. In contrast to free enviroxime, the liposome preparation was readily and successfully delivered by small‐particle aerosol to the upper and lower respiratory tract of mice. In another attempt to overcome the disadvantages of enviroxime, several benzimidazole as well as nonbenzimidazole analogs were synthesized and studied.252, 253, 254, 255 Even though some compounds were better bioavailable and could be administered orally,254 none of these compounds was tested in clinical studies.
Figure 4.
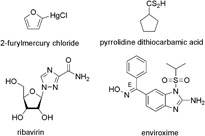
Chemical structures of most active inhibitors of HRV replication.
Figure 5.
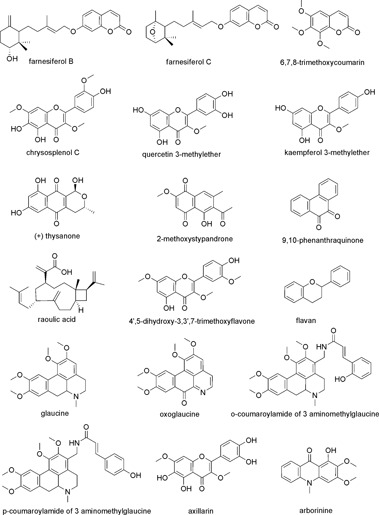
Chemical structures of natural anti‐HRV compounds.
Table III.
Reported Antiviral Activities of Natural Compounds Described in the Review (Chemical Structures Are Given in Fig. 5)
| Antiviral activity in vitro | |||||
|---|---|---|---|---|---|
| Natural product | IC50 (µM) | Assay/virus tested | Positive control IC50 (µM) | Proposed mechanism of action | Citation |
| Farnesiferol B | 2.61 | Cell protection assay HRV‐2 | Pleconaril 0.03 | Capsid binder | 144 |
| Farnesiferol C | 2.51 | Cell protection assay HRV‐2 | Pleconaril 0.03 | Capsid binder | 144 |
| 6,7,8‐Trimethoxy‐coumarin | 11.98 | Cell protection assay HRV‐2 | Pleconaril 0.03 | Capsid binder | 318 |
| Flavan | 0.05 | Plaque reduction assay HRV‐1B | – | Capsid binder | 149 |
| Arborinine | 3.19 | Cell protection assay HRV‐2 | Pleconaril 0.03 | Capsid binder | 318 |
| (+)‐Thysanone | 47.1 | 3C protease assay HRV‐14 | – | Inhibitor of 3C protease | 323 |
| 2‐Methoxy‐stypandrone | 4.6 | 3C protease assay HRV‐14 | – | Inhibitor of 3C protease | 323 |
| 9,10‐Phenanthra‐quinone | 1.4 | 3C protease assay HRV‐14 | – | Inhibitor of 3C protease | 324 |
| Chrysosplenol C | 0.75 | Cell protection assay poliovirus type 1 | Guanidine HCl 310–1250 | Inhibitor of virus replication | 317 |
| Quercetin 3‐methylether | 0.95a | Titer reduction assay poliovirus 1A/S3 | – | Inhibitor of virus | 320 |
| 0.03b | Virus yield reduction assay HRV‐15 | – | Replication | 236 | |
| Kaempferol 3‐methylether | 0.67a | Titer reduction assay poliovirus 1A/S3 | – | Inhibitor of virus replication | 320 |
| 4′,5‐Dihydroxy‐3,3′,7‐trimethoxyflavone | ∼0.3 | Cell protection assay 20 HRV serotypes | – | Inhibitor of virus replication | 192 |
| Glaucine | 22.0 | Cell protection assay HRV‐14 | Disoxaril 1.5 | Inhibitor of virus replication | 327 |
| Oxoglaucine | 0.3 | Cell protection assay HRV‐14 | Disoxaril 1.5 | Inhibitor of virus replication | 327 |
| o‐Coumaroylamide of 3 aminomethylglaucine | 15.0 | Cell protection assay HRV‐14 | Disoxaril 1.5 | Inhibitor of virus replication | 327 |
| p‐Coumaroylamide of 3 aminomethylglaucine | 13.0 | Cell protection assay HRV‐14 | Disoxaril 1.5 | Inhibitor of virus replication | 327 |
| Axillarin | 1.82 | Cell protection assay HRV‐2 | – | Inhibitor of virus replication | 319 |
| Raoulic acid | <0.27 0.51 | Cell protection assay HRV‐2 Cell protection assay HRV‐3 | Ribavirin 356.5 (HRV‐2) ‐ (HRV‐3) | – | 325 |
a99% effective dose.
bMaximum tested concentration.
4. APPLICATION OF INTERFERONS
Besides virus‐specific targets, cellular inhibitors like interferons may represent a therapeutical approach. Among other activities, interferons exhibit antiviral activity. The advantages of interferon application include the broad spectrum of activity and low risk of resistance development. Human leukocyte and fibroblast as well as recombinant human α interferons prevent the HRV‐induced CPE in cell culture whereas a variation in sensitivity was observed.256, 257, 258 Intranasally applied recombinant interferon α and interferon β have been shown to be effective in humans when provided prophylactically both in experimental and natural rhinovirus colds.259, 260, 261, 262, 263, 264 Significant reductions in illness frequency, mean symptom score, nasal secretion weights, and frequency of virus isolation were observed. In contrast, recombinant interferon γ did not prevent HRV infection or illness and may enhance the symptoms.265 Little to no therapeutic effect was found in patients with common cold after interferon treatment.150, 266 Moreover, blood‐tinged mucus and nasal bleeding were described as side effects.263, 267 Combining interferons with dichloroflavan, enviroxime, chalcone Ro‐09‐0410 produced synergistic increases in antiviral activity in vitro against HRV‐2 and HRV‐9.268 An attempt to demonstrate synergy between the anti‐HRV effect of recombinant human rHuIFN α and enviroxime in HRV‐9 and HRV‐14‐infected volunteers failed.269 According to the authors, the main reason for this failure may be the rapid removal of enviroxime from the nose when given intranasally.
5. ANTI‐HRV AGENTS FROM NATURE AND PROPOSED MECHANISM OF ACTION
A. Impact of Natural Products
Nature provides an astonishing pool of secondary metabolites biosynthesized from living organisms such as plants, fungi, protozoan, insects, and other animal sources. In contrast to synthetic compounds, natural products are characterized by an overwhelming chemical diversity. Previously, the chemical diversity space between these two groups was evaluated with respect to drug substances by Feher and Schmidt.270 It is shown that combinatorial compounds densely populate a small area, whereas natural products cover a wider range quite similar to the chemical space occupied by drug substances. The authors accordingly suggest that combinatorial libraries that mimic the distribution properties of natural products might be more biologically relevant. One may assume that secondary metabolites evolved as reaction to their target receptors related to defence, protection, attraction, and signalling. These adaptation processes have enriched not only the metabolites' structural diversity but have also optimized drug‐like metabolic traits likely to have favorable pharmacokinetic properties.271, 272 It is this evolutionary concept that gives the pool of natural products the greatest source of scaffold diversity with molecules of biological relevance.
Newman and Cragg analyzed the number of drugs approved between 1981 and 2006 and circumstantiated that especially the anti‐infective area is strongly dependent on natural products and structures derived from natural scaffolds.273 The anti‐infectives including the antiviral vaccines are with 22.8% or 230 launched drugs by far the major category with only about 30% being synthetic in origin. From 1981 to 2006, 78 vaccines and antiviral drugs have been approved. Excluding the high number of vaccines (25) and biologicals (12), most of the 41 small antiviral molecules are based on nucleoside structures or on peptidomimetics; only 16.7% are classified as totally synthetic drugs. However, till now, neither a synthetic nor a naturally derived anti‐HRV drug substance has been approved for the treatment or prevention of HRV infections.
Intensive research and development efforts in the field of natural products revealed several inhibitors of viral attachment and entry, and inhibitors of viral protease from natural sources. The efficacy of natural products is not only reflected by statistics of launched drugs but also by empirical knowledge gained over centuries by successful application of natural‐based ethnomedicinal products such as plants, culinary herbs, and spices. Phytochemical and pharmacological work performed with ethnomedicinal anti‐HRVs mainly from plants revealed a high number of active metabolites from different chemical classes, e.g. coumarins, flavonoids, alkaloids, quinones, terpenoids, polyphenols, and polysaccharides.
Natural products include complex extracts and their chemical entities, which are biosynthesized by nature. For an unambiguous presentation of anti‐HRV natural products it is of prior importance to first distinguish between a single chemical entity from nature, i.e. an isolated, purified natural compound, on one hand, and a natural preparation comprising hundreds to thousands of constituents, mainly secondary metabolites, on the other hand. If natural preparations are derived from plants, they might also be labelled as botanicals, phytoceuticals, or phytotherapeutic agents. These multicomponent preparations might show a varying profile of their constituents depending on the used species, origin, collection time, plant parts, extraction procedures, preparation methods, and manufacturing processes, just to mention a few important elements. These parameters affect the final product in terms of the qualitative and quantitative composition of chemical constituents, which may have an impact in biological activity. Accordingly, studies performed with phytochemically not specified extracts or nonstandardized preparations often suffer from irreproducible and incomparable results
B. Anti‐HRV Natural Preparations
A wide variety of natural preparations showed to be acting therapeutically in HRV and other viral infections with often complementary and overlapping antiviral mechanisms of action.274, 275, 276 Most of these remedies are described in ethnopharmacological sources or handed down for generations. They usually consist of simply prepared natural items whose chemical composition is complex. Many of the contained secondary metabolites, possibly active principles, have never been examined chemically or biochemically using modern medical knowledge. They are however components of plant medicines, which have stood the test of time and as such may offer clues of great interest to medicinal chemists. A clear advantage of the application of these products is their absent or relatively low toxicity due to a usually long‐term empirical trial.
Although the knowledge of the immuno‐pathogenesis of RV‐induced diseases remains limited, the host defense function of the airway epithelium plays an important role in the innate‐immune response to HRV‐infection.277 Host cells respond by the production of mediators with antiviral activity such as type I interferons and nitric oxide, and produce cytokines and chemokines that influence the subsequent induced innate‐ and specific‐immune response. These processes are beneficial in facilitating clearance of virus from the respiratory tract, but also cause immuno‐pathology. Following HRV‐infection, disease severity is dependent on direct, harmful effects of the virus as well as tissue damage as a result of the host antiviral immune response. Accordingly, a number of agents with phenomenological effects against common cold have shown to exert their activity more in the field of regenerating tissue damage than on a direct anti‐HRV effect.
Several herbal remedies consisting of a multitude of secondary metabolites from different chemical classes may attribute in a beneficial way for the treatment of common cold by reducing symptom severity and duration due to their immune‐modulating, anti‐oxidative, and anti‐inflammatory properties. Beside these commonly observed bioactivities of natural products, multicomponent mixtures like botanicals often show overlapping symptomatic effects as well as synergistic and/or additive properties. Thus, it is a challenging endeavor to track down an observed phenomenological effect of a complex mixture on a molecular level.
The following section explores the significance and current knowledge of selected botanicals for the prevention and therapy of common cold. Questions about (1) clinical evidence of efficacy, (2) the constituents or at least the chemical classes that are involved in the observed anti‐HRV effect, and (3) the involved pharmacological targets were covered as far as possible.
1. Echinacea (E. angustifolia, E. purpurea, E. pallida)
Echinacea preparations include expressed juice from aerial parts as well as extracts of roots or aerial parts, or both, from one or more species of the genus Echinacea (E. angustifolia, E. purpurea, and E. pallida). They are the most recognized botanicals for prevention and treatment of common cold and flu, and account for the second top‐selling herbal products in the US‐market.278 Accordingly, Echinacea has come under much scientific scrutiny. The high number of studies dealing with the effectiveness of Echinacea for preventing and treating the common cold from clinical trials was recently reviewed by Woelkart et al.279 The authors summarized the findings of the meta‐analyses regarding the 16 randomized controlled trials evaluated in the Cochrane database,280 the 14 randomized clinical trials analyzed by Shah et al.,281 and the experimental HRV‐infection studies pooled by Schoop et al.282 To sum up, the clinical data on Echinacea so far are not fully consistent, mainly based on problems inherent in assessing the efficacy of Echinacea preparations, such as lack of comparability of available preparations, study design, and outcome. Nevertheless, the meta‐analyses showed some evidence that preparations based on the aerial parts of Echinacea purpurea might be effective for the early treatment of colds in adults.280 Echinacea showed to decrease the odds of developing the common cold by 58% and the duration of a cold by 1.4 days.281 Similarly, the evaluation of three induced rhinovirus prevention studies revealed the odds of experiencing a clinical cold were 55% higher with placebo than with Echinacea.282
Stepping into a molecular level, several constituents found in Echinacea species could potentially affect the symptoms of common cold. Chemically identified substances include polysaccharides and glycoproteins, caffeic acid derivatives (especially cichoric acid and echinacoside), and lipophilic polyacetylenes and alkamides. Pharmacological studies have shown that cichoric acid, alkamides, glycoproteins, and polysaccharides possess immunomodulatory activity. Additionally, alkamides have been reported to exert not only anti‐inflammatory effects but also cannabinomimetic properties, which are suggested as molecular mode of action of Echinacea alkamides as immunomodulatory agents.283 Raduner et al. showed that some Echinacea alkamides exert cannabinoid type 2 receptor‐dependent and independent immunomodulatory effects on cytokine expression.284 Different Echinacea constituents were evaluated for their anti‐oxidative effects measuring the inhibition of in vitro Cu(II)‐catalyzed oxidation of human low‐density lipoprotein. Thereby, the major caffeic acid derivatives, cichoric acid and echinacoside, showed the highest anti‐oxidative effects, which was even higher when combined with a natural mixture of alkamides.285 Sharma et al. used cytokine antibody arrays to investigate the changes in the pro‐inflammatory cytokines and chemokines released from human bronchial epithelial cells exposed to HRV 14.286 Application of two chemically characterized Echinacea extracts showed a reversion of the stimulated release of numerous pro‐inflammatory cytokine‐related molecules, e.g. for the cytokine IL‐6, and the chemokines IL‐8 and eotaxin. In a similar study, an Echinacea extract rich in polysaccharides and another rich in alkamides and caffeic acid derivatives were as well able to neutralize the effects of HRV‐infected epithelial cells.287 Using gene expression analysis both studies revealed the anti‐HRV benefit of Echinacea preparations being involved in multiple immune response signaling pathways. Taken together, the numerous pharmacological findings from literature, the potential of Echinacea preparations, and their constituents to combat or prevent common cold can be deduced to immune modulating, anti‐inflammatory, and anti‐oxidative properties that may also act in some combination of these event, rather than acting directly on HRV.
2. Garlic (Allium sativum)
Garlic cloves have been used traditionally to treat a number of infectious diseases. However, only few confirmatory studies have been published regarding the traditional antiviral uses. The clinical effectiveness of garlic on the prevention of common cold was investigated by Josling in 2005, who published a double‐blind, placebo controlled study assessing 146 patients more than a 12‐week treatment period with an allicin‐containing garlic supplement.288 Common cold infections and symptoms were recorded in a daily diary. Patients in the treatment group had significantly fewer colds than patients in the placebo group (24 vs. 65, P<0.001) who also had a longer duration of symptoms (5.01 vs. 1.52 days, P<0.001).
As soon as the garlic is chewed, cut, or pressed, its main ingredient, the sulphur containing alliin, is broken down by the enzyme alliinase to the thiosulfinate allicin. By steam distillation allicin is transformed to diallyl disulfide and diallyl trisulfide that are responsible for the distinctive smell of garlic. Further, allicin transformation compounds, such as E‐ and Z‐ajoene, are not found in fresh garlic, but in lipophilic extracts. By investigation of different garlic extracts and isolates against a number of different human pathological viruses, Weber et al. could show that allicin was the most active virucidal component from fresh garlic and fresh extracts.289 Results of the direct pre‐HRV‐2‐infection incubation assay let suggest allicin to bind to the viral protein capsid, leading to a subsequent inhibition of viral adsorption and penetration. Although the garlic thiosulfinates are endowed with significant cytotoxicity, the antiviral effects were obtained in nontoxic concentrations.289 Beside the direct anti‐HRV effect of fresh garlic extract and allicin, a number of human immune functions were found to be enhanced in vitro by aqueous garlic extract, its polar, and thiosulfinate fractions.290
3. North American Ginseng (Panax quinquefolium)
In North America, Panax quinquefolium, the ginseng species indigenous to both Canada and the United States, has been a popular herbal remedy to combat stress, and to modulate both natural and acquired immune responses. American ginseng root extracts, rich in poly‐furanosyl‐pyranosyl‐saccharides, have been found efficacious in the prevention of upper respiratory infections in immunocompetent healthy adults.291, 292
In a randomized, double‐blind, placebo controlled trial, 200 mg of a proprietary American ginseng root extract was given to 43 community‐dwelling elderly adults (age >65 year) twice a day more than a “cold and flu” season period of 4 months. One month into the study, all participants received an influenza vaccination. During the first two months, no significant differences in duration and incidence were observed when compared to placebo. However, during the last two months significantly fewer subjects of the ginseng group reported acute respiratory syndromes than the placebo group (32 vs. 62%). Additionally, the duration of respiratory symptoms was reduced by 55% in the ginseng group.291
In a similarly arranged trial, 323 healthy adults (ages 18–65 years) with a history of at least two colds the previous year commenced a 4 month study at the beginning of a cold and flu season.292 They received two 200 mg capsules daily of standardized American ginseng root extract or a placebo. Outcomes measured were number of colds including symptom severity and total number of symptomatic days. A therapeutic effect was reported regarding symptom severity and fewer symptom days that were 31 and 34.5% lower in the ginseng group than in the placebo group.
A phase II randomized, controlled trial of 2 dosing schedules of American ginseng root extracts, rich in poly‐furanosyl‐pyranosyl‐saccharides, evaluated the safety, tolerability, and efficacy in a pediatric population already suffering from an upper respiratory tract infection. The results showed no serious adverse events and a good tolerability of both ginseng doses; however, frequency and severity of symptoms were not significantly different among each of the three treatment groups, i.e. standard dose, low dose, and placebo.293
The most prominent constituents of the genus Panax are the triterpene saponins ginsenosides. They are known to have numerous pharmacological activities such as anti‐cancer, anti‐diabetes, antiviral, and anti‐atherosclerosis effects. Some compounds of this chemical class showed to be responsible for the immunostimulant activity of ginseng.294 On the other hand, the efficacy of a polysaccharide‐rich extract of American ginseng was compared with an extract rich in ginsenosides on systemic and gut‐associated immune function. The authors of this study investigated the lymphocytes isolated from spleen, mesenteric lymph nodes and Peyer's patches, and immune cell proportions and cytokine production from Sprague–Dawley rats. They could show that the polysaccharide‐rich ginseng extract modifies the rats' systemic immune responses and affects the gut‐associated immunity in a manner distinct from that of the ginsenoside‐containing extract of American ginseng.295
A direct antiviral activity of ginseng constituents could be attested for the polysaccharides on rotavirus infection in MA104 cells. The triterpene saponins, however, did not exhibit any rotavirus infection‐inhibitory activity in this study.296 A moderate in vitro virucidal effect (ID50 62 µM) of the ginseng saponin chikusetsusaponin III against herpes simplex virus type I was detected by Fukushima et al. This compound exhibited an intracellular inhibitory activity, but could only marginally affect the viral proteins postinfection.297
4. Bu‐zhong‐yi‐qi‐tang/Hochu‐ekki‐to
The ancient Chinese formula Bu‐zhong‐yi‐qi‐tang (Japanese name Hochu‐ekki‐to) is a traditional herbal medicine in China and Japan that is composed of ten species of medicinal plants, namely Astragali radix, Atractylodis lanceae rhizoma, Ginseng radix, Angelicae radix, Bupleuri radix, Zizyphi fructus, Aurantii nobilis pericarpium, Glycyrrhizae radix, Cimicifugae rhizoma, and Zingiberis rhizoma. This formula is reported to have various immunomodulatory,298, 299, 300 and anti‐inflammatory activities.301 Yamaya et al. recently investigated the effects of Hochu‐ekki‐to in cultures of human airway epithelial cells infected with HRV‐14.302 The output of virus, associated levels of viral RNA, and the production of ICAM‐1, cytokines and acidic endosomes in cells were measured. In airway epithelial cells Hochu‐ekki‐to was able to decrease virus output and susceptibility to HRV infection by decreasing ICAM‐1 and by blocking the entry of viral RNA into the cytoplasm from the endosomes. Glycyrrhizin, a major component of one herbal ingredient of Hochu‐ekki‐to, i.e. Glycyrrhiza glabra, was able to reduce supernatant virus titers dose‐dependently, with a maximum effect between 0.12 and 0.6 µM. However, no clinical trials with representative numbers of subjects are published so far.
5. Umckaloabo (Pelargonium sidoides)
P. sidoides and P. reniforme form the origin of the popular drug Umckaloabo. This herbal remedy from South Africa has found entrance in Western medicine mainly as aqueous ethanolic root extract from P. sidoides for the treatment of infections of the respiratory tract. The efficacy of Umckaloabo compared with placebo has been evaluated in 103 adults suffering from common cold by Lizogub et al.303 The applied herbal preparation was well tolerated by the patients. The study demonstrated only a weak efficacy of Umckaloabo compared to placebo after 5 days. After 10 days, however, the P. sidoides extract significantly reduced the severity of symptoms and shortened the duration of the common cold compared with placebo. Just recently, Timmer et al. selected randomized controlled trials examining the efficacy of P. sidoides preparations for the treatment of various acute respiratory infections and analyzed their efficacy and safety.304 The authors concluded that Umckaloabo may be effective in alleviating symptoms of acute rhino‐sinusitis and the common cold in adults. It may be effective in relieving symptoms in acute bronchitis in adults and children, and sinusitis in adults. Reliable data on the treatment for other acute respiratory infections however were not obtained.
Identification of the metabolites from Umckaloabo revealed a high number of different chemical classes, such as phenolic and cinnamic acids, tannins, flavonoids, and coumarins. Antibacterial activities of Umckaloabo against different pathogens have been reported. Phenols, coumarins, and tannins have been identified to contribute with moderate antibacterial activities, however, cannot explain the effect of the whole extract (reviewed by Kolodziej305, 306). Additionally, P. sidoides extracts have been reported to significantly activate the nonspecific immune system by induction of TNF and NO‐release, and IFN‐like activities.305 These effects are assumed to contribute to the controversially discussed potential of P. sidoides extract for the treatment of upper respiratory tract infections. Only one study reports a direct antiviral effect, i.e. a clear dose‐dependent anti‐herpes simplex virus acitivity for the aqueous root extract of P. sidoides.307 Further pharmacological studies are needed to elucidate potential direct anti‐HRV properties of Umckaloaba and its constituents.
6. Carrageenan (Sulphated Polysaccharides)
Carrageenan, a mixture of different polysaccharides, which is mainly extracted from red seaweeds, has been extensively used in food, cosmetic and pharmaceutical industry as a thickener and gelling agent. It has previously shown an antiviral efficacy against several viruses.308, 309 In a recently published study, Lambda‐, Kappa, and Iota‐carrageenan were investigated for their anti‐HRV inhibiting potential. At a concentration of 200 µg/mL Iota‐carragenan, a sulphated polysaccharide, was able to fully inhibit virus‐induced cell death in HRV‐2 infected HeLa cells.310 Based on their studies, Grassauer et al. concluded that Iota‐carrageenan is effective against different HRV‐serotypes on primary human epithelial cells. It is hypothesized by the authors that Iota‐carrageenan might create a hostile environment for HRV and thereby block viral entry and replication. Because of its safe application and proved in vitro efficacy, Iota‐carrageenan deserves consideration as a candidate for clinical trials for prevention and therapy of HRV‐induced common cold.
7. Difficulties for the Development of Botanicals Combating Common Cold
The level of knowledge on the impact of the six botanicals on HRV‐infection discussed above is different and heterogeneous. The best studied herbal remedy associated with common cold, i.e. Echinacea, showcases the innate problem connected with multi‐component mixtures: starting from the late nineties till June 2009 some 100 original articles have been published to this topic and tried to elucidate questions concerning efficacy, molecular mechanism, and bioactive ingredients of Echinacea. Although some evidence is provided for the effects of extracts, chemical classes as well as well‐defined constituents on specific targets and pathways, the findings cannot be deduced to a common denominator. Further, results from clinical trials often suffer from lack of comparability, because of using different study designs, outcome measures, and overall the application of different preparations.279 A proper quality analysis and characterization of the preparation under investigation is mandatory and should follow the recommendations and guidelines for reporting clinical trials for herbal medicine.311 As underlined before (Sections 5.1. and 5.2.), the chemical complexity of a natural preparation might be beneficial in terms of synergistic, additive, and overlapping effects caused by the multitude of evolutionary trimmed metabolites, which may attribute with modulating multi‐target effects. On the other side, exactly this fact is hardly compatible with the proper assignment of an activity to a defined chemical entity according to Western medical practise. In contrast to a single compound (synthetic or naturally based), the chemistry of a botanical is not only complex but also varying. The analytical profile and in term the pharmacological profile of the investigated samples can differ substantially. Accordingly, the quantitative and qualitative comparison of different studies resulting from botanicals is by far more complex than those performed with pure single compounds, and may also explain why so little emphasis from pharmaceutical industry has been put into the further development of even promising natural preparations.
6. STRATEGIES FOR THE DISCOVERY OF ANTI‐HRV LEAD STRUCTURES
In general, the search for potent, selective, nontoxic compounds that might be developed further to a drug substance is a multidisciplinary, time‐ and cost‐consuming process. Therefore, strategies for a target‐oriented discovery of lead‐structures either from nature or synthesis are in high demand. Some of them will be discussed in the following section, providing examples from anti‐HRV research.
A. Ethnopharmacological Approach
As already mentioned, nature provides an extremely rich pool of bioactive natural products. It is however a challenging endeavor to find exactly those compounds that show an activity on the focused target. In natural product research hints from folk medicine are a valuable starting point to dig for lead structures of certain interest. The majority of active principles from higher plants has been discovered as a result of ethnopharmacologically directed pharmacognostic research.312, 313 Oral or written indications for a beneficial application of a natural material first need a critical evaluation as to the selection of the correct material, its medicinal preparation, the kind of application, and overall the pharmacological profile. A rational criticism of often anecdotal efficacy from traditional medicine is a mandatory attitude to avoid overinterpretation of handed‐down information.314 In case of pretended anti‐HRV remedies, the reported biological efficacy obviously suffers from being restricted to respiratory diseases, which might be caused by a panel of bacterial or viral infections. Thus, an approved remedy may affect the microbes, or show an immune modulating or even anti‐inflammatory effect, or a combination of these. The holistic access is an innate character of ethnopharmacology and needs to be tracked down to the specifically involved target/s.
In a recently published study focusing on the discovery of HRV‐capsid binders from nature using a pharmacophore‐based virtual screening of an ethnopharmacologically biased 3D‐multiconformational database, some 30 secondary metabolites were predicted to act as capsid‐binding inhibitors of HRV.144 For an in depth phytochemical and pharmacological investigation, it was mandatory to focus on one promising natural material. Thus, we consulted the ethnobotanical source Materia medica, which was written by Pedanius Dioscorides in the 1st century AD. The treatise consisting of five books comprises some 1,000 or so drugs derived from minerals, animals, and the majority of them from plants (∼800). It represents a great repository of botanical, medical, and pharmacological lore.
Scrutinizing the natural materials underlying the obtained virtual hits we came across asafetida, which is a gum resin gained from the roots of a variety of foul‐smelling Ferula species from the Apiaceae family. Based on the descriptions given in the Materia medica there is strong evidence that the juice (i.e. resin) of the popular ancient silphion originating from Media and Syria corresponds to asafetida (book III, cap. 80).315 Yielded by incision of the root and stalk and frequently mixed with sagapenon, i.e. the resin of F. persica Willd (book III, cap. 81), it is reported to be effective in the context of upper respiratory diseases, e.g. “for chronic harshness of the throat,” “it clears the voice,” “shrinks the uvulas,” “suitable for a cough,” “for pleurisy,” “for chest pain;” sagapenon is described as follows: “it clears thick matters from the lungs,” “given to those who are chilled.”315 These descriptions finally helped to prioritize those virtual hits, which have been reported to be constituents of asafetida.316 The pharmacological investigation of asafetida and its constituents farnesiferol B and C (Fig. 5; Table III) revealed a distinct anti‐HRV‐2 effect in the low micromolar range using a CPE inhibitory assay.143 The results of this study provided a rationale for the ancient usage of asafetida for upper respiratory tract infections. On the other hand, the traditionally manifested evidence for asafetida for the treatment of common cold symptoms substantially helped in the selection of this plant material in the search for anti‐HRV capsid binders.144
Typically, an ethnopharmacologically based discovery of an active (anti‐HRV) extract is followed by a bioassay guided fractionation, and the isolation of those constituents that are responsible for the extracts' bioactivity. The concept of a bioassay‐guided approach is the fractionation accompanied by simultaneous detection of the activity during the separation steps, which results in a continuous enrichment, and finally in the isolation of the active ingredient/s. In this way a large number of anti‐HRV agents from natural sources have already been discovered.
Semple et al. investigated the active principle of the Asteraceae plant Pterocaulon sphacelatum, which has been used in traditional medicine of Aboriginal people of Australia as a favored treatment for respiratory infections, especially colds. By means of an antiviral activity‐guided fractionation measuring the poliovirus‐induced CPE assay, the authors identified the flavonoid 3,7,3′‐trimethoxy‐5,6,4′‐trihydroxyflavone, i.e. chrysosplenol C (Fig. 5; Table III) as potent and specific inhibitor of the picornaviral replication. 6,7,8‐Trimethoxycoumarin (Fig. 5; Table III) was also isolated as a major constituent from the ethanolic extract of P. sphacelatum, but showed no activity against poliovirus.317 Interestingly, this coumarin exhibited a significant effect in the HRV‐2‐induced CPE assay in a recently performed virtual parallel screening study performed in our laboratory.318 In contrast to the isolated coumarin, chrysosplenol C was already known as a member of the 4′‐hydroxy‐3‐methoxyflavones, which represent potent and specific inhibitors of picornaviral, especially rhinoviral replication.319, 320, 321 In 1993, Vanden Berghe, Haemers, and Vlietinck provided a profound survey of antiviral agents from higher plants, and demonstrated the impact of ethnobotanical knowledge in their search for antiviral compounds from African medicinal plants. The selection of investigated plant species was mainly based upon their use in the treatment of viral diseases by African traditional healers.320 The antiviral activity of 100 different plant species belonging to 33 families was investigated; thereof 21 species exhibited prominent antiviral properties against one or more of the tested viruses. The most pronounced activity against picornaviruses was recorded within the genus Euphorbia. All compounds detected as antiviral constituents from the respective extracts were identified as 3‐methoxyflavone derivatives, especially 3‐methylethers of quercetin and kaempferol (Fig. 5; Table III). They showed no significant cytotoxicity and were highly active in tissue culture against all human picornaviruses. In tissue culture, cells infected with different picornaviruses (among them also HRV) no CPE was observed, when cells had been treated with 2 µg/mL of these 3‐methoxyflavones.320
B. Activity Screening
The selection of natural materials based on ethnopharmacology is a profound rationale for lead structure discovery and highly superior to random selection. This however rarely applies to marine organisms, fungi, and microbes. Although these organisms are esteemed as highly valuable source for bioactive metabolites,322 hardly any records from folk medicine are given for them.
A medium‐sized activity screening using a robust cell‐based or in vitro‐assay with reasonable effort as to time and costs is a strategy to get a first insight into the antiviral activities of extracts, fractions, and compounds from synthesis or nature.
For the discovery of novel naturally based HRV 3C‐protease inhibitors, Singh et al. used a small peptide containing Q‐G scissile bond as substrate for the in vitro screening of extracts. The authors isolated the novel benzoisochromanquinone (+)‐thysanone (Fig. 5; Table III) from an extract of the fungus Thysanophora penicilloides with potent HRV 3C‐protease inhibitory activity.323 A continued screening revealed a pronounced HRV 3C‐protease inhibitory activity for the extract of the Chinese herb Polygonum cuspidatum. By bioassay‐guided fractionation 2‐methoxystypandrone (Fig. 5; Table III), a naphthoquinone, with an IC50 value of 4.6 µM, was isolated from the plant material. The total syntheses of this natural compound and further analogues allowed for a structure—activity relationship, and particularly the comparison between activities of ortho‐ vs. para‐quinones. To measure the selectivity of the compound series against cystein proteases other than HRV 3C‐protease, the compounds were evaluated against papain. The simple 9,10‐phenanthraquinone (Fig. 5; Table III) was the most active compound of the series and showed an IC50 value of 1.4 µM with a distinctly higher degree of selectivity than 2‐methoxystypandrone.324
A CPE reduction assay was applied for the identification of raoulic acid, a bicyclic C25 terpene acid (Fig. 5; Table III) isolated from Raoulia australis (Asteraceae). Raoulic acid exerted an antiviral activity against coxsackie virus B3, B4, enterovirus 71, and HRV‐2 and ‐3 with IC50 values in the submicromolar range. No activity was recorded against influenza A and B viruses.325
In the course of the systematic screening of microbial and natural products for anti‐HRV activity, Ishitsuka et al. identified 4′,5‐dihydroxy‐3,3′,7‐trimethoxyflavone (Fig. 5; Table III) from the leaves of the Chinese medicinal plant Agastache rugosa (Lamiaceae) as natural compound with high activity against all picornaviruses except mengovirus. The authors synthesized the orally active 4′,5‐diacetyloxy‐3,3′,7‐trimethoxyflavone and performed investigations in tissue cultures and in mice to determine the compound's mode of action. This was assumed to be the process of viral replication, thus located between viral uncoating and initiation of viral RNA synthesis.326 The activity of flavan (Fig. 5; Table III) was discovered serendipitously during a screening program by using a plaque inhibition assay.194 Based on these results, Bauer et al. synthesized 4′,6‐dichloroflavan (BW863C, Fig. 2; Table II), which revealed an activity against a number of HRV serotypes in the range between 0.007 and 10 µM.
C. Structure–Activity Relationship
As soon as an active lead compound is identified it is of utmost importance to scrutinize the derivatives' activity to (i) obtain insight into the chemical requirements mandatory for the focused biological activity, (ii) improve the compound's pharmacological profile in terms of potency, selectivity, cytotoxicity, etc., and to (iii) improve its bioavailability.
The nonphenolic aporphine alkaloid glaucine is a prominent constituent of the aerial parts of Gaucium flavum (Papaveraveae). In a recently published study, Spasova et al. investigated the antiviral potential of glaucine (Fig. 5; Table III), glaucine derivatives, and its semi‐synthesized 3‐aminoethylglaucine cinnamoyl‐ and hydroxycinnamoyl amides. Beside the anti‐oxidative potential of the newly synthesized compounds, they all exerted an antiviral activity against the replication of HRV‐14. The best anti‐HRV activity was observed for oxoglaucine (Fig. 5; Table III; IC50 1.5 µM, SI 170), and the o‐coumaroylamide and p‐coumaroylamide of 3 aminomethylglaucine (Fig. 5; Table III; IC50 15 µM, SI 10.3 and IC50 13 µM, SI 11, respectively).327
The early findings of the anti‐HRV active naturally derived 3‐methoxyflavone inspired chemists and pharmacologists in the synthesis and the pharmacological evaluation of derivatives thereof decorated with various substitution patterns. Several reports published during the last two decades reviewed the findings of antiviral flavonoid research.320, 328, 329
By investigating the antiviral activity of a wide variety of naturally occurring flavonoids, Tsuchiya et al. found chrysosplenol B and C (Fig. 5; Table III) contained in Chrysosplenium plants, and axillarin (Fig. 5; Table III) as potent anti‐HRV agents. Based on their findings, the flavone skeleton decorated with a methoxy group in position 3 and a 5‐hydroxyl group revealed as mandatory for an anti‐HRV activity.319
A series of antipicornaviral 4′‐hydroxy‐3‐methoxyflavone derivatives was synthesized by De Meyer et al. in order to establish a structure–activity relationship. Thereby, different substitution patterns of the A‐ring system with methyl, hydroxy, methoxy, halo, nitro, and amino was performed. Their activity against polio and HRV was compared with those of naturally occurring flavonoids. Further, the importance of the 4′ hydroxyl‐group and of the 3‐methoxyl‐group was confirmed by investigation of different derivatives lacking these features.321 The results showed that 4′‐hydroxy‐3‐methoxyflavones with a monosubstituted A‐ring are less active than the corresponding compounds having a polysubstituted A‐ring. Within the tested series of compounds, 4′,7‐dihydroxy‐3‐methoxy‐5,6‐dimethylflavone emerged not only as noncytotoxic but also as most potent substance in both antiviral test systems. The lowest concentrations for this compound that protect 50% of the cells from CPE of 12 HRV serotypes were in the range from 0.016 to 0.5 µg/mL. In contrast to quercetin, this flavone was also reported to have no mutagenic properties (measured up to 2.5 mg/plate) in a short‐term microbial assay.321
The mechanism studies performed with 3‐methoxyflavones have shown an interference with an early stage in the viral RNA synthesis; no induction of resistance was observed.320 In contrast to this mode of action, the anti‐HRV chalcones and flavans are reported to interact directly with specific sites on the viral capsid proteins, thereby preventing uncoating and the consequent liberation of viral RNA.193, 330, 331
Due to its anti‐HRV potency, low toxicity, and promising bioavailability, dichloroflavan was evaluated for its protective efficacy against experimental HRV‐infection in two clinical, double‐blind, placebo‐controlled trials. However, the drug candidate failed either when administered orally or intranasally.332, 333 Unfortunately, till now no clinical trials evaluating the efficacy of 3‐methoxyflavones in common cold have been performed.
D. Computational Approaches
The common idea of all computational approaches is to extract knowledge from a more or less large set of data in order to make predictions of new events.334 Within the lead discovery process, computational approaches, such as virtual screening, docking, quantitative structure–activity relationship, have largely enhanced the impact of computational chemistry and nowadays chemoinformatics plays a predominant role in drug research.335 The key goal of the use of such methods is to reduce the overall cost associated to the discovery and development of a new drug by identifying the most promising candidates to focus the experimental efforts on. A number of books and reviews on the impact of computational chemistry for lead structure determination highlight these efforts.336, 337, 338, 339
In general, in silico methods can be divided into (i) ligand‐based approaches, which rely on known active compounds. Based on their physicochemical properties crucial for biological affinity, activities are predicted by extrapolation on not‐yet tested substances, e.g. machine learning techniques and classical quantitative structure–activity relationship (QSAR). Ligand‐based approaches are invaluable tools in cases where no structural information about the pharmacological targets is available. (ii) On the other hand, structure‐based approaches use experimentally determined 3D structures of the targets, such as molecular docking or structure‐based pharmacophore modeling for virtual screening. These methods allow for gaining insight into protein–ligand interactions at an experimentally determined (static) level (however not considering flexibility). A unique platform containing 3D coordinates of experimentally solved protein structures (by X‐ray crystallography or NMR) is the Protein Data Bank (PDB) currently comprising more than 50,000 structures of biomolecules and protein–ligand complexes.340 Additionally, there are several PDB‐related web services and tools, which enable to use the PDB‐portal in a rich diversity of information services for students and scientists.341
Particularly in the early stage of drug development, such as lead discovery and lead optimization, computational approaches allow for a target‐oriented and rationalized proceeding, and thus may substantially help to maximize the success rate.338 A recently published review on the impact of computer‐assisted approaches in antiviral research thoroughly describes underlying in silico techniques, and highlights the benefits of computational approaches for the discovery of antiviral lead structures.342
In anti‐HRV research the capsid protein and the protease 3C revealed to be promising targets (as described before). Inhibitors are assumed to have a major impact for the treatment of HRV‐infections. Additionally, these targets are structurally elucidated, and some potent ligands are known as well. These facts enable the performance of sensible computer‐assisted approaches, both ligand‐ and structure‐based. Some studies using an in silico approach for the discovery of potential anti‐HRV agents focusing on the mentioned targets will be reported in the following paragraphs.
For compounds acting as potential HRV‐capsid binders, some classical QSAR and 3D QSAR studies have been performed. While in classical QSAR, the relationship between 2D calculated properties derived from chemical structures and measured biological activities are explored statistically, 3D QSAR techniques are aimed at deriving a correlation and in turn activity prediction based on spatial arrangements of chemical properties and atoms. Applying the 3D QSAR technique CoMFA (Comparative Molecular Field Analysis) statistical models are derived, which are visualized in color‐coded contours around the molecule. Therein spots indicate where electrostatic properties and spatial arrangements are favorable for biological activity.343
In the studies of Diana et al., Artico et al., and Verma et al., QSAR techniques helped on one hand to analyze and rationalize the structural features of active compounds essential for the interaction to the HRV canyon's binding pocket, and on the other hand to search for new classes of capsid binders, thus to narrow the synthetic challenges for specific anti‐HRV agents, respectively.344, 345, 346, 347, 348 In all these investigations hydrophobicity was found to be one of the most important determinants of substance activity. QSAR combined with simplex representation of molecular structure was applied by Kuz'min et al. based on the selectivity index (CC50/IC50) and the HRV‐2 inhibitory concentration of a set of [(biphenyloxy)propyl]isoxazole derivatives.188 On the basis of QSAR analysis and computational design, three new isoxazoles with high activity prediction were selected and synthesized. They all revealed a strong coincidence between experimental and predicted anti‐HRV activity and selectivity index. Terminal benzene substituents with negative electrostatic potential and a molecule length of approximately 5.5–5.6 Å have been suggested as mandatory features within this chemical class for a HRV‐2 inhibitory activity.
HRV‐serotypes show a high level of conservation at the protease 3C binding site; Sequence alignment and secondary structure predictions suggested an overall architecture and mechanism of HRV‐3C proteases that correlates with cellular cystein‐ and serine proteases, such as chymotrypsin and trypsin.349, 350 The identity among 3C proteases from different families is however modest and provides space for the development of specific inhibitors for HRV‐3C protease.350 In a recently published review on selective inhibitors of picornavirus replication, De Palma et al. summarized all currently known chemical structures acting as peptidic or nonpeptidic inhibitors of this viral target.156 In 2000, Reich et al., used the HRV 3C protease co‐crystal structure information to rationalize the target‐oriented synthesis of HRV 3C protease inhibitors from the class of substituted benzamides. Activity data and subsequent crystallographic studies pointed out important requirements for the inhibition of the 3C protease.351 Similarly, Maugeri et al. used a structure‐based approach, and performed docking studies based on the 3C protease crystal structure with a virtual library consisting of benzamide derivatives. Quantum‐mechanic calculation proposed substituents with most promising biological activities. This workflow guided the design and synthesis of substances virtually assumed to act as substrate analogues. Synthesis of some of these compounds and biological testing confirmed the underlying hypothesis.219 Quantitative molecular modeling studies were performed to better define and predict interactions between bicyclic 2‐pyridone derivatives that showed to be irreversible inhibitors of the 3C protease. In these studies molecular mechanics simulations to evaluate chemical rate of covalent bond formation and free energy calculation combined with crystallographic studies were applied to explain the differences in activity of some irreversible peptidomimetic inhibitors. These data were used as a basis for further optimization of these compounds.224, 352
In a recent study performed by Kuo et al. some 6,800 compounds were subjected to a high troughput screening in the search for novel inhibitors for both 3C and 3CL proteases from picornavirus and coronavirus, respectively.217 Five nonpeptides were identified with IC50 values ≤10 µM against severe acute respiratory syndrome—coronavirus 3CL‐protease; one molecule was found to additionally inhibit the 3C proteases of coxsackievirus, enterovirus, and rhinovirus. This compound (ID 43146) contains a dihydropyrazole ring decorated with two phenyl groups and a lengthy N‐butyl‐benzimidazolylamino‐toluene. It was used as starting point for the selection of further four analogs showing IC50 values in the range of 0.5–10 µM against the tested viral proteases. By means of docking‐based computer modeling, the authors tried to rationalize the binding discrepancies responsible for individual and common protease inhibitors, thus to provide a rational base for the development of nonpeptide multiple‐function inhibitors against coronaviruses and piciornaviruses.
E. Pharmacophore Models in the Search for Anti‐HRV Agents
In anti‐HRV research, first application scenarios have been conducted using pharmacophore models. According to the official IUPAC definition by Wermuth et al.,353 a pharmacophore describes the 3D arrangement of steric and electronic features necessary to trigger or block a biological response. Pharmacophores can be represented by three‐dimensional chemical features, which include hydrogen bond donors and acceptors, aromatic rings, hydrophobic groups, as well as positive and negative ionisable moieties. Additionally, the shape of ligands can be represented by shape features, which essentially describe the van der Waals radii of the ligand atoms. The pharmacophore concept has proven to be successful, not only in rationalizing structure–activity relationships but also by its large impact in developing appropriate 3D‐tools for efficient virtual screening.336, 354 Steindl et al. elaborated ligand‐ and structure‐based pharmacophore models implementing the essential feature of the covalent binding to the cysteine 147 in the active site of the HRV‐2 3C protease. Thus, the in silico approach focused on defining a new pharmacophore feature representing a target structure for nucleophilic addition in the ligands, which is a crucial step for protease inactivation. The generated hypotheses retrieved known 3C protease inhibitors in the virtual screening cycle, and proposed potential (unconfirmed) ligands of the 3D protease binding site from available databases.355
The viral capsid of several HRVs has been elucidated by crystallization and resolution of the 3D‐structure. The HRV coat protein complexed with its highly active inhibitor WIN 61209 was used as starting point for the generation of structure‐based pharmacophore models by Steindl et al. in 2005. The models were used for virtual screening of a large commercially available 3D database. For final selection of virtual hits worth to be subjected to biological testing, docking studies and principal component analysis were performed. Six candidates were tested for their ability to inhibit HRV serotype 39 by multiple‐cycle CPE inhibition assay. Although all of them showed a certain antiviral potential, one longitudinal piperazine derivative inhibited the virus at a concentration below 10 µM. Some of the test candidates showed difficulties in the interpretation of experimental results due to their relatively high cytotoxicity and bad solubility. This circumstance asks for more cautious estimation of molecular properties for compound selection as stated by the authors.356 In a subsequently performed study, the best validated pharmacophore model was used for the identification of naturally derived HRV coat protein inhibitors.144 For virtual screening experiments the in‐house generated 3D‐database DIOS was used (as described before). Based on the virtually predicted ligands and considering knowledge from traditional use, sesquiterpene umbelliferons from the gum resin of Ferula sp., i.e. asafetida, were finally selected as most promising candidates. For biological evaluation, the antiviral activities of asafetida and its isolated constituents were assessed by an exploratory determination of the inhibition of the CPE induced by HRV serotypes 1A, 2, 14, and 16. The results revealed a dose‐dependent and selective anti‐HRV activity against serotype 2 for asafetida and its virtually predicted constituents, farnesiferols B and C (Fig. 5; Table III; IC50: 2.6 and 2.5 µM, respectively). To scrutinize the selectivity of these two compounds against HRV‐2 in comparison to the other tested serotypes, the amino acid sequences of HRV‐2 and HRV‐16 VP1 were aligned. Since all amino acid residues involved in ligand binding showed 100% match in both serotypes, the experimentally determined selectivity profile could not be explained by different binding pockets. The serotype alignment does however not reflect potential protein flexibility during binding, which might differ between the HRV serotypes. Additionally, off‐target effects could be a reason for the observed selectivity.
In a recently performed virtual parallel screening approach, we tried to identify potential targets of human pathological relevance for 16 constituents isolated and identified from the medicinal plant Ruta graveolens.318 Using the screening platform Pipline Pilot357 included in Discovery Studio,358 low‐energy conformers of the 16 identified molecules from R. graveolens were subjected to parallel screening. For this purpose, the Inte:Ligand pharmacophore model collection was used.359 It currently comprises 2.208 models covering 280 unique pharmacological targets. Based on the predicted ligand‐target interactions, the authors focused on three biological targets, namely acetylcholinesterase, the HRV coat protein, and the cannabinoid receptor type 2. Virtual hits and nonhits were assayed on their respective targets for a critical evaluation of the performed target‐fishing approach. Beside other predicted bioactivities, determination of their CPEs on HRV‐2 revealed the virtual hit arborinine (Fig. 5; Table III; IC50: 3.2 µM) and the nonpredicted hit 6,7,8‐trimethoxycoumarin (Fig. 5; Table III; IC50: 12.9 µM) as the most active anti‐HRV constituents. It could be shown that the applied in silico strategy has the capacity of catalyzing drug discovery profoundly for all those diseases where molecular targets or molecular ligands are well defined to create reliable pharmacophore models.
7. CONCLUSION
HRV, a prominent member of the Picornaviridae family, infects humans more frequently than any other virus. Infections with HRV mainly lead to upper respiratory diseases, such as the common cold, but may also cause more severe lower respiratory tract disorders. Although the symptomology and severity of the common cold is relatively mild and the course of disease self‐limiting, the socio‐economic impact is tremendous in terms of recouping lost productivity due to sick leave.
In the last decades, a number of anti‐HRV agents from different origin—synthetics, natural compounds, biologicals, botanicals, and nutritionals—have been discovered. Different concepts have been used as strategy for their discovery either starting from a phenomenological effect, e.g. empirical knowledge from folk medicine, or from a target‐based molecular level, e.g. ICAMs, capsid binders, and HRV protease inhibitors. Some of the outcomes revealed potent and promising activities that partly have been evaluated for the management of HRV‐induced common colds in clinical trials mainly with sobering benefit. Since the HRV infection is not life‐threatening in most cases, a potential therapy has to be safe and effective with an almost unrecognizable level of side effects. These preconditions render the search for anti‐HRV‐agents into a high challenging endeavor and explains why no antiviral agent is approved for the prevention or treatment of HRV‐infection until today, despite the significant efforts. Moreover, the high variability of rhinoviruses suggests the need of more than one active principle covering different modes of action for an effective treatment. Therefore, there is a high need for an ongoing search for new synthetic as well as natural compounds on a molecular level.
The increasing knowledge about the HRV life cycle, gene, and protein sequence combined with the improved technologies in the field of experimental and computational methods have continuously enabled further insights into the spectacular world of HRV. Only with this fundamental research, virtual screening approaches, such as QSAR, pharmacophore‐ and docking‐based screening cycles, or computational compound design, are applicable on a rational base. With the gained expertise from different disciplines and an adequate infrastructure for further research, it is encouraging to hope that discovery and clinical development efforts will continue in the search for agents that may treat or prevent the annoying common cold.
Biographical Information
Judith Maria Rollinger (1964) is an Associate Professor of Pharmacognosy at the Institute of Pharmacy, University of Innsbruck. She studied Pharmacy at the University of Innsbruck, Austria, and then received training in microbiology and pharmacy at the University Hospital in Innsbruck. In 1999 she received her Ph.D. in Pharmacognosy dealing with research in crystal polymorphism (University of Innsbruck). In postdoctoral time, Judith M. Rollinger extended her studies to the fields of phytochemistry, ethnopharmacology, molecular modelling and its application in natural product science completing the habilitation thesis entitled “The search for bioactive natural products” in 2006 and the venia legendi as Professor of Pharmacognosy in 2007.
Judith M. Rollinger is a project leader and an associate in various national projects, as well as executive guest editor for Current Pharmaceutical Design. She has been awarded the PHOENIX Science award (2005) and received several additional awards. Her research focuses on the interdisciplinary field of integrating computational techniques in classical pharmacognostic research as strategy for the discovery of bioactive natural products for treating neurodegenerative diseases, viral infections, and inflammation.
Michaela Schmidtke (1961) is an Associate Professor at the Institute of Virology and Antiviral Therapy, University Clinical Center, Jena, Germany. She studied biology and received her diploma in microbiology at the University of Chisinau, Moldowa. In 1992 she obtained her Ph.D. degree at the faculty of biology, University of Jena, Germany dealing with “Antiviral investigation with selected picornaviruses in vitro.” Until 1994 she was a postdoctoral research fellow at the Institute of Immunology, University Marburg, where she studied picornavirus–immune cell interactions and the virus‐induced cytokine induction. Thereafter, she was a postdoctoral fellow at the Institute of Virology and Antiviral Therapy, University Clinical Center, Jena, Germany, where she received the venia legendi in Virology in 2009 with the habilitation thesis dealing with the “Development of selective antiviral compounds against enteroviruses” (2008). Her research interests focus on the development of novel inhibitors against respiratory viruses, particularly enteroviruses and influenza viruses.
REFERENCES
- 1. Le Gall O, Christian P, Fauquet CM, King AM, Knowles NJ, Nakashima N, Stanway G, Gorbalenya AE. Picornavirales, a proposed order of positive‐sense single‐stranded RNA viruses with a pseudo‐T=3 virion architecture. Arch Virol 2008;153:715–727. [DOI] [PubMed] [Google Scholar]
- 2. Briese T, Renwick N, Venter M, Jarman RG, Ghosh D, Kondgen S, Shrestha SK, Hoegh AM, Casas I, Adjogoua EV, Akoua‐Koffi C, Myint KS, Williams DT, Chidlow G, van den Berg R, Calvo C, Koch O, Palacios G, Kapoor V, Villari J, Dominguez SR, Holmes KV, Harnett G, Smith D, Mackenzie JS, Ellerbrok H, Schweiger B, Schonning K, Chadha MS, Leendertz FH, Mishra AC, Gibbons RV, Holmes EC, Lipkin WI. Global distribution of novel rhinovirus genotype. Emerg Infect Dis 2008;14:944–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kistler AL, Webster DR, Rouskin S, Magrini V, Credle JJ, Schnurr DP, Boushey HA, Mardis ER, Li H, DeRisi JL. Genome‐wide diversity and selective pressure in the human rhinovirus. Virol J 2007;4:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lau SK, Yip CC, Tsoi HW, Lee RA, So LY, Lau YL, Chan KH, Woo PC, Yuen KY. Clinical features and complete genome characterization of a distinct human rhinovirus (HRV) genetic cluster, probably representing a previously undetected HRV species, HRV‐C, associated with acute respiratory illness in children. J Clin Microbiol 2007;45:3655–3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. McErlean P, Shackelton LA, Lambert SB, Nissen MD, Sloots TP, Mackay IM. Characterisation of a newly identified human rhinovirus, HRV‐QPM, discovered in infants with bronchiolitis. J Clin Virol 2007;39:67–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. McErlean P, Shackelton LA, Andrews E, Webster DR, Lambert SB, Nissen MD, Sloots TP, Mackay IM. Distinguishing molecular features and clinical characteristics of a putative new rhinovirus species, human rhinovirus C (HRV C). PLoS ONE 2008;3:e1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Palmenberg AC, Spiro D, Kuzmickas R, Wang S, Djikeng A, Rathe JA, Fraser‐Liggett CM, Liggett SB. Sequencing and analyses of all known human rhinovirus genomes reveal structure and evolution. Science 2009;324:55–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Oberste MS, Maher K, Flemister MR, Marchetti G, Kilpatrick DR, Pallansch MA. Comparison of classic and molecular approaches for the identification of untypeable enteroviruses. J Clin Microbiol 2000;38:1170–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Oberste MS, Maher K, Kilpatrick DR, Pallansch MA. Molecular evolution of the human enteroviruses: Correlation of serotype with VP1 sequence and application to picornavirus classification. J Virol 1999;73:1941–1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Oberste MS, Maher K, Kilpatrick DR, Flemister MR, Brown BA, Pallansch MA. Typing of human enteroviruses by partial sequencing of VP1. J Clin Microbiol 1999;37:1288–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Horsnell C, Gama RE, Hughes PJ, Stanway G. Molecular relationships between 21 human rhinovirus serotypes. J Gen Virol 1995;76:2549–2555. [DOI] [PubMed] [Google Scholar]
- 12. Ledford RM, Patel NR, Demenczuk TM, Watanyar A, Herbertz T, Collett MS, Pevear DC. VP1 sequencing of all human rhinovirus serotypes: Insights into genus phylogeny and susceptibility to antiviral capsid‐binding compounds. J Virol 2004;78:3663–3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Callahan PL, Mizutani S, Colonno RJ. Molecular cloning and complete sequence determination of RNA genome of human rhinovirus type 14. Proc Natl Acad Sci USA 1985;82:732–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Duechler M, Skern T, Sommergruber W, Neubauer C, Gruendler P, Fogy I, Blaas D, Kuechler E. Evolutionary relationships within the human rhinovirus genus: Comparison of serotypes 89, 2, and 14. Proc Natl Acad Sci USA 1987;84:2605–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Harris JR, Racaniello VR. Amino acid changes in proteins 2B and 3A mediate rhinovirus type 39 growth in mouse cells. J Virol 2005;79:5363–5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hughes PJ, North C, Jellis CH, Minor PD, Stanway G. The nucleotide sequence of human rhinovirus 1B: Molecular relationships within the rhinovirus genus. J Gen Virol 1988;69:49–58. [DOI] [PubMed] [Google Scholar]
- 17. Lee WM, Wang W, Rueckert RR. Complete sequence of the RNA genome of human rhinovirus 16, a clinically useful common cold virus belonging to the ICAM‐1 receptor group. Virus Genes 1995;9:177–181. [DOI] [PubMed] [Google Scholar]
- 18. Skern T, Sommergruber W, Blaas D, Gruendler P, Fraundorfer F, Pieler C, Fogy I, Kuechler E. Human rhinovirus 2: Complete nucleotide sequence and proteolytic processing signals in the capsid protein region. Nucleic Acids Res 1985;13:2111–2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Stanway G, Hughes PJ, Mountford RC, Minor PD, Almond JW. The complete nucleotide sequence of a common cold virus: Human rhinovirus 14. Nucleic Acids Res 1984;12:7859–7875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tapparel C, Junier T, Gerlach D, Cordey S, Van Belle S, Perrin L, Zdobnov EM, Kaiser L. New complete genome sequences of human rhinoviruses shed light on their phylogeny and genomic features. BMC Genomics 2007;8:224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Savolainen C, Blomqvist S, Mulders MN, Hovi T. Genetic clustering of all 102 human rhinovirus prototype strains: Serotype 87 is close to human enterovirus 70. J Gen Virol 2002;83:333–340. [DOI] [PubMed] [Google Scholar]
- 22. Heikkinen T, Jarvinen A. The common cold. Lancet 2003;361:51–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Savolainen C, Mulders MN, Hovi T. Phylogenetic analysis of rhinovirus isolates collected during successive epidemic seasons. Virus Res 2002;85:41–46. [DOI] [PubMed] [Google Scholar]
- 24. Lee WM, Kiesner C, Pappas T, Lee I, Grindle K, Jartti T, Jakiela B, Lemanske RF, Jr. , Shult PA, Gern JE. A diverse group of previously unrecognized human rhinoviruses are common causes of respiratory illnesses in infants. PLoS ONE 2007;2:e966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Peltola V, Waris M, Osterback R, Susi P, Ruuskanen O, Hyypia T. Rhinovirus transmission within families with children: incidence of symptomatic and asymptomatic infections. J Infect Dis 2008;197:382–389. [DOI] [PubMed] [Google Scholar]
- 26. Renwick N, Schweiger B, Kapoor V, Liu Z, Villari J, Bullmann R, Miething R, Briese T, Lipkin WI. A recently identified rhinovirus genotype is associated with severe respiratory‐tract infection in children in Germany. J Infect Dis 2007;196:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Monto AS, Sullivan KM. Acute respiratory illness in the community. Frequency of illness and the agents involved. Epidemiol Infect 1993;110:145–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Puhakka T, Makela MJ, Alanen A, Kallio T, Korsoff L, Arstila P, Leinonen M, Pulkkinen M, Suonpaa J, Mertsola J, Ruuskanen O. Sinusitis in the common cold. J Allergy Clin Immunol 1998;102:403–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ruohola A, Waris M, Allander T, Ziegler T, Heikkinen T, Ruuskanen O. Viral etiology of common cold in children, Finland. Emerg Infect Dis 2009;15:344–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. van Gageldonk‐Lafeber AB, Heijnen ML, Bartelds AI, Peters MF, van der Plas SM, Wilbrink B. A case‐control study of acute respiratory tract infection in general practice patients in The Netherlands. Clin Infect Dis 2005;41:490–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Versteegh FG, Weverling GJ, Peeters MF, Wilbrink B, Veenstra‐van Schie MT, van Leeuwen‐Gerritsen JM, Mooi‐Kokenberg EA, Schellekens JF, Roord JJ. Community‐acquired pathogens associated with prolonged coughing in children: a prospective cohort study. Clin Microbiol Infect 2005;11:801–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Arruda E, Pitkaranta A, Witek TJ, Jr. , Doyle CA, Hayden FG. Frequency and natural history of rhinovirus infections in adults during autumn. J Clin Microbiol 1997;35:2864–2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Makela MJ, Puhakka T, Ruuskanen O, Leinonen M, Saikku P, Kimpimaki M, Blomqvist S, Hyypia T, Arstila P. Viruses and bacteria in the etiology of the common cold. J Clin Microbiol 1998;36:539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Monto AS. The seasonality of rhinovirus infections and its implications for clinical recognition. Clin Ther 2002;24:1987–1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zalm MM, van Ewijk BE, Wilbrink B, Uiterwaal CS, Wolfs TF, van der Ent CK. Respiratory pathogens in children with and without respiratory symptoms. J Pediatr 2009;154:396–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Blomqvist S, Roivainen M, Puhakka T, Kleemola M, Hovi T. Virological and serological analysis of rhinovirus infections during the first two years of life in a cohort of children. J Med Virol 2002;66:263–268. [DOI] [PubMed] [Google Scholar]
- 37. Savolainen‐Kopra C, Blomqvist S, Kilpi T, Roivainen M, Hovi T. Novel species of human rhinoviruses in acute otitis media. Pediatr Infect Dis J 2009;28:59–61. [DOI] [PubMed] [Google Scholar]
- 38. El‐Sahly HM, Atmar RL, Glezen WP, Greenberg SB. Spectrum of clinical illness in hospitalized patients with “common cold” virus infections. Clin Infect Dis 2000;31:96–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Garbino J, Gerbase MW, Wunderli W, Kolarova L, Nicod LP, Rochat T, Kaiser L. Respiratory viruses and severe lower respiratory tract complications in hospitalized patients. Chest 2004;125:1033–1039. [DOI] [PubMed] [Google Scholar]
- 40. Garbino J, Inoubli S, Mossdorf E, Weber R, Tamm M, Soccal P, Aubert JD, Bridevaux PO, Tapparel C, Kaiser L. Respiratory viruses in HIV‐infected patients with suspected respiratory opportunistic infection. AIDS 2008;22:701–705. [DOI] [PubMed] [Google Scholar]
- 41. Gern JE, Calhoun W, Swenson C, Shen G, Busse WW. Rhinovirus infection preferentially increases lower airway responsiveness in allergic subjects. Am J Respir Crit Care Med 1997;155:1872–1876. [DOI] [PubMed] [Google Scholar]
- 42. Ghosh S, Champlin R, Couch R, Englund J, Raad I, Malik S, Luna M, Whimbey E. Rhinovirus infections in myelosuppressed adult blood and marrow transplant recipients. Clin Infect Dis 1999;29:528–532. [DOI] [PubMed] [Google Scholar]
- 43. Ison MG, Hayden FG, Kaiser L, Corey L, Boeckh M. Rhinovirus infections in hematopoietic stem cell transplant recipients with pneumonia. Clin Infect Dis 2003;36:1139–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jacques J, Bouscambert‐Duchamp M, Moret H, Carquin J, Brodard V, Lina B, Motte J, Andreoletti L. Association of respiratory picornaviruses with acute bronchiolitis in French infants. J Clin Virol 2006;35:463–466. [DOI] [PubMed] [Google Scholar]
- 45. Kaiser L, Aubert JD, Pache JC, Deffernez C, Rochat T, Garbino J, Wunderli W, Meylan P, Yerly S, Perrin L, Letovanec I, Nicod L, Tapparel C, Soccal PM. Chronic rhinoviral infection in lung transplant recipients. Am J Respir Crit Care Med 2006;174:1392–1399. [DOI] [PubMed] [Google Scholar]
- 46. Arruda LK, Sole D, Baena‐Cagnani CE, Naspitz CK. Risk factors for asthma and atopy. Curr Opin Allergy Clin Immunol 2005;5:153–159. [DOI] [PubMed] [Google Scholar]
- 47. Grissell TV, Powell H, Shafren DR, Boyle MJ, Hensley MJ, Jones PD, Whitehead BF, Gibson PG. Interleukin‐10 gene expression in acute virus‐induced asthma. Am J Respir Crit Care Med 2005;172:433–439. [DOI] [PubMed] [Google Scholar]
- 48. Johnston SL, Pattemore PK, Sanderson G, Smith S, Lampe F, Josephs L, Symington P, O'Toole S, Myint SH, Tyrrell DA, et al. Community study of role of viral infections in exacerbations of asthma in 9–11 year old children. BMJ 1995;310:1225–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wark PA, Johnston SL, Bucchieri F, Powell R, Puddicombe S, Laza‐Stanca V, Holgate ST, Davies DE. Asthmatic bronchial epithelial cells have a deficient innate immune response to infection with rhinovirus. J Exp Med 2005;201:937–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Camargo CA, Jr. , Ginde AA, Clark S, Cartwright CP, Falsey AR, Niewoehner DE. Viral pathogens in acute exacerbations of chronic obstructive pulmonary disease. Intern Emerg Med 2008;3:355–359. [DOI] [PubMed] [Google Scholar]
- 51. McManus TE, Marley AM, Baxter N, Christie SN, O'Neill HJ, Elborn JS, Coyle PV, Kidney JC. Respiratory viral infection in exacerbations of COPD. Respir Med 2008;102:1575–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rohde G, Schlosser B, Arinir U, Kronsbein J, Knoop H, Ringshausen F, Schultze‐Werninghaus G. The role of respiratory infections in chronic obstructive pulmonary disease. Med Klin 2007;102:893–898. [DOI] [PubMed] [Google Scholar]
- 53. Schaller M, Hogaboam CM, Lukacs N, Kunkel SL. Respiratory viral infections drive chemokine expression and exacerbate the asthmatic response. J Allergy Clin Immunol 2006;118:295–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Seemungal T, Harper‐Owen R, Bhowmik A, Moric I, Sanderson G, Message S, Maccallum P, Meade TW, Jeffries DJ, Johnston SL, Wedzicha JA. Respiratory viruses, symptoms, and inflammatory markers in acute exacerbations and stable chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2001;164:1618–1623. [DOI] [PubMed] [Google Scholar]
- 55. Seemungal TA, Harper‐Owen R, Bhowmik A, Jeffries DJ, Wedzicha JA. Detection of rhinovirus in induced sputum at exacerbation of chronic obstructive pulmonary disease. Eur Respir J 2000;16:677–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Crowcroft NS, Zambon M, Harrison TG, Mok Q, Heath P, Miller E. Respiratory syncytial virus infection in infants admitted to paediatric intensive care units in London, and in their families. Eur J Pediatr 2008;167:395–399. [DOI] [PubMed] [Google Scholar]
- 57. Smyth AR, Smyth RL, Tong CY, Hart CA, Heaf DP. Effect of respiratory virus infections including rhinovirus on clinical status in cystic fibrosis. Arch Dis Child 1995;73:117–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wat D, Gelder C, Hibbitts S, Cafferty F, Bowler I, Pierrepoint M, Evans R, Doull I. The role of respiratory viruses in cystic fibrosis. J Cyst Fibros 2008;7:320–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Khetsuriani N, Lu X, Teague WG, Kazerouni N, Anderson LJ, Erdman DD. Novel human rhinoviruses and exacerbation of asthma in children. Emerg Infect Dis 2008;14:1793–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Miller EK, Edwards KM, Weinberg GA, Iwane MK, Griffin MR, Hall CB, Zhu Y, Szilagyi PG, Morin LL, Heil LH, Lu X, Williams JV. A novel group of rhinoviruses is associated with asthma hospitalizations. J Allergy Clin Immunol 2009;123:98–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gwaltney JM, Hendley JO. Rhinovirus transmission: one if by air, two if by hand. Trans Am Clin Climatol Assoc 1978;89:194–200. [PMC free article] [PubMed] [Google Scholar]
- 62. Musher DM. How contagious are common respiratory tract infections? N Engl J Med 2003;348:1256–1266. [DOI] [PubMed] [Google Scholar]
- 63. Winther B, McCue K, Ashe K, Rubino JR, Hendley JO. Environmental contamination with rhinovirus and transfer to fingers of healthy individuals by daily life activity. J Med Virol 2007;79:1606–1610. [DOI] [PubMed] [Google Scholar]
- 64. Dick EC, Jennings LC, Mink KA, Wartgow CD, Inhorn SL. Aerosol transmission of rhinovirus colds. J Infect Dis 1987;156:442–448. [DOI] [PubMed] [Google Scholar]
- 65. Gwaltney JM, Jr. , Moskalski PB, Hendley JO. Hand‐to‐hand transmission of rhinovirus colds. Ann Intern Med 1978;88:463–467. [DOI] [PubMed] [Google Scholar]
- 66. Gwaltney JM, Jr. , Hendley JO. Transmission of experimental rhinovirus infection by contaminated surfaces. Am J Epidemiol 1982;116:828–833. [DOI] [PubMed] [Google Scholar]
- 67. Gwaltney JM, Jr. , Moskalski PB, Hendley JO. Interruption of experimental rhinovirus transmission. J Infect Dis 1980;142:811–815. [DOI] [PubMed] [Google Scholar]
- 68. Wutzler P, Sauerbrei A, Klocking R, Brogmann B, Reimer K. Virucidal activity and cytotoxicity of the liposomal formulation of povidone‐iodine. Antiviral Res 2002;54:89–97. [DOI] [PubMed] [Google Scholar]
- 69. Turner RB, Hendley JO. Virucidal hand treatments for prevention of rhinovirus infection. J Antimicrob Chemother 2005;56:805–807. [DOI] [PubMed] [Google Scholar]
- 70. Arruda E, Boyle TR, Winther B, Pevear DC, Gwaltney JM, Jr. , Hayden FG. Localization of human rhinovirus replication in the upper respiratory tract by in situ hybridization. J Infect Dis 1995;171:1329–1333. [DOI] [PubMed] [Google Scholar]
- 71. Bardin PG, Sanderson G, Robinson BS, Holgate ST, Tyrrell DA. Experimental rhinovirus infection in volunteers. Eur Respir J 1996;9:2250–2255. [DOI] [PubMed] [Google Scholar]
- 72. Gern JE, Galagan DM, Jarjour NN, Dick EC, Busse WW. Detection of rhinovirus RNA in lower airway cells during experimentally induced infection. Am J Respir Crit Care Med 1997;155:1159–1161. [DOI] [PubMed] [Google Scholar]
- 73. Gern JE, Vrtis R, Grindle KA, Swenson C, Busse WW. Relationship of upper and lower airway cytokines to outcome of experimental rhinovirus infection. Am J Respir Crit Care Med 2000;162:2226–2231. [DOI] [PubMed] [Google Scholar]
- 74. Winther B, Farr B, Turner RB, Hendley JO, Gwaltney JM, Jr. , Mygind N. Histopathologic examination and enumeration of polymorphonuclear leukocytes in the nasal mucosa during experimental rhinovirus colds. Acta Otolaryngol Suppl 1984;413:19–24. [DOI] [PubMed] [Google Scholar]
- 75. Winther B, Gwaltney JM, Jr. , Mygind N, Turner RB, Hendley JO. Sites of rhinovirus recovery after point inoculation of the upper airway. JAMA 1986;256:1763–1767. [PubMed] [Google Scholar]
- 76. Harris JM 2nd, Gwaltney JM, Jr . Incubation periods of experimental rhinovirus infection and illness. Clin Infect Dis 1996;23:1287–1290. [DOI] [PubMed] [Google Scholar]
- 77. Mosser AG, Vrtis R, Burchell L, Lee WM, Dick CR, Weisshaar E, Bock D, Swenson CA, Cornwell RD, Meyer KC, Jarjour NN, Busse WW, Gern JE. Quantitative and qualitative analysis of rhinovirus infection in bronchial tissues. Am J Respir Crit Care Med 2005;171:645–651. [DOI] [PubMed] [Google Scholar]
- 78. Papadopoulos NG, Bates PJ, Bardin PG, Papi A, Leir SH, Fraenkel DJ, Meyer J, Lackie PM, Sanderson G, Holgate ST, Johnston SL. Rhinoviruses infect the lower airways. J Infect Dis 2000;181:1875–1884. [DOI] [PubMed] [Google Scholar]
- 79. Jennings LC, Anderson TP, Beynon KA, Chua A, Laing RT, Werno AM, Young SA, Chambers ST, Murdoch DR. Incidence and characteristics of viral community‐acquired pneumonia in adults. Thorax 2008;63:42–48. [DOI] [PubMed] [Google Scholar]
- 80. Juven T, Mertsola J, Waris M, Leinonen M, Meurman O, Roivainen M, Eskola J, Saikku P, Ruuskanen O. Etiology of community‐acquired pneumonia in 254 hospitalized children. Pediatr Infect Dis J 2000;19:293–298. [DOI] [PubMed] [Google Scholar]
- 81. Templeton KE, Scheltinga SA, van den Eeden WC, Graffelman AW, van den Broek PJ, Claas EC. Improved diagnosis of the etiology of community‐acquired pneumonia with real‐time polymerase chain reaction. Clin Infect Dis 2005;41:345–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Tsolia MN, Psarras S, Bossios A, Audi H, Paldanius M, Gourgiotis D, Kallergi K, Kafetzis DA, Constantopoulos A, Papadopoulos NG. Etiology of community‐acquired pneumonia in hospitalized school‐age children: Evidence for high prevalence of viral infections. Clin Infect Dis 2004;39:681–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Turner RB, Weingand KW, Yeh CH, Leedy DW. Association between interleukin‐8 concentration in nasal secretions and severity of symptoms of experimental rhinovirus colds. Clin Infect Dis 1998;26:840–846. [DOI] [PubMed] [Google Scholar]
- 84. Pappas DE, Hendley JO, Hayden FG, Winther B. Symptom profile of common colds in school‐aged children. Pediatr Infect Dis J 2008;27:8–11. [DOI] [PubMed] [Google Scholar]
- 85. Gwaltney JM, Jr. , Scheld WM, Sande MA, Sydnor A. The microbial etiology and antimicrobial therapy of adults with acute community‐acquired sinusitis: A fifteen‐year experience at the University of Virginia and review of other selected studies. J Allergy Clin Immunol 1992;90:457–462. [DOI] [PubMed] [Google Scholar]
- 86. Lehtinen P, Jartti T, Virkki R, Vuorinen T, Leinonen M, Peltola V, Ruohola A, Ruuskanen O. Bacterial coinfections in children with viral wheezing. Eur J Clin Microbiol Infect Dis 2006;25:463–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Bardin PG, Fraenkel DJ, Sanderson G, Dorward M, Lau LC, Johnston SL, Holgate ST. Amplified rhinovirus colds in atopic subjects. Clin Exp Allergy 1994;24:457–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Calhoun WJ, Swenson CA, Dick EC, Schwartz LB, Lemanske RF, Jr. , Busse WW. Experimental rhinovirus 16 infection potentiates histamine release after antigen bronchoprovocation in allergic subjects. Am Rev Respir Dis 1991;144:1267–1273. [DOI] [PubMed] [Google Scholar]
- 89. Fraenkel DJ, Bardin PG, Sanderson G, Lampe F, Johnston SL, Holgate ST. Lower airways inflammation during rhinovirus colds in normal and in asthmatic subjects. Am J Respir Crit Care Med 1995;151:879–886. [DOI] [PubMed] [Google Scholar]
- 90. Gern JE, Busse WW. The effects of rhinovirus infections on allergic airway responses. Am J Respir Crit Care Med 1995;152:S40–S45. [DOI] [PubMed] [Google Scholar]
- 91. Lemanske RF, Jr. , Dick EC, Swenson CA, Vrtis RF, Busse WW. Rhinovirus upper respiratory infection increases airway hyperreactivity and late asthmatic reactions. J Clin Invest 1989;83:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Mallia P, Message SD, Kebadze T, Parker HL, Kon OM, Johnston SL. An experimental model of rhinovirus induced chronic obstructive pulmonary disease exacerbations: A pilot study. Respir Res 2006;7:116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Message SD, Laza‐Stanca V, Mallia P, Parker HL, Zhu J, Kebadze T, Contoli M, Sanderson G, Kon OM, Papi A, Jeffery PK, Stanciu LA, Johnston SL. Rhinovirus‐induced lower respiratory illness is increased in asthma and related to virus load and Th1/2 cytokine and IL‐10 production. Proc Natl Acad Sci USA 2008;105:13562–13567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Fendrick AM, Monto AS, Nightengale B, Sarnes M. The economic burden of non‐influenza‐related viral respiratory tract infection in the United States. Arch Intern Med 2003;163:487–494. [DOI] [PubMed] [Google Scholar]
- 95. Patick AK. Rhinovirus chemotherapy. Antiviral Res 2006;71:391–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Gonzales R, Malone DC, Maselli JH, Sande MA. Excessive antibiotic use for acute respiratory infections in the United States. Clin Infect Dis 2001;33:757–762. [DOI] [PubMed] [Google Scholar]
- 97. Hayden FG. Influenza virus and rhinovirus‐related otitis media: Potential for antiviral intervention. Vaccine 2000;19:S66–S70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Racaniello VR. Picornaviridae: The viruses and their replication. In: Knipe DM, Howley PM, editors. Fields virology, 5th ed. Volume 1. Philadelphia: Lippincott Williams & Wilkins; 2007. pp 796–839. [Google Scholar]
- 99. Arnold E, Rossmann MG. Analysis of the structure of a common cold virus, human rhinovirus 14, refined at a resolution of 3.0 A. J Mol Biol 1990;211:763–801. [DOI] [PubMed] [Google Scholar]
- 100. Hadfield AT, Lee W, Zhao R, Oliveira MA, Minor I, Rueckert RR, Rossmann MG. The refined structure of human rhinovirus 16 at 2.15 A resolution: Implications for the viral life cycle. Structure 1997;5:427–441. [DOI] [PubMed] [Google Scholar]
- 101. Kim SS, Smith TJ, Chapman MS, Rossmann MC, Pevear DC, Dutko FJ, Felock PJ, Diana GD, McKinlay MA. Crystal structure of human rhinovirus serotype 1A (HRV1A). J Mol Biol 1989;210:91–111. [DOI] [PubMed] [Google Scholar]
- 102. Oliveira MA, Zhao R, Lee WM, Kremer MJ, Minor I, Rueckert RR, Diana GD, Pevear DC, Dutko FJ, McKinlay MA, et al. The structure of human rhinovirus 16. Structure 1993;1:51–68. [DOI] [PubMed] [Google Scholar]
- 103. Rossmann MG, Arnold E, Erickson JW, Frankenberger EA, Griffith JP, Hecht HJ, Johnson JE, Kamer G, Luo M, Mosser AG, et al. Structure of a human common cold virus and functional relationship to other picornaviruses. Nature 1985;317:145–153. [DOI] [PubMed] [Google Scholar]
- 104. Verdaguer N, Blaas D, Fita I. Structure of human rhinovirus serotype 2 (HRV2). J Mol Biol 2000;300:1179–1194. [DOI] [PubMed] [Google Scholar]
- 105. Zhao R, Pevear DC, Kremer MJ, Giranda VL, Kofron JA, Kuhn RJ, Rossmann MG. Human rhinovirus 3 at 3.0 A resolution. Structure 1996;4:1205–1220. [DOI] [PubMed] [Google Scholar]
- 106. Rossmann MG. The canyon hypothesis. Viral Immunol 1989;2:143–161. [DOI] [PubMed] [Google Scholar]
- 107. Rossmann MG. Viral cell recognition and entry. Protein Sci 1994;3:1712–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Rossmann MG, Olson NH, Kolatkar PR, Oliveira MA, Cheng RH, Greve JM, McClelland A, Baker TS. Crystallographic and cryo EM analysis of virion‐receptor interactions. Arch Virol Suppl 1994;9:531–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Staunton DE, Merluzzi VJ, Rothlein R, Barton R, Marlin SD, Springer TA. A cell adhesion molecule, ICAM‐1, is the major surface receptor for rhinoviruses. Cell 1989;56:849–853. [DOI] [PubMed] [Google Scholar]
- 110. Vlasak M, Roivainen M, Reithmayer M, Goesler I, Laine P, Snyers L, Hovi T, Blaas D. The minor receptor group of human rhinovirus (HRV) includes HRV23 and HRV25, but the presence of a lysine in the VP1 HI loop is not sufficient for receptor binding. J Virol 2005;79:7389–7395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Hofer F, Gruenberger M, Kowalski H, Machat H, Huettinger M, Kuechler E, Blaas D. Members of the low density lipoprotein receptor family mediate cell entry of a minor‐group common cold virus. Proc Natl Acad Sci USA 1994;91:1839–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Rankl C, Kienberger F, Wildling L, Wruss J, Gruber Hermann J, Blaas D, Hinterdorfer P. Multiple receptors involved in human rhinovirus attachment to live cells. Proc Natl Acad Sci USA 2008;105:17778–17783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Kolatkar PR, Bella J, Olson NH, Bator CM, Baker TS, Rossmann MG. Structural studies of two rhinovirus serotypes complexed with fragments of their cellular receptor. EMBO J 1999;18:6249–6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Hewat EA, Neumann E, Conway JF, Moser R, Ronacher B, Marlovits TC, Blaas D. The cellular receptor to human rhinovirus 2 binds around the 5‐fold axis and not in the canyon: A structural view. EMBO J 2000;19:6317–6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Uncapher CR, DeWitt CM, Colonno RJ. The major and minor group receptor families contain all but one human rhinovirus serotype. Virology 1991;180:814–817. [DOI] [PubMed] [Google Scholar]
- 116. Khan AG, Pichler J, Rosemann A, Blaas D. Human rhinovirus type 54 infection via heparan sulfate is less efficient and strictly dependent on low endosomal pH. J Virol 2007;81:4625–4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Vlasak M, Goesler I, Blaas D. Human rhinovirus type 89 variants use heparan sulfate proteoglycan for cell attachment. J Virol 2005;79:5963–5970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Grunert HP, Wolf KU, Langner KD, Sawitzky D, Habermehl KO, Zeichhardt H. Internalization of human rhinovirus 14 into HeLa and ICAM‐1‐transfected BHK cells. Med Microbiol Immunol 1997;186:1–9. [DOI] [PubMed] [Google Scholar]
- 119. Reischl A, Reithmayer M, Winsauer G, Moser R, Gosler I, Blaas D. Viral evolution toward change in receptor usage: Adaptation of a major group human rhinovirus to grow in ICAM‐1‐negative cells. J Virol 2001;75:9312–9319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Zauner W, Blaas D, Kuechler E, Wagner E. Rhinovirus‐mediated endosomal release of transfection complexes. J Virol 1995;69:1085–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Schober D, Kronenberger P, Prchla E, Blaas D, Fuchs R. Major and minor receptor group human rhinoviruses penetrate from endosomes by different mechanisms. J Virol 1998;72:1354–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Hewat EA, Blaas D. Cryoelectron microscopy analysis of the structural changes associated with human rhinovirus type 14 uncoating. J Virol 2004;78:2935–2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Hoover‐Litty H, Greve JM. Formation of rhinovirus‐soluble ICAM‐1 complexes and conformational changes in the virion. J Virol 1993;67:390–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Brabec M, Baravalle G, Blaas D, Fuchs R. Conformational changes, plasma membrane penetration, and infection by human rhinovirus type 2: Role of receptors and low pH. J Virol 2003;77:5370–5377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Prchla E, Kuechler E, Blaas D, Fuchs R. Uncoating of human rhinovirus serotype 2 from late endosomes. J Virol 1994;68:3713–3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Verdaguer N, Fita I, Reithmayer M, Moser R, Blaas D. X‐ray structure of a minor group human rhinovirus bound to a fragment of its cellular receptor protein. Nat Struct Mol Biol 2004;11:429–434. [DOI] [PubMed] [Google Scholar]
- 127. Knott JA, Orr DC, Montgomery DS, Sullivan CA, Weston A. The expression and purification of human rhinovirus protease 3C. Eur J Biochem 1989;182:547–555. [DOI] [PubMed] [Google Scholar]
- 128. Sommergruber W, Zorn M, Blaas D, Fessl F, Volkmann P, Maurer‐Fogy I, Pallai P, Merluzzi V, Matteo M, Skern T, et al. Polypeptide 2A of human rhinovirus type 2: Identification as a protease and characterization by mutational analysis. Virology 1989;169:68–77. [DOI] [PubMed] [Google Scholar]
- 129. Haghighat A, Svitkin Y, Novoa I, Kuechler E, Skern T, Sonenberg N. The eIF4G‐eIF4E complex is the target for direct cleavage by the rhinovirus 2A proteinase. J Virol 1996;70:8444–8450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Liebig HD, Ziegler E, Yan R, Hartmuth K, Klump H, Kowalski H, Blaas D, Sommergruber W, Frasel L, Lamphear B, et al. Purification of two picornaviral 2A proteinases: Interaction with eIF‐4 gamma and influence on in vitro translation. Biochemistry 1993;32:7581–7588. [DOI] [PubMed] [Google Scholar]
- 131. Sommergruber W, Ahorn H, Klump H, Seipelt J, Zoephel A, Fessl F, Krystek E, Blaas D, Kuechler E, Liebig HD, et al. 2A proteinases of coxsackie‐and rhinovirus cleave peptides derived from eIF‐4 gamma via a common recognition motif. Virology 1994;198:741–745. [DOI] [PubMed] [Google Scholar]
- 132. Binford SL, Maldonado F, Brothers MA, Weady PT, Zalman LS, Meador JW 3rd, Matthews DA, Patick AK. Conservation of amino acids in human rhinovirus 3C protease correlates with broad‐spectrum antiviral activity of rupintrivir, a novel human rhinovirus 3C protease inhibitor. Antimicrob Agents Chemother 2005;49:619–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Turner RB, Couch RB. Rhinoviruses. In: Knipe DM, Howley PM, editors. Fields virology, 5th ed. Volume 1. Philadelphia: Lippincott Williams & Wilkins; 2007. pp 895–909. [Google Scholar]
- 134. Ghildyal R, Dagher H, Donninger H, de Silva D, Li X, Freezer NJ, Wilson JW, Bardin PG. Rhinovirus infects primary human airway fibroblasts and induces a neutrophil chemokine and a permeability factor. J Med Virol 2005;75:608–615. [DOI] [PubMed] [Google Scholar]
- 135. Jakiela B, Brockman‐Schneider R, Amineva S, Lee WM, Gern JE. Basal cells of differentiated bronchial epithelium are more susceptible to rhinovirus infection. Am J Respir Cell Mol Biol 2008;38:517–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Lopez‐Souza N, Dolganov G, Dubin R, Sachs LA, Sassina L, Sporer H, Yagi S, Schnurr D, Boushey HA, Widdicombe JH. Resistance of differentiated human airway epithelium to infection by rhinovirus. Am J Physiol 2004;286:L373–L381. [DOI] [PubMed] [Google Scholar]
- 137. Conti C, Orsi N, Stein ML. Effect of isoflavans and isoflavenes on rhinovirus 1B and its replication in HeLa cells. Antiviral Res 1988;10:117–127. [DOI] [PubMed] [Google Scholar]
- 138. Diana GD, Otto MJ, Treasurywala AM, McKinlay MA, Oglesby RC, Maliski EG, Rossmann MG, Smith TJ. Enantiomeric effects of homologues of disoxaril on the inhibitory activity against human rhinovirus‐14. J Med Chem 1988;31:540–544. [DOI] [PubMed] [Google Scholar]
- 139. Otto MJ, Fox MP, Fancher MJ, Kuhrt MF, Diana GD, McKinlay MA. In vitro activity of WIN 51711, a new broad‐spectrum antipicornavirus drug. Antimicrob Agents Chemother 1985;27:883–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Andries K, Dewindt B, Snoeks J, Wouters L, Moereels H, Lewi PJ, Janssen PA. Two groups of rhinoviruses revealed by a panel of antiviral compounds present sequence divergence and differential pathogenicity. J Virol 1990;64:1117–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Watson KG, Brown RN, Cameron R, Chalmers DK, Hamilton S, Jin B, Krippner GY, Luttick A, McConnell DB, Reece PA, Ryan J, Stanislawski PC, Tucker SP, Wu WY, Barnard DL, Sidwell RW. An orally bioavailable oxime ether capsid binder with potent activity against human rhinovirus. J Med Chem 2003;46:3181–3184. [DOI] [PubMed] [Google Scholar]
- 142. Barnard DL, Hubbard VD, Smee DF, Sidwell RW, Watson KG, Tucker SP, Reece PA. In vitro activity of expanded‐spectrum pyridazinyl oxime ethers related to pirodavir: novel capsid‐binding inhibitors with potent antipicornavirus activity. Antimicrob Agents Chemother 2004;48:1766–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Makarov VA, Riabova OB, Granik VG, Wutzler P, Schmidtke M. Novel [(biphenyloxy)propyl]isoxazole derivatives for inhibition of human rhinovirus 2 and coxsackievirus B3 replication. J Antimicrob Chemother 2005;55:483–488. [DOI] [PubMed] [Google Scholar]
- 144. Rollinger JM, Steindl TM, Schuster D, Kirchmair J, Anrain K, Ellmerer EP, Langer T, Stuppner H, Wutzler P, Schmidtke M. Structure‐based virtual screening for the discovery of natural inhibitors for human rhinovirus coat protein. J Med Chem 2008;51:842–851. [DOI] [PubMed] [Google Scholar]
- 145. Binford SL, Weady PT, Maldonado F, Brothers MA, Matthews DA, Patick AK. In vitro resistance study of rupintrivir, a novel inhibitor of human rhinovirus 3C protease. Antimicrob Agents Chemother 2007;51:4366–4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Yin FH, Lomax NB. Establishment of a mouse model for human rhinovirus infection. J Gen Virol 1986;67:2335–2340. [DOI] [PubMed] [Google Scholar]
- 147. Newcomb DC, Sajjan US, Nagarkar DR, Wang Q, Nanua S, Zhou Y, McHenry CL, Hennrick KT, Tsai WC, Bentley JK, Lukacs NW, Johnston SL, Hershenson MB. Human rhinovirus 1B exposure induces phosphatidylinositol 3‐kinase‐dependent airway inflammation in mice. Am J Respir Crit Care Med 2008;177:1111–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Bartlett NW, Walton RP, Edwards MR, Aniscenko J, Caramori G, Zhu J, Glanville N, Choy KJ, Jourdan P, Burnet J, Tuthill TJ, Pedrick MS, Hurle MJ, Plumpton C, Sharp NA, Bussell JN, Swallow DM, Schwarze J, Guy B, Almond JW, Jeffery PK, Lloyd CM, Papi A, Killington RA, Rowlands DJ, Blair ED, Clarke NJ, Johnston SL. Mouse models of rhinovirus‐induced disease and exacerbation of allergic airway inflammation. Nat Med 2008;14:199–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Hayden FG, Gwaltney JM, Jr . Prophylactic activity of intranasal enviroxime against experimentally induced rhinovirus type 39 infection. Antimicrob Agents Chemother 1982;21:892–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Hayden FG, Gwaltney JM, Jr . Intranasal interferon‐alpha 2 treatment of experimental rhinoviral colds. J Infect Dis 1984;150:174–180. [DOI] [PubMed] [Google Scholar]
- 151. Levandowski RA, Pachucki CT, Rubenis M, Jackson GG. Topical enviroxime against rhinovirus infection. Antimicrob Agents Chemother 1982;22:1004–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Phillpotts RJ, Wallace J, Tyrrell DA, Tagart VB. Therapeutic activity of enviroxime against rhinovirus infection in volunteers. Antimicrob Agents Chemother 1983;23:671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Turner RB, Dutko FJ, Goldstein NH, Lockwood G, Hayden FG. Efficacy of oral WIN 54954 for prophylaxis of experimental rhinovirus infection. Antimicrob Agents Chemother 1993;37:297–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Turner RB, Riker DK, Gangemi JD. Ineffectiveness of echinacea for prevention of experimental rhinovirus colds. Antimicrob Agents Chemother 2000;44:1708–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Barnard DL. Current status of anti‐picornavirus therapies. Curr Pharm Des 2006;12:1379–1390. [DOI] [PubMed] [Google Scholar]
- 156. De Palma AM, Vliegen I, De Clercq E, Neyts J. Selective inhibitors of picornavirus replication. Med Res Rev 2008;28:823–884. [DOI] [PubMed] [Google Scholar]
- 157. Lall MS, Jain RP, Vederas JC. Inhibitors of 3C cysteine proteinases from Picornaviridae. Curr Top Med Chem 2004;4:1239–1253. [DOI] [PubMed] [Google Scholar]
- 158. Colonno RJ, Callahan PL, Long WJ. Isolation of a monoclonal antibody that blocks attachment of the major group of human rhinoviruses. J Virol 1986;57:7–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Hodits RA, Nimpf J, Pfistermueller DM, Hiesberger T, Schneider WJ, Vaughan TJ, Johnson KS, Haumer M, Kuechler E, Winter G, et al. An antibody fragment from a phage display library competes for ligand binding to the low density lipoprotein receptor family and inhibits rhinovirus infection. J Biol Chem 1995;270:24078–24085. [DOI] [PubMed] [Google Scholar]
- 160. Hayden FG, Gwaltney JM, Jr. , Colonno RJ. Modification of experimental rhinovirus colds by receptor blockade. Antiviral Res 1988;9:233–247. [DOI] [PubMed] [Google Scholar]
- 161. Fang F, Yu M. Viral receptor blockage by multivalent recombinant antibody fusion proteins: Inhibiting human rhinovirus (HRV) infection with CFY196. J Antimicrob Chemother 2004;53:23–25. [DOI] [PubMed] [Google Scholar]
- 162. Crump CE, Arruda E, Hayden FG. Comparative antirhinoviral activities of soluble intercellular adhesion molecule‐1 (sICAM‐1) and chimeric ICAM‐1/immunoglobulin A molecule. Antimicrob Agents Chemother 1994;38:1425–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Marlin SD, Staunton DE, Springer TA, Stratowa C, Sommergruber W, Merluzzi VJ. A soluble form of intercellular adhesion molecule‐1 inhibits rhinovirus infection. Nature 1990;344:70–72. [DOI] [PubMed] [Google Scholar]
- 164. Ohlin A, Hoover‐Litty H, Sanderson G, Paessens A, Johnston SL, Holgate ST, Huguenel E, Greve JM. Spectrum of activity of soluble intercellular adhesion molecule‐1 against rhinovirus reference strains and field isolates. Antimicrob Agents Chemother 1994;38:1413–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Marlovits TC, Abrahamsberg C, Blaas D. Soluble LDL minireceptors. Minimal structure requirements for recognition of minor group human rhinovirus. J Biol Chem 1998;273:33835–33840. [DOI] [PubMed] [Google Scholar]
- 166. Moser R, Snyers L, Wruss J, Angulo J, Peters H, Peters T, Blaas D. Neutralization of a common cold virus by concatemers of the third ligand binding module of the VLDL‐receptor strongly depends on the number of modules. Virology 2005;338:259–269. [DOI] [PubMed] [Google Scholar]
- 167. Nicodemou A, Petsch M, Konecsni T, Kremser L, Kenndler E, Casasnovas JM, Blaas D. Rhinovirus‐stabilizing activity of artificial VLDL‐receptor variants defines a new mechanism for virus neutralization by soluble receptors. FEBS Lett 2005;579:5507–5511. [DOI] [PubMed] [Google Scholar]
- 168. Arruda E, Crump CE, Marlin SD, Merluzzi VJ, Hayden FG. In vitro studies of the antirhinovirus activity of soluble intercellular adhesion molecule‐1. Antimicrob Agents Chemother 1992;36:1186–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Greve JM, Forte CP, Marlor CW, Meyer AM, Hoover‐Litty H, Wunderlich D, McClelland A. Mechanisms of receptor‐mediated rhinovirus neutralization defined by two soluble forms of ICAM‐1. J Virol 1991;65:6015–6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Martin S, Casasnovas JM, Staunton DE, Springer TA. Efficient neutralization and disruption of rhinovirus by chimeric ICAM‐1/immunoglobulin molecules. J Virol 1993;67:3561–3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Marlovits TC, Zechmeister T, Gruenberger M, Ronacher B, Schwihla H, Blaas D. Recombinant soluble low density lipoprotein receptor fragment inhibits minor group rhinovirus infection in vitro. FASEB J 1998;12:695–703. [DOI] [PubMed] [Google Scholar]
- 172. Huguenel ED, Cohn D, Dockum DP, Greve JM, Fournel MA, Hammond L, Irwin R, Mahoney J, McClelland A, Muchmore E, Ohlin AC, Scuderi P. Prevention of rhinovirus infection in chimpanzees by soluble intercellular adhesion molecule‐1. Am J Respir Crit Care Med 1997;155:1206–1210. [DOI] [PubMed] [Google Scholar]
- 173. Turner RB, Wecker MT, Pohl G, Witek TJ, McNally E, St George R, Winther B, Hayden FG. Efficacy of tremacamra, a soluble intercellular adhesion molecule 1, for experimental rhinovirus infection: A randomized clinical trial. J Am Med Assoc 1999;281:1797–1804. [DOI] [PubMed] [Google Scholar]
- 174. Diana GD. Inhibitors of picornavirus replication. Curr Med Chem 2003;2:1–12. [Google Scholar]
- 175. Rossmann MG. The structure of antiviral agents that inhibit uncoating when complexed with viral capsids. Antiviral Res 1989;11:3–13. [DOI] [PubMed] [Google Scholar]
- 176. Zhang Y, Simpson AA, Ledford RM, Bator CM, Chakravarty S, Skochko GA, Demenczuk TM, Watanyar A, Pevear DC, Rossmann MG. Structural and virological studies of the stages of virus replication that are affected by antirhinovirus compounds. J Virol 2004;78:11061–11069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Badger J, Minor I, Oliveira MA, Smith TJ, Rossmann MG. Structural analysis of antiviral agents that interact with the capsid of human rhinoviruses. Proteins 1989;6:1–19. [DOI] [PubMed] [Google Scholar]
- 178. Pevear DC, Fancher MJ, Felock PJ, Rossmann MG, Miller MS, Diana G, Treasurywala AM, McKinlay MA, Dutko FJ. Conformational change in the floor of the human rhinovirus canyon blocks adsorption to HeLa cell receptors. J Virol 1989;63:2002–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Smith TJ, Kremer MJ, Luo M, Vriend G, Arnold E, Kamer G, Rossmann MG, McKinlay MA, Diana GD, Otto MJ. The site of attachment in human rhinovirus 14 for antiviral agents that inhibit uncoating. Science 1986;233:1286–1293. [DOI] [PubMed] [Google Scholar]
- 180. Kim KH, Willingmann P, Gong ZX, Kremer MJ, Chapman MS, Minor I, Oliveira MA, Rossmann MG, Andries K, Diana GD, et al. A comparison of the anti‐rhinoviral drug binding pocket in HRV14 and HRV1A. J Mol Biol 1993;230:206–227. [DOI] [PubMed] [Google Scholar]
- 181. Hadfield AT, Diana GD, Rossmann MG. Analysis of three structurally related antiviral compounds in complex with human rhinovirus 16. Proc Natl Acad Sci USA 1999;96:14730–14735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Dewindt B, van Eemeren K, Andries K. Antiviral capsid‐binding compounds can inhibit the adsorption of minor receptor rhinoviruses. Antiviral Res 1994;25:67–72. [DOI] [PubMed] [Google Scholar]
- 183. Kaiser L, Crump CE, Hayden FG. In vitro activity of pleconaril and AG7088 against selected serotypes and clinical isolates of human rhinoviruses. Antiviral Res 2000;47:215–220. [DOI] [PubMed] [Google Scholar]
- 184. Pevear DC, Tull TM, Seipel ME, Groarke JM. Activity of pleconaril against enteroviruses. Antimicrob Agents Chemother 1999;43:2109–2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Schmidtke M, Hammerschmidt E, Schuler S, Zell R, Birch‐Hirschfeld E, Makarov VA, Riabova OB, Wutzler P. Susceptibility of coxsackievirus B3 laboratory strains and clinical isolates to the capsid function inhibitor pleconaril: Antiviral studies with virus chimeras demonstrate the crucial role of amino acid 1092 in treatment. J Antimicrob Chemother 2005;56:648–656. [DOI] [PubMed] [Google Scholar]
- 186. Andries K, Dewindt B, De Brabander M, Stokbroekx R, Janssen PA. In vitro activity of R 61837, a new antirhinovirus compound. Arch Virol 1988;101:155–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Andries K, Dewindt B, Snoeks J, Willebrords R, van Eemeren K, Stokbroekx R, Janssen PA. In vitro activity of pirodavir (R 77975), a substituted phenoxy‐pyridazinamine with broad‐spectrum antipicornaviral activity. Antimicrob Agents Chemother 1992;36:100–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Kuz'min VE, Artemenko AG, Muratov EN, Volineckaya IL, Makarov VA, Riabova OB, Wutzler P, Schmidtke M. Quantitative structure–activity relationship studies of [(biphenyloxy)propyl]isoxazole derivatives. Inhibitors of human rhinovirus 2 replication. J Med Chem 2007;50:4205–4213. [DOI] [PubMed] [Google Scholar]
- 189. Buontempo PJ, Cox S, Wright‐Minogue J, DeMartino JL, Skelton AM, Ferrari E, Albin R, Rozhon EJ, Girijavallabhan V, Modlin JF, O'Connell JF. SCH 48973: A potent, broad‐spectrum, antienterovirus compound. Antimicrob Agents Chemother 1997;41:1220–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Cox S, Buontempo PJ, Wright‐Minogue J, DeMartino JL, Skelton AM, Ferrari E, Schwartz J, Rozhon EJ, Linn CC, Girijavallabhan V, O'Connell JF. Antipicornavirus activity of SCH 47802 and analogs: In vitro and in vivo studies. Antiviral Res 1996;32:71–79. [DOI] [PubMed] [Google Scholar]
- 191. Rozhon E, Cox S, Buontempo P, O'Connell J, Slater W, De Martino J, Schwartz J, Miller G, Arnold E, Zhang A, et al. SCH 38057: A picornavirus capsid‐binding molecule with antiviral activity after the initial stage of viral uncoating. Antiviral Res 1993;21:15–35. [DOI] [PubMed] [Google Scholar]
- 192. Ishitsuka H, Ninomiya YT, Ohsawa C, Fujiu M, Suhara Y. Direct and specific inactivation of rhinovirus by chalcone Ro 09‐0410. Antimicrob Agents Chemother 1982;22:617–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Ninomiya Y, Shimma N, Ishitsuka H. Comparative studies on the antirhinovirus activity and the mode of action of the rhinovirus capsid‐binding agents, chalcone amides. Antiviral Res 1990;13:61–74. [DOI] [PubMed] [Google Scholar]
- 194. Bauer DJ, Selway JW, Batchelor JF, Tisdale M, Caldwell IC, Young DA. 4′,6‐Dichloroflavan (BW683C), a new anti‐rhinovirus compound. Nature 1981;292:369–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Bargar TM, Dulworth JK, Kenny MT, Massad R, Daniel JK, Wilson T, Sargent RN. 3,4‐Dihydro‐2‐phenyl‐2H‐pyrano[2,3‐b]pyridines with potent antirhinovirus activity. J Med Chem 1986;29:1590–1595. [DOI] [PubMed] [Google Scholar]
- 196. Kenny MT, Dulworth JK, Bargar TM, Torney HL, Graham MC, Manelli AM. In vitro antiviral activity of the 6‐substituted 2‐(3′,4′‐dichlorophenoxy)‐2H‐pyrano[2,3‐b]pyridines MDL 20,610, MDL 20,646, and MDL 20,957. Antimicrob Agents Chemother 1986;30:516–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Markley LD, Tong YC, Dulworth JK, Steward DL, Goralski CT, Johnston H, Wood SG, Vinogradoff AP, Bargar TM. Antipicornavirus activity of substituted phenoxybenzenes and phenoxypyridines. J Med Chem 1986;29:427–433. [DOI] [PubMed] [Google Scholar]
- 198. Powers RD, Gwaltney JM, Jr. , Hayden FG. Activity of 2‐(3,4‐dichlorophenoxy)‐5‐nitrobenzonitrile (MDL‐860) against picornaviruses in vitro. Antimicrob Agents Chemother 1982;22:639–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Rossmann MG, McKinlay MA. Application of crystallography to the design of antiviral agents. Infect Agents Dis 1992;1:3–10. [PubMed] [Google Scholar]
- 200. Schiff GM, Sherwood JR. Clinical activity of pleconaril in an experimentally induced coxsackievirus A21 respiratory infection. J Infect Dis 2000;181:20–26. [DOI] [PubMed] [Google Scholar]
- 201. Hayden FG, Coats T, Kim K, Hassman HA, Blatter MM, Zhang B, Liu S. Oral pleconaril treatment of picornavirus‐associated viral respiratory illness in adults: Efficacy and tolerability in phase II clinical trials. Antivir Ther 2002;7:53–65. [PubMed] [Google Scholar]
- 202. Hayden FG, Herrington DT, Coats TL, Kim K, Cooper EC, Villano SA, Liu S, Hudson S, Pevear DC, Collett M, McKinlay M. Efficacy and safety of oral pleconaril for treatment of colds due to picornaviruses in adults: Results of 2 double‐blind, randomized, placebo‐controlled trials. Clin Infect Dis 2003;36:1523–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Pevear DC, Hayden FG, Demenczuk TM, Barone LR, McKinlay MA, Collett MS. Relationship of pleconaril susceptibility and clinical outcomes in treatment of common colds caused by rhinoviruses. Antimicrob Agents Chemother 2005;49:4492–4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Ma JD, Nafziger AN, Rhodes G, Liu S, Gartung AM, Bertino JS, Jr . The effect of oral pleconaril on hepatic cytochrome P450 3A activity in healthy adults using intravenous midazolam as a probe. J Clin Pharmacol 2006;46:103–108. [DOI] [PubMed] [Google Scholar]
- 205. Hayden FG, Andries K, Janssen PA. Safety and efficacy of intranasal pirodavir (R77975) in experimental rhinovirus infection. Antimicrob Agents Chemother 1992;36:727–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Krenn BM, Holzer B, Gaudernak E, Triendl A, van Kuppeveld FJ, Seipelt J. Inhibition of polyprotein processing and RNA replication of human rhinovirus by pyrrolidine dithiocarbamate involves metal ions. J Virol 2005;79:13892–13899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Badorff C, Fichtlscherer B, Rhoads RE, Zeiher AM, Muelsch A, Dimmeler S, Knowlton KU. Nitric oxide inhibits dystrophin proteolysis by coxsackieviral protease 2A through S‐nitrosylation: A protective mechanism against enteroviral cardiomyopathy. Circulation 2000;102:2276–2281. [DOI] [PubMed] [Google Scholar]
- 208. Saura M, Zaragoza C, McMillan A, Quick RA, Hohenadl C, Lowenstein JM, Lowenstein CJ. An antiviral mechanism of nitric oxide: Inhibition of a viral protease. Immunity 1999;10:21–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Zaragoza C, Ocampo CJ, Saura M, McMillan A, Lowenstein CJ. Nitric oxide inhibition of coxsackievirus replication in vitro. J Clin Invest 1997;100:1760–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Zell R, Markgraf R, Schmidtke M, Gorlach M, Stelzner A, Henke A, Sigusch HH, Gluck B. Nitric oxide donors inhibit the coxsackievirus B3 proteinases 2A and 3C in vitro, virus production in cells, and signs of myocarditis in virus‐infected mice. Med Microbiol Immunol 2004;193:91–100. [DOI] [PubMed] [Google Scholar]
- 211. Kaul P, Singh I, Turner RB. Effect of nitric oxide on rhinovirus replication and virus‐induced interleukin‐8 elaboration. Am J Respir Crit Care Med 1999;159:1193–1198. [DOI] [PubMed] [Google Scholar]
- 212. Molla A, Hellen CU, Wimmer E. Inhibition of proteolytic activity of poliovirus and rhinovirus 2A proteinases by elastase‐specific inhibitors. J Virol 1993;67:4688–4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. Deszcz L, Cencic R, Sousa C, Kuechler E, Skern T. An antiviral peptide inhibitor that is active against picornavirus 2A proteinases but not cellular caspases. J Virol 2006;80:9619–9627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Deszcz L, Seipelt J, Vassilieva E, Roetzer A, Kuechler E. Antiviral activity of caspase inhibitors: Effect on picornaviral 2A proteinase. FEBS Lett 2004;560:51–55. [DOI] [PubMed] [Google Scholar]
- 215. Wang QM, Johnson RB, Jungheim LN, Cohen JD, Villarreal EC. Dual inhibition of human rhinovirus 2A and 3C proteases by homophthalimides. Antimicrob Agents Chemother 1998;42:916–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Matthews DA, Smith WW, Ferre RA, Condon B, Budahazi G, Sisson W, Villafranca JE, Janson CA, McElroy HE, Gribskov CL, et al. Structure of human rhinovirus 3C protease reveals a trypsin‐like polypeptide fold, RNA‐binding site, and means for cleaving precursor polyprotein. Cell 1994;77:761–771. [DOI] [PubMed] [Google Scholar]
- 217. Kuo C‐J, Liu H‐G, Lo Y‐K, Seong C‐M, Lee K‐I, Jung Y‐S, Liang P‐H. Individual and common inhibitors of coronavirus and picornavirus main proteases. FEBS Lett 2009;583:549–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Matthews DA, Dragovich PS, Webber SE, Fuhrman SA, Patick AK, Zalman LS, Hendrickson TF, Love RA, Prins TJ, Marakovits JT, Zhou R, Tikhe J, Ford CE, Meador JW, Ferre RA, Brown EL, Binford SL, Brothers MA, Delisle DM, Worland ST. Structure‐assisted design of mechanism‐based irreversible inhibitors of human rhinovirus 3C protease with potent antiviral activity against multiple rhinovirus serotypes. Proc Natl Acad Sci USA 1999;96:11000–11007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219. Maugeri C, Alisi MA, Apicella C, Cellai L, Dragone P, Fioravanzo E, Florio S, Furlotti G, Mangano G, Ombrato R, Luisi R, Pompei R, Rincicotti V, Russo V, Vitiello M, Cazzolla N. New anti‐viral drugs for the treatment of the common cold. Bioorg Med Chem 2008;16:3091–3107. [DOI] [PubMed] [Google Scholar]
- 220. Dragovich PS, Webber SE, Babine RE, Fuhrman SA, Patick AK, Matthews DA, Lee CA, Reich SH, Prins TJ, Marakovits JT, Littlefield ES, Zhou R, Tikhe J, Ford CE, Wallace MB, Meador JW 3rd, Ferre RA, Brown EL, Binford SL, Harr JE, DeLisle DM, Worland ST. Structure‐based design, synthesis, and biological evaluation of irreversible human rhinovirus 3C protease inhibitors. 1. Michael acceptor structure‐activity studies. J Med Chem 1998;41:2806–2818. [DOI] [PubMed] [Google Scholar]
- 221. Dragovich PS, Webber SE, Babine RE, Fuhrman SA, Patick AK, Matthews DA, Reich SH, Marakovits JT, Prins TJ, Zhou R, Tikhe J, Littlefield ES, Bleckman TM, Wallace MB, Little TL, Ford CE, Meador JW 3rd, Ferre RA, Brown EL, Binford SL, DeLisle DM, Worland ST. Structure‐based design, synthesis, and biological evaluation of irreversible human rhinovirus 3C protease inhibitors. 2. Peptide structure‐activity studies. J Med Chem 1998;41:2819–2834. [DOI] [PubMed] [Google Scholar]
- 222. Dragovich PS, Prins TJ, Zhou R, Brown EL, Maldonado FC, Fuhrman SA, Zalman LS, Tuntland T, Lee CA, Patick AK, Matthews DA, Hendrickson TF, Kosa MB, Liu B, Batugo MR, Gleeson JP, Sakata SK, Chen L, Guzman MC, Meador JW 3rd, Ferre RA, Worland ST. Structure‐based design, synthesis, and biological evaluation of irreversible human rhinovirus 3C protease inhibitors. 6. Structure‐activity studies of orally bioavailable, 2‐pyridone‐containing peptidomimetics. J Med Chem 2002;45:1607–1623. [DOI] [PubMed] [Google Scholar]
- 223. Dragovich PS, Prins TJ, Zhou R, Fuhrman SA, Patick AK, Matthews DA, Ford CE, Meador JW 3rd, Ferre RA, Worland ST. Structure‐based design, synthesis, and biological evaluation of irreversible human rhinovirus 3C protease inhibitors. 3. Structure‐activity studies of ketomethylene‐containing peptidomimetics. J Med Chem 1999;42:1203–1212. [DOI] [PubMed] [Google Scholar]
- 224. Dragovich PS, Prins TJ, Zhou R, Johnson TO, Brown EL, Maldonado FC, Fuhrman SA, Zalman LS, Patick AK, Matthews DA, Hou X, Meador JW, Ferre RA, Worland ST. Structure‐based design, synthesis, and biological evaluation of irreversible human rhinovirus 3C protease inhibitors. Part 7: Structure‐activity studies of bicyclic 2‐pyridone‐containing peptidomimetics. Bioorg Med Chem Lett 2002;12:733–738. [DOI] [PubMed] [Google Scholar]
- 225. Dragovich PS, Prins TJ, Zhou R, Johnson TO, Hua Y, Luu HT, Sakata SK, Brown EL, Maldonado FC, Tuntland T, Lee CA, Fuhrman SA, Zalman LS, Patick AK, Matthews DA, Wu EY, Guo M, Borer BC, Nayyar NK, Moran T, Chen L, Rejto PA, Rose PW, Guzman MC, Dovalsantos EZ, Lee S, McGee K, Mohajeri M, Liese A, Tao J, Kosa MB, Liu B, Batugo MR, Gleeson JP, Wu ZP, Liu J, Meador JW 3rd, Ferre RA. Structure‐based design, synthesis, and biological evaluation of irreversible human rhinovirus 3C protease inhibitors. 8. Pharmacological optimization of orally bioavailable 2‐pyridone‐containing peptidomimetics. J Med Chem 2003;46:4572–4585. [DOI] [PubMed] [Google Scholar]
- 226. Dragovich PS, Prins TJ, Zhou R, Webber SE, Marakovits JT, Fuhrman SA, Patick AK, Matthews DA, Lee CA, Ford CE, Burke BJ, Rejto PA, Hendrickson TF, Tuntland T, Brown EL, Meador JW 3rd, Ferre RA, Harr JE, Kosa MB, Worland ST. Structure‐based design, synthesis, and biological evaluation of irreversible human rhinovirus 3C protease inhibitors. 4. Incorporation of P1 lactam moieties as l‐glutamine replacements. J Med Chem 1999;42:1213–1224. [DOI] [PubMed] [Google Scholar]
- 227. Patick AK, Binford SL, Brothers MA, Jackson RL, Ford CE, Diem MD, Maldonado F, Dragovich PS, Zhou R, Prins TJ, Fuhrman SA, Meador JW, Zalman LS, Matthews DA, Worland ST. In vitro antiviral activity of AG7088, a potent inhibitor of human rhinovirus 3C protease. Antimicrob Agents Chemother 1999;43:2444–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228. Zalman LS, Brothers MA, Dragovich PS, Zhou R, Prins TJ, Worland ST, Patick AK. Inhibition of human rhinovirus‐induced cytokine production by AG7088, a human rhinovirus 3C protease inhibitor. Antimicrob Agents Chemother 2000;44:1236–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229. Hsyu PH, Pithavala YK, Gersten M, Penning CA, Kerr BM. Pharmacokinetics and safety of an antirhinoviral agent, ruprintrivir, in healthy volunteers. Antimicrob Agents Chemother 2002;46:392–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230. Hayden FG, Turner RB, Gwaltney JM, Chi‐Burris K, Gersten M, Hsyu P, Patick AK, Smith GJ 3rd, Zalman LS. Phase II, randomized, double‐blind, placebo‐controlled studies of ruprintrivir nasal spray 2‐percent suspension for prevention and treatment of experimentally induced rhinovirus colds in healthy volunteers. Antimicrob Agents Chemother 2003;47:3907–3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231. Patick AK, Brothers MA, Maldonado F, Binford S, Maldonado O, Fuhrman S, Petersen A, Smith GJ 3rd, Zalman LS, Burns‐Naas LA, Tran JQ. In vitro antiviral activity and single‐dose pharmacokinetics in humans of a novel, orally bioavailable inhibitor of human rhinovirus 3C protease. Antimicrob Agents Chemother 2005;49:2267–2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232. Schubert S, Furste JP, Werk D, Grunert HP, Zeichhardt H, Erdmann VA, Kurreck J. Gaining target access for deoxyribozymes. J Mol Biol 2004;339:355–363. [DOI] [PubMed] [Google Scholar]
- 233. Stone JK, Rijnbrand R, Stein DA, Ma Y, Yang Y, Iversen PL, Andino R. A morpholino oligomer targeting highly conserved internal ribosome entry site sequence is able to inhibit multiple species of picornavirus. Antimicrob Agents Chemother 2008;52:1970–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234. Phipps KM, Martinez A, Lu J, Heinz BA, Zhao G. Small interfering RNA molecules as potential anti‐human rhinovirus agents: In vitro potency, specificity, and mechanism. Antiviral Res 2004;61:49–55. [DOI] [PubMed] [Google Scholar]
- 235. Verheyden B, Andries K, Rombaut B. Mode of action of 2‐furylmercury chloride, an anti‐rhinovirus compound. Antiviral Res 2004;61:189–194. [DOI] [PubMed] [Google Scholar]
- 236. Van Hoof L, Vanden Berghe DA, Hatfield GM, Vlietinck AJ. Plant antiviral agents; V. 3‐Methoxyflavones as potent inhibitors of viral‐induced block of cell synthesis. Planta Med 1984;50:513–517. [PubMed] [Google Scholar]
- 237. Gaudernak E, Seipelt J, Triendl A, Grassauer A, Kuechler E. Antiviral effects of pyrrolidine dithiocarbamate on human rhinoviruses. J Virol 2002;76:6004–6015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238. De Clercq E, Murase J, Marquez VE. Broad‐spectrum antiviral and cytocidal activity of cyclopentenylcytosine, a carbocyclic nucleoside targeted at CTP synthetase. Biochem Pharmacol 1991;41:1821–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239. Andersen DO, Murray BK, Robins RK, North JA. In vitro antiviral activity of ribavirin against picornaviruses. Antivir Chem Chemother 1992;6:361–370. [Google Scholar]
- 240. De Clercq E. Another ten stories in antiviral drug discovery (part C): “Old” and “new” antivirals, strategies, and perspectives. Med Res Rev 2009;29:611–645. [DOI] [PubMed] [Google Scholar]
- 241. Crotty S, Maag D, Arnold JJ, Zhong W, Lau JY, Hong Z, Andino R, Cameron CE. The broad‐spectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen. Nat Med 2000;6:1375–1379. [DOI] [PubMed] [Google Scholar]
- 242. Reyes GR. Ribavirin: Recent insights into antiviral mechanisms of action. Curr Opin Drug Disc 2001;4:651–656. [PubMed] [Google Scholar]
- 243. DeLong DC, Reed SE. Inhibition of rhinovirus replication in in organ culture by a potential antiviral drug. J Infect Dis 1980;141:87–91. [DOI] [PubMed] [Google Scholar]
- 244. Wikel JH, Paget CJ, DeLong DC, Nelson JD, Wu CY, Paschal JW, Dinner A, Templeton RJ, Chaney MO, Jones ND, Chamberlin JW. Synthesis of syn and anti isomers of 6‐[[(hydroxyimino)phenyl]methyl]‐1‐[(1‐methylethyl)sulfonyl]‐1H‐benzimidaz ol‐2‐amine. Inhibitors of rhinovirus multiplication. J Med Chem 1980;23:368–372. [DOI] [PubMed] [Google Scholar]
- 245. Ninomiya Y, Aoyama M, Umeda I, Suhara Y, Ishitsuka H. Comparative studies on the modes of action of the antirhinovirus agents Ro 09‐0410, Ro 09‐0179, RMI‐15,731, 4′,6‐dichloroflavan, and enviroxime. Antimicrob Agents Chemother 1985;27:595–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246. Heinz BA, Vance LM. The antiviral compound enviroxime targets the 3A coding region of rhinovirus and poliovirus. J Virol 1995;69:4189–4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247. Heinz BA, Vance LM. Sequence determinants of 3A‐mediated resistance to enviroxime in rhinoviruses and enteroviruses. J Virol 1996;70:4854–4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248. Brown‐Augsburger P, Vance LM, Malcolm SK, Hsiung H, Smith DP, Heinz BA. Evidence that enviroxime targets multiple components of the rhinovirus 14 replication complex. Arch Virol 1999;144:1569–1585. [DOI] [PubMed] [Google Scholar]
- 249. Miller FD, Monto AS, DeLong DC, Exelby A, Bryan ER, Srivastava S. Controlled trial of enviroxime against natural rhinovirus infections in a community. Antimicrob Agents Chemother 1985;27:102–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250. Phillpotts RJ, Jones RW, Delong DC, Reed SE, Wallace J, Tyrrell DA. The activity of enviroxime against rhinovirus infection in man. Lancet 1981;1:1342–1344. [DOI] [PubMed] [Google Scholar]
- 251. Wyde PR, Six HR, Wilson SZ, Gilbert BE, Knight V. Activity against rhinoviruses, toxicity, and delivery in aerosol of enviroxime in liposomes. Antimicrob Agents Chemother 1988;32:890–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252. Hamdouchi C, de Blas J, del Prado M, Gruber J, Heinz BA, Vance L. 2‐Amino‐3‐substituted‐6‐[(E)‐1‐phenyl‐2‐(N‐methylcarbamoyl)vinyl]imid azo[1,2‐a]pyridines as a novel class of inhibitors of human rhinovirus: Stereospecific synthesis and antiviral activity. J Med Chem 1999;42:50–59. [DOI] [PubMed] [Google Scholar]
- 253. Hamdouchi C, Ezquerra J, Vega JA, Vaquero JJ, Alvarez‐Builla J, Heinz BA. Short synthesis and anti‐rhinoviral activity of imidazo[1,2‐a]pyridines: The effect of acyl groups at 3‐position. Bioorg Med Chem Lett 1999;9:1391–1394. [DOI] [PubMed] [Google Scholar]
- 254. Victor F, Brown TJ, Campanale K, Heinz BA, Shipley LA, Su KS, Tang J, Vance LM, Spitzer WA. Synthesis, antiviral activity, and biological properties of vinylacetylene analogs of enviroxime. J Med Chem 1997;40:1511–1518. [DOI] [PubMed] [Google Scholar]
- 255. Victor F, Loncharich R, Tang J, Spitzer WA. Synthesis and antiviral activity of C2 analogs of enviroxime: An exploration of the role of critical functionality. J Med Chem 1997;40:3478–3483. [DOI] [PubMed] [Google Scholar]
- 256. Came PE, Schafer TW, Silver GH. Sensitivity of rhinoviruses to human leukocyte and fibroblast interferons. J Infect Dis 1976;133:A136–A139. [DOI] [PubMed] [Google Scholar]
- 257. Kramer MJ, Dennin R, Kramer C, Jones G, Connell E, Rolon N, Gruarin A, Kale R, Trown PW. Cell and virus sensitivity studies with recombinant human alpha interferons. J Interferon Res 1983;3:425–435. [DOI] [PubMed] [Google Scholar]
- 258. Sperber SJ, Hayden FG. Comparative susceptibility of respiratory viruses to recombinant interferons‐alpha 2b and ‐beta. J Interferon Res 1989;9:285–293. [DOI] [PubMed] [Google Scholar]
- 259. Farr BM, Gwaltney JM, Jr. , Adams KF, Hayden FG. Intranasal interferon‐alpha 2 for prevention of natural rhinovirus colds. Antimicrob Agents Chemother 1984;26:31–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260. Hayden FG, Gwaltney JM, Jr . Intranasal interferon alpha 2 for prevention of rhinovirus infection and illness. J Infect Dis 1983;148:543–550. [DOI] [PubMed] [Google Scholar]
- 261. Higgins PG, Al‐Nakib W, Willman J, Tyrrell DA. Interferon‐beta ser as prophylaxis against experimental rhinovirus infection in volunteers. J Interferon Res 1986;6:153–159. [DOI] [PubMed] [Google Scholar]
- 262. Phillpotts RJ, Scott GM, Higgins PG, Wallace J, Tyrrell DA, Gauci CL. An effective dosage regimen for prophylaxis against rhinovirus infection by intranasal administration of HuIFN‐alpha 2. Antiviral Res 1983;3:121–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263. Samo TC, Greenberg SB, Couch RB, Quarles J, Johnson PE, Hook S, Harmon MW. Efficacy and tolerance of intranasally applied recombinant leukocyte A interferon in normal volunteers. J Infect Dis 1983;148:535–542. [DOI] [PubMed] [Google Scholar]
- 264. Samo TC, Greenberg SB, Palmer JM, Couch RB, Harmon MW, Johnson PE. Intranasally applied recombinant leukocyte A interferon in normal volunteers. II. Determination of minimal effective and tolerable dose. J Infect Dis 1984;150:181–188. [DOI] [PubMed] [Google Scholar]
- 265. Higgins PG, Al‐Nakib W, Barrow GI, Tyrrell DA. Recombinant human interferon‐gamma as prophylaxis against rhinovirus colds in volunteers. J Interferon Res 1988;8:591–596. [DOI] [PubMed] [Google Scholar]
- 266. Monto AS, Schwartz SA, Albrecht JK. Ineffectiveness of postexposure prophylaxis of rhinovirus infection with low‐dose intranasal alpha 2b interferon in families. Antimicrob Agents Chemother 1989;33:387–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267. Farr BM, Gwaltney JM, Jr. , Hendley JO, Hayden FG, Naclerio RM, McBride T, Doyle WJ, Sorrentino JV, Riker DK, Proud D. A randomized controlled trial of glucocorticoid prophylaxis against experimental rhinovirus infection. J Infect Dis 1990;162:1173–1177. [DOI] [PubMed] [Google Scholar]
- 268. Ahmad AL, Tyrrell DA. Synergism between anti‐rhinovirus antivirals: Various human interferons and a number of synthetic compounds. Antiviral Res 1986;6:241–252. [DOI] [PubMed] [Google Scholar]
- 269. Higgins PG, Barrow GI, Al‐Nakib W, Tyrrell DA, DeLong DC, Lenox‐Smith I. Failure to demonstrate synergy between interferon‐alpha and a synthetic antiviral, enviroxime, in rhinovirus infections in volunteers. Antiviral Res 1988;10:141–149. [DOI] [PubMed] [Google Scholar]
- 270. Feher M, Schmidt JM. Property distributions: Differences between drugs, natural products, and molecules from combinatorial chemistry. J Chem Inf Comput Sci 2003;43:218–227. [DOI] [PubMed] [Google Scholar]
- 271. Zheng S, Luo X, Chen G, Zhu W, Shen J, Chen K, Jiang H. A new rapid and effective chemistry space filter in recognizing a druglike database. J Chem Inf Model 2005;45:856–862. [DOI] [PubMed] [Google Scholar]
- 272. Samiulla DS, Vaidyanathan VV, Arun PC, Balan G, Blaze M, Bondre S, et al. Rational selection of structurally diverse natural product scaffolds with favorable ADME properties for drug discovery. Mol Divers 2005;9:131–139. [DOI] [PubMed] [Google Scholar]
- 273. Newman DJ, Cragg GM. Natural products as sources of new drugs over the last 25 years. J Nat Prod 2007;70:461–477. [DOI] [PubMed] [Google Scholar]
- 274. Chattopadhyay D. Ethnomedicinal antivirals: Scope and opportunity. In: Ahmad I, Aqil F, Owais M, editors. Modern phytomedicine: Turning medicinal plants into drugs. Weinheim: Wiley‐VCH; 2006. pp 313–339. [Google Scholar]
- 275. Cos P, Vanden Berghe D, De Bruyne T, Vlietinck AJ. Plant substances as antiviral agents: An update (1997–2001). Curr Org Chem 2003;7:1163–1180. [Google Scholar]
- 276. Vlietinck AJ, De Bruyne T, Vanden Berghe DA. Plant substances as antiviral agents. Curr Org Chem 1997;1:307–344. [Google Scholar]
- 277. Message SD, Johnston SL. Host defense function of the airway epithelium in health and disease: Clinical background. J Leukocyte Biol 2004;75:5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278. Blumenthal M. Herb sales down 6 percent in mainstream market. Herbal Gram 2005;66:63. [Google Scholar]
- 279. Woelkart K, Linde K, Bauer R. Echinacea for preventing and treating the common cold. Planta Med 2008;74:633–637. [DOI] [PubMed] [Google Scholar]
- 280. Linde K, Barrett B, Wölkart K, Bauer R, Melchart D. Echinacea for preventing and treating the common cold. The Cochrane Database of Systematic Reviews 2006:Issue 1. [DOI] [PubMed] [Google Scholar]
- 281. Shah SA, Sander S, White CM, Rinaldi M, Coleman C. Evaluation of Echinacea for the prevention and treatment of the common cold: A meta‐analysis. Lancet Infect Dis 2007;7:473–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282. Schoop R, Klein P, Suter A, Johnston SL. Echinacea in the prevention of induced rhinovirus colds: A meta‐analysis. Clin Ther 2006;28:174–183. [DOI] [PubMed] [Google Scholar]
- 283. Woelkart K, Bauer R. The role of alkamides as an active principle of Echinacea. Planta Med 2007;73:615–623. [DOI] [PubMed] [Google Scholar]
- 284. Raduner S, Majewska A, Chen J‐Z, Xie X‐Q, Hamon J, Faller B, Altmann K‐H, Gertsch J. Alkylamides from Echinacea are a new class of cannabinomimetics: Cannabinoid type 2 receptor‐dependent and ‐independent immunomodulatory effects. J Biol Chem 2006;281:14192–14206. [DOI] [PubMed] [Google Scholar]
- 285. Dalby‐Brown L, Barsett H, Landbo R, Meyer AS, Molgaard P. Synergistic anti‐oxidative effects of alkamides, caffeic acid derivatives, and polysaccharide fractions from Echinacea purpurea on in vitro oxidation of human low‐density lipoproteins. J Agric Food Chem 2005;53:9413–9423. [DOI] [PubMed] [Google Scholar]
- 286. Sharma M, Arnason JT, Burt A, Hudson JB. Echinacea extracts modulate the pattern of chemokine and cytokine secretion in rhinovirus‐infected and uninfected epithelial cells. Phytother Res 2006;20:147–152. [DOI] [PubMed] [Google Scholar]
- 287. Altamirano‐Dimas M, Hudson JB, Cochrane D, Nelson C, Arnason JT. Modulation of immune response gene expression by echinacea extracts: Results of a gene array analysis. Can J Physiol Pharmacol 2007;85:1091–1098. [DOI] [PubMed] [Google Scholar]
- 288. Josling P. Preventing the common cold with a garlic supplement: A double‐blind, placebo‐controlled survey. Adv Ther 2001;18:189–193. [DOI] [PubMed] [Google Scholar]
- 289. Weber ND, Andersen DO, North JA, Murray BK, Lawson LD, Hughes BG. In vitro virucidal effects of Allium sativum (garlic) extract and compounds. Planta Med 1992;58:417–423. [DOI] [PubMed] [Google Scholar]
- 290. Burger RA, Warren RP, Lawson LD, Hughes BG. Enhancement of in vitro human immune function by Allium sativum L. (garlic) fractions. Int J Pharmacog 1993;31:169–174. [Google Scholar]
- 291. McElhaney Janet E, Goel V, Toane B, Hooten J, Shan Jacqueline J. Efficacy of COLD‐fX in the prevention of respiratory symptoms in community‐dwelling adults: A randomized, double‐blinded, placebo controlled trial. J Altern Complement Med 2006;12:153–157. [DOI] [PubMed] [Google Scholar]
- 292. Predy GN, Goel V, Lovlin R, Donner A, Stitt L, Basu TK. Efficacy of an extract of North American ginseng containing poly‐furanosyl‐pyranosyl‐saccharides for preventing upper respiratory tract infections: A randomized controlled trial. Can Medic Assoc J 2005;173:1043–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293. Vohra S, Johnston BC, Laycock KL, Midodzi WK, Dhunnoo I, Harris E, Baydala L. Safety and tolerability of North American ginseng extract in the treatment of pediatric upper respiratory tract infection: A phase II randomized, controlled trial of 2 dosing schedules. Pediatrics 2008;122:e402–e410. [DOI] [PubMed] [Google Scholar]
- 294. Cho JY. Inhibitory effect of ginsenoside Rg3 and its derivative ginsenoside Rg3‐2H on NO production and lymphocyte proliferation. J Ginseng Res 2008;32:264–269. [Google Scholar]
- 295. Biondo PD, Goruk S, Ruth MR, O'Connell E, Field CJ. Effect of CVT‐E002 (COLD‐fX) versus a ginsenoside extract on systemic and gut‐associated immune function. Int Immunopharmacol 2008;8:1134–1142. [DOI] [PubMed] [Google Scholar]
- 296. Bae E‐A, Shin J‐E, Park S‐H, Kim D‐H. Inhibitory effect of ginseng polysaccharides on rotavirus infection. J Microbiol Biotechnol 2004;14:202–204. [Google Scholar]
- 297. Fukushima Y, Bastow KF, Ohba T, Lee KH. Antiviral activity of dammarane saponins against herpes simplex virus I. Int J Pharmacog 1995;33:2–6. [Google Scholar]
- 298. Kuroiwa A, Liou SY, Yan H, Eshita A, Naitoh S, Nagayama A. Effect of a traditional Japanese herbal medicine, Hochu‐ekki‐to (Bu‐Zhong‐Yi‐Qi Tang), on immunity in elderly persons. Int Immunopharmacol 2004;4:317–324. [DOI] [PubMed] [Google Scholar]
- 299. Iwama H, Amagaya S, Ogihara Y. Effects of five kampohozais on the mitogenic activity of lipopolysaccharide, concanavalin A, phorbol myristate acetate and phytohemagglutinin in vivo. J Ethnopharmacol 1986;18:193–204. [DOI] [PubMed] [Google Scholar]
- 300. Utsuyama M, Seidlar H, Kitagawa M, Hirokawa K. Immunological restoration and anti‐tumor effect by Japanese herbal medicine in aged mice. Mech Ageing Dev 2001;122:341–352. [DOI] [PubMed] [Google Scholar]
- 301. Yang S‐H, Yu C‐L. Antiinflammatory effects of Bu‐zhong‐yi‐qi‐tang in patients with perennial allergic rhinitis. J Ethnopharmacol 2008;115:104–109. [DOI] [PubMed] [Google Scholar]
- 302. Yamaya M, Sasaki T, Yasuda H, Inoue D, Suzuki T, Asada M, Yoshida M, Seki T, Iwasaki K, Nishimura H, Nakayama K. Hochu‐ekki‐to inhibits rhinovirus infection in human tracheal epithelial cells. Br J Pharmacol 2007;150:702–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303. Lizogub Viktor G, Riley David S, Heger M. Efficacy of a pelargonium sidoides preparation in patients with the common cold: A randomized, double blind, placebo‐controlled clinical trial. Explore‐NY 2007;3:573–584. [DOI] [PubMed] [Google Scholar]
- 304. Timmer A, Gunther J, Rucker G, Motschall E, Antes G, Kern Winfried V. Pelargonium sidoides extract for acute respiratory tract infections. The Cochrane Database of Systematic Reviews 2008:Issue 3. [DOI] [PubMed] [Google Scholar]
- 305. Kolodziej H. Traditionally used pelargonium species: Chemistry and biological activity of umckaloabo extracts and their constituents. Curr Top Phytochem 2000;3:77–93. [Google Scholar]
- 306. Kolodziej H. Aqueous ethanolic extract of the roots of Pelargonium sidoides—New scientific evidence for an old anti‐infective phytopharmaceutical. Planta Med 2008;74:661–666. [DOI] [PubMed] [Google Scholar]
- 307. Schnitzler P, Schneider S, Stintzing FC, Carle R, Reichling J. Efficacy of an aqueous Pelargonium sidoides extract against herpesvirus. Phytomedicine 2008;15:1108–1116. [DOI] [PubMed] [Google Scholar]
- 308. Carlucci MJ, Scolaro LA, Noseda MD, Cerezo AS, Damonte EB. Protective effect of a natural carrageenan on genital herpes simplex virus infection in mice. Antiviral Res 2004;64:137–141. [DOI] [PubMed] [Google Scholar]
- 309. Gonzalez ME, Alarcon B, Carrasco L. Polysaccharides as antiviral agents: Antiviral activity of carrageenan. Antimicrob Agents Chemother 1987;31:1388–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310. Grassauer A, Weinmuellner R, Meier C, Pretsch A, Prieschl‐Grassauer E, Unger H. Iota‐Carrageenan is a potent inhibitor of rhinovirus infection. Virol J 2008;5:107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311. Gagnier Joel J, Boon H, Rochon P, Moher D, Barnes J, Bombardier C. Recommendations for reporting randomized controlled trials of herbal interventions: Explanation and elaboration. J Clin Epidemiol 2006;59:1134–1149. [DOI] [PubMed] [Google Scholar]
- 312. Farnsworth NR. Ethnopharmacology and drug development. In: Chadwick DJ, Marsh J, editors. Ethnobotany and the search for new drugs: Ciba Foundation Symposium 185. New York: Wiley; 1994. pp 42–59. [DOI] [PubMed] [Google Scholar]
- 313. Heinrich M. Ethnobotany and natural products: The search for new molecules, new treatments of old diseases or a better understanding of indigenous cultures? Curr Top Med Chem 2003;3:141–154. [DOI] [PubMed] [Google Scholar]
- 314. Gertsch J. How scientific is the science in ethnopharmacology? Historical perspectives and epistemological problems. J Ethnopharmacol 2009;122:177–183. [DOI] [PubMed] [Google Scholar]
- 315. Beck LYt. Pedanius Dioscorides of Anazarbus—De materia medica. Hildesheim: Olms; 2005. [Google Scholar]
- 316. Nassar MI, Abu‐Mustafa EA, Ahmed AA. Sesquiterpene coumarins from Ferula assafoetida L. Pharmazie 1995;50:766–767. [Google Scholar]
- 317. Semple SJ, Nobbs SF, Pyke SM, Reynolds GD, Flower RLP. Antiviral flavonoid from Pterocaulon sphacelatum, an Australian Aboriginal medicine. J Ethnopharmacol 1999;68:283–288. [DOI] [PubMed] [Google Scholar]
- 318. Rollinger JM, Schuster D, Danzl B, Schwaiger S, Markt P, Schmidtke M, Gertsch J, Raduner S, Wolber G, Langer T, Stuppner H. In silico target fishing for rationalized ligand discovery exemplified on constituents of Ruta graveolens L. Planta Med 2009;75:195–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319. Tsuchiya Y, Shimizu M, Hiyama Y, Itoh K, Hashimoto Y, Nakayama M, Horie T, Morita N. Antiviral activity of natural occurring flavonoids in vitro. Chem Pharm Bull 1985;33:3881–3886. [DOI] [PubMed] [Google Scholar]
- 320. Vanden Berghe DAR, Haemers A, Vlietinck AJ. Antiviral agents from higher plants and example of structure‐activity relationship of 3‐methoxyflavones. In: Colegate SM, Molyneux RJ, editors. Bioactive Natural Products: Detection, Isolation and Structural Determination. Boca Raton: CRC; 1993. pp 405–440. [Google Scholar]
- 321. De Meyer N, Haemers A, Mishra L, Pandey HK, Pieters LAC, Vanden Berghe DA, Vlietinck AJ. 4′‐Hydroxy‐3‐methoxyflavones with potent antipicornavirus activity. J Med Chem 1991;34:736–746. [DOI] [PubMed] [Google Scholar]
- 322. Cragg GM, Newman DJ. Biodiversity: A continuing source of novel drug leads. Pure Appl Chem 2005;77:7–24. [Google Scholar]
- 323. Singh SB, Cordingley MG, Ball RG, Smith JL, Dombrowski AW, Goetz MA. Structure and stereochemistry of thysanone: A novel human rhinovirus 3C‐protease inhibitor from Thysanophora penicilloides . Tetrahedron Lett 1991;32:5279–5282. [Google Scholar]
- 324. Singh SB, Graham PL, Reamer RA, Cordingley MG. Discovery, total synthesis, HRV 3C‐protease inhibitory activity, and structure‐activity relationships of 2‐methoxystypandrone and its analogues. Bioorg Med Chem Lett 2001;11:3143–3146. [DOI] [PubMed] [Google Scholar]
- 325. Choi HJ, Lim CH, Song JH, Baek SH, Kwon DH. Antiviral activity of raoulic acid from Raoulia australis against Picornaviruses. Phytomedicine 2009;16:35–39. [DOI] [PubMed] [Google Scholar]
- 326. Ishitsuka H, Ohsawa C, Ohiwa T, Umeda I, Suhara Y. Antipicornavirus flavone Ro 09‐0179. Antimicrob Agents Chemother 1982;22:611–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327. Spasova M, Philipov S, Nikolaeva‐Glomb L, Galabov AS, Milkova T. Cinnamoyl‐and hydroxycinnamoyl amides of glaucine and their antioxidative and antiviral activities. Bioorg Med Chem 2008;16:7457–7461. [DOI] [PubMed] [Google Scholar]
- 328. Selway JWT. Antiviral activity of flavones and flavans. Prog Clin Biol Res 1986;213:521–536. [PubMed] [Google Scholar]
- 329. Vlietinck AJ, Vanden Berghe DA, Haemers A. Present status and prospects of flavonoids as anti‐viral agents. Prog Clin Biol Res 1988;280:283–299. [PubMed] [Google Scholar]
- 330. Tisdale M, Selway JWT. Inhibition of an early stage of rhinovirus replication by dichloroflavan (BW683C). J Gen Virol 1983;64:795–803. [DOI] [PubMed] [Google Scholar]
- 331. Tisdale M, Selway JWT. Effect of dichloroflavan (BW683C) on the stability and uncoating of rhinovirus type 1B. J Antimicrob Chemother 1984;14:97–105. [DOI] [PubMed] [Google Scholar]
- 332. Al‐Nakib W, Willman J, Higgins PG, Tyrrell DA, Shepherd WM, Freestone DS. Failure of intranasally administered 4′, 6‐dichloroflavan to protect against rhinovirus infection in man. Arch Virol 1987;92:255–260. [DOI] [PubMed] [Google Scholar]
- 333. Phillpotts RJ, Wallace J, Tyrrell DA, Freestone DS, Shepherd WM. Failure of oral 4′,6‐dichloroflavan to protect against rhinovirus infection in man. Arch Virol 1983;75:115–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334. Gasteiger J, Teckentrup A, Terfloth L, Spycher S. Neural networks as data mining tools in drug design. J Phys Org Chem 2003;16:232–245. [Google Scholar]
- 335. Xu J, Hagler A. Chemoinformatics and drug discovery. Molecules 2002;7:566–600. [Google Scholar]
- 336. Langer T, Hoffmann RD. Pharmacophores and pharmacophore searches. In: Langer T, Hoffmann RD, editors. Methods and principles in medicinal chemistry. Volume 32. Weinheim: Wiley‐VCH; 2006. [Google Scholar]
- 337. Manly CJ, Louise‐May S, Hammer JD. The impact of informatics and computational chemistry on synthesis and screening. Drug Discov Today 2001;6:1101–1110. [DOI] [PubMed] [Google Scholar]
- 338. Kirchmair J, Distinto S, Schuster D, Spitzer G, Langer T, Wolber G. Enhancing drug discovery through in silico screening: Strategies to increase true positives retrieval rates. Curr Med Chem 2008;15:2040–2053. [DOI] [PubMed] [Google Scholar]
- 339. Zupan J, Gasteiger J. Neural networks for chemists. Weinheim: Wiley‐VCH; 2000. [Google Scholar]
- 340. Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res 2000;28:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341. Kirchmair J, Markt P, Distinto S, Schuster D, Spitzer GM, Liedl KR, Langer T, Wolber G. The Protein Data Bank (PDB), its related services and software tools as key components for in silico guided drug discovery. J Med Chem 2008;51:7021–7040. [DOI] [PubMed] [Google Scholar]
- 342. Kirchmair J, Distinto S, Liedl KR, Markt P, Rollinger JM, Schuster D, Spitzer GM, Wolber G. Development of antiviral agents using molecular modeling and virtual screening techniques. J Infect Dis 2009; in press. [DOI] [PubMed] [Google Scholar]
- 343. Cramer R III, Patterson D, Bunce J. Comparative molecular field analysis (CoMFA). 1. Effect of shape on binding of steroids to carrier proteins. J Am Chem Soc 1988;110:5959–5967. [DOI] [PubMed] [Google Scholar]
- 344. Artico M, Botta M, Corelli F, Mai A, Massa S, Ragno R. Investigation on QSAR and binding mode of a new class of human rhinovirus‐14 inhibitors by CoMFA and docking experiments. Bioorg Med Chem 1996;4:1715–1724. [DOI] [PubMed] [Google Scholar]
- 345. Diana GD, Cutcliffe D, Oglesby RC, Otto MJ, Mallamo JP, Akullian V, McKinlay MA. Synthesis and structure‐activity studies of some disubstituted phenylisoxazoles against human picornavirus. J Med Chem 1989;32:450–455. [DOI] [PubMed] [Google Scholar]
- 346. Diana GD, Kowalczyk P, Treasurywala AM, Oglesby RC, Pevear DC, Dutko FJ. CoMFA analysis of the interactions of antipicornavirus compounds in the binding pocket of human rhinovirus‐14. J Med Chem 1992;35:1002–1008. [DOI] [PubMed] [Google Scholar]
- 347. Diana GD, Oglesby RC, Akullian V, Carabateas PM, Cutcliffe D, Mallamo JP, Otto MJ, McKinlay MA, Maliski EG, Michalec SJ. Structure‐activity studies of 5‐[[4‐(4,5‐dihydro‐2‐oxazolyl)phenoxy]alkyl]‐3‐methylisoxazoles: Inhibitors of picornavirus uncoating. J Med Chem 1987;30:383–388. [DOI] [PubMed] [Google Scholar]
- 348. Verma RP, Hansch C. Understanding human rhinovirus infections in terms of QSAR. Virology 2007;359:152–161. [DOI] [PubMed] [Google Scholar]
- 349. Cheah KC, Leong LEC, Porter AG. Site‐directed mutagenesis suggests close functional relationship between a human rhinovirus 3C cysteine protease and cellular trypsin‐like serine proteases. J Biol Chem 1990;265:7180–7187. [PubMed] [Google Scholar]
- 350. Malcolm BA. The picornaviral 3C proteinases: Cysteine nucleophiles in serine proteinase folds. Protein Sci 1995;4:1439–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351. Reich SH, Johnson T, Wallace MB, Kephart SE, Fuhrman SA, Worland ST, Matthews DA, Hendrickson TF, Chan F, Meador J, Ferre RA, Brown EL, DeLisle DM, Patick AK, Binford SL, Ford CE. Substituted benzamide inhibitors of human rhinovirus 3C protease: Structure‐based design, synthesis, and biological evaluation. J Med Chem 2000;43:1670–1683. [DOI] [PubMed] [Google Scholar]
- 352. Dragovich PS, Prins TJ, Zhou R, Brown EL, Maldonado FC, Fuhrman SA, Zalman LS, Tuntland T, Lee CA, Patick AK, Matthews DA, Hendrickson TF, Kosa MB, Liu B, Batugo MR, Gleeson J‐PR, Sakata SK, Chen L, Guzman MC, Meador JW III, Ferre RA, Worland ST. Structure‐based design, synthesis, and biological evaluation of irreversible human rhinovirus 3C protease inhibitors. 6. Structure‐activity studies of orally bioavailable, 2‐pyridone‐containing peptidomimetics. J Med Chem 2002;45:1607–1623. [DOI] [PubMed] [Google Scholar]
- 353. Wermuth C‐G, Ganellin CR, Lindberg P, Mitscher LA. Glossary of terms used in medicinal chemistry (IUPAC recommendations 1997). Annu Rep Med Chem 1998;33:385–395. [Google Scholar]
- 354. Wolber G, Kosara R. Pharmacophores and pharmacophore searches. In: Langer T, Hoffmann RD, editors. Methods and principles in medicinal chemistry. Volume 32. Weinheim: Wiley‐VCH; 2006. pp 131–150. [Google Scholar]
- 355. Steindl T, Laggner C, Langer T. Human rhinovirus 3C protease: Generation of pharmacophore models for peptidic and nonpeptidic inhibitors and their application in virtual screening. J Chem Inf Model 2005;45:716–724. [DOI] [PubMed] [Google Scholar]
- 356. Steindl T, Langer T. Docking versus pharmacophore model generation: A comparison of high‐throughput virtual screening strategies for the search of human rhinovirus coat protein inhibitors. QSAR Comb Sci 2005;24:470–479. [Google Scholar]
- 357. Pipeline Pilot. San Diego, CA: Accelrys. [Google Scholar]
- 358. Discovery Studio. San Diego, CA: Accelrys. [Google Scholar]
- 359. Inte:Ligand Pharmacophore Database. Vienna, Austria: Inte:Ligand. [Google Scholar]


